Introduction
In insects, odorant reception occurs mainly in the antennae via hair-like structures called olfactory sensilla. These morphofunctional units enclose olfactory receptor neurons, which are surrounded by a protein-enriched lymph. Detection of olfactory molecules (compounds) at the peripheral level is a complex process involving numerous molecular actors (Durand et al., Reference Durand, Carot-Sans, Chertemps, Montagné, Jacquin-Joly, Debernard and Maïbèche-Coisne2010; Zhou, Reference Zhou2010; Zhou et al., Reference Zhou, Field and He2010). This includes the binding and transport of hydrophobic odorant molecules by odorant-binding proteins (OBPs), their recognition by odorant receptors (ORs) and their inactivation through hydrolysis by specific enzymes such as the odorant-degrading enzymes (ODEs) (Rützler & Zwiebel, Reference Rützler and Zwiebel2005). The rapid inactivation of signals by ODEs plays an integral role in insect chemoreception, which prevents the accumulation of stimulants and subsequent sensory adaptation (Vogt & Riddiford, Reference Vogt and Riddiford1981), and allows insects to rapidly respond to changes in chemical volatiles in the environment. Several enzymes are known to be involved in such mechanisms, including cytochrome P450s, certain esterases, glutathione S-transferases (GSTs), members of the short-chain dehydrogenase/reductase family (aldehyde oxidases and alcohol dehydrogenases) and uridine diphosphate (UDP)-glycosyltransferases (UGTs) (Younus et al., Reference Younus, Chertemps, Pearce, Pandey, Bozzolan, Coppin, Russell, Maïbèche-Coisne and Oakeshott2014). Compared to other ODE families, such as esterases, there are limited published reports on UGTs that are involved in insect olfaction.
Glycoside conjugation is an important metabolic pathway for the biotransformation of a variety of lipophilic xenobiotics and endobiotics (Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). UGTs can convert lipophilic aglycones into more hydrophilic glycosides by catalyzing the conjugation of a glycosyl group donated by a UDP-glycoside to various small hydrophobic molecules. Through this process, UGTs facilitate the excretion of hydrophobic compounds and protect the cell from being damaged by toxic hydrophobic compounds to maintain proper intracellular regulation (Ahn et al., Reference Ahn, Badenes-Pérez, Reichelt, Svatoš, Schneider, Gershenzon and Heckel2011, Reference Ahn, Vogel and Heckel2012). In insects, the significance of the glycosylation of small hydrophobic compounds has been overlooked for many years, although recently, insect UGTs have been suggested to play multiple roles in the detoxification and sequestration of a variety of plant allelochemicals and insecticides (Kojima et al., Reference Kojima, Fujii, Suwa, Miyazawa and Ishikawa2010; Ahn et al., Reference Ahn, Badenes-Pérez, Reichelt, Svatoš, Schneider, Gershenzon and Heckel2011). Of note, recent findings have revealed that insect UGTs are implicated in the termination of olfactory signals (Ahn et al., Reference Ahn, Vogel and Heckel2012; Younus et al., Reference Younus, Chertemps, Pearce, Pandey, Bozzolan, Coppin, Russell, Maïbèche-Coisne and Oakeshott2014).
The role of UGTs in vertebrate olfaction is well established (Heydel et al., Reference Heydel, Leclerc, Bernard, Pelczar, Gradinaru, Magdalou, Minn, Artur and Goudonnet2001). UGT2A1, which is highly expressed in the rat olfactory epithelium (Zhang et al., Reference Zhang, Zhang, Liu, Su, Weng, Ling, Chen, Gu, Schilling and Ding2005), can conjugate odorants, and terminate the odorant signals (Lazard et al., Reference Lazard, Zupko, Poria, Nef, Lazarovits, Horn, Khen and Lancet1991). However, in insects, evidence on UGT expression in the antennae is limited to three species, Drosophila melanogaster (Wang et al., Reference Wang, Hasan and Pikielny1999), Bombyx mori (Huang et al., Reference Huang, Chai, Zhang, Liu, Dai, Lu and Xiang2008) and Manduca sexta (Robertson et al., Reference Robertson, Martos, Sears, Todres, Walden and Nardi1999), suggesting a possible role in olfactory processing. Two UGTs, UGT35a, and UGT35b, have previously been shown to be preferentially expressed in the third antennal segment of D. melanogaster, and the latter was suggested to be possibly involved in odorant turnover (Wang et al., Reference Wang, Hasan and Pikielny1999). Because of the rich diversity and the variety of the possible functions of detoxification enzymes expressed in each species, the identification and characterization of individual members of these enzyme families, which are each specialized in odorant degradation within the antennae, are still challenging (Younus et al., Reference Younus, Chertemps, Pearce, Pandey, Bozzolan, Coppin, Russell, Maïbèche-Coisne and Oakeshott2014).
The dark black chafer, Holotrichia parallela Motschulsky (Coleoptera: Scarabaeidae), is one of the most important soil pests worldwide (Ju et al., Reference Ju, Li, Jiang, Qu, Guo, Han and Li2014). In China, H. parallela has caused a significant loss in crop yields and great economic damage by attacking crops, vegetables and economically important trees (Ju et al., Reference Ju, Li, Jiang, Qu, Guo, Han and Li2014; Zhang et al., Reference Zhang, Wang, Yang, Yi, Liu and Xi2016). Because H. parallela larvae live in the soil, it is difficult to control them using traditional pesticides. Chemical communication is so crucial for the interaction of H. parallela with their environment that olfactory-related gene products could be effectively utilized as new targets to reduce insect populations (Ju et al., Reference Ju, Qu, Wang, Jiang, Li, Dong and Han2012).
In this study, 20 putative UGT genes were identified from the H. parallela antennae transcriptome. We describe the sequence and phylogenetic analyses of H. parallela antennae UGTs and predict the corresponding UGT protein structures. This study also elucidates UGT expression patterns in different tissues. These findings serve as an important basis for the identification of H. parallela UGTs that participate in odorant degradation.
Method
Insect and tissue collection
H. parallela strains used in this study were collected in Cangzhou, China. The antennae, heads, thoraces, abdomens, legs, and wings of male and female adults were dissected and promptly immersed in liquid nitrogen and stored at −80°C until use (Wang et al., Reference Wang, Yi, Yang, Liu, Zhang and Xi2017).
Identification of UDP-glycosyltransferases genes from H. parallela
H. parallela antennae transcriptome data used in this study are from our laboratory (data not shown). UDP-glycosyltransferase genes were selected by searching the sequences in the antennal transcriptome database and annotations for keywords (UDP-glycosyltransferases). Subsequently, UGT conserved domains (UDPGT) of the selected H. parallela UGT genes were further predicted using the Pfam database (http://pfam.xfam.org). For possible alternatively spliced genes, only the longest coding transcript was selected.
Bioinformatic analyses
Open reading frames (ORFs) of genes were predicted using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Theoretical isoelectric points and molecular weights of deduced proteins were calculated using the ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/) (Gasteiger et al., Reference Gasteiger, Gattiker, Hoogland, Ivanyi, Appel and Bairoch2003). Signal peptides and transmembrane domains were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., Reference Petersen, Brunak, von Heijne and Nielsen2011) and TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM), respectively. Amino acid sequence multiple alignments were analyzed by using DNAMAN software (http://www.lynnon.com/pc/alignm.html) with default parameters (Duan et al., Reference Duan, Xiong, Lu, Wang and Ai2016). SWISS-MODEL (http://swissmodel.expasy.org/), which is available within ExPASy, were used to predict tertiary structures (Biasini et al., Reference Biasini, Bienert, Waterhouse, Arnold, Studer, Schmidt, Kiefer, Cassarino, Bertoni, Bordoli and Schwede2014). BLASTX best hits were found using the BLASTX program, provided by NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., Reference Altschul, Madden, Schäffer, Zhang, Zhang, Miller and Lipman1997). The neighbor-joining phylogenetic tree was constructed by MEGA6 using the p-distance metric at bootstrap 1000 (Ahn et al., Reference Ahn, Vogel and Heckel2012; Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
RNA isolation and cDNA synthesis
Total RNA was extracted from different tissues (antennae, heads, thoraces, abdomens, legs, and wings) using RNAiso Plus (Takara, Dalian, China) following the manufacturer's protocol. Total RNA was quantified and checked for purity and integrity using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and gel electrophoresis. PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A, Takara, Dalian, China) was used for cDNA synthesis.
Quantitative real-time polymerase chain reaction (qPCR)
Primer pairs for qPCR were designed using Primer 5 software and are listed in Supplementary table 2. GAPDH was used as a reference gene (Zhang et al., Reference Zhang, Wang, Yang, Yi, Liu and Xi2016). mRNA levels were measured by qPCR using the SYBR® Premix Ex Taq™ (Takara, Dalian, China). Each amplification reaction contained 1 µl of cDNA, 10 µl of SYBR Premix Ex Taq, 0.4 µl of 10 µM of forward primer, 0.4 µl of reverse primer, 0.4 µl of ROX Reference Dye II and 7.8 µl water in a 20 µl total volume. qPCR was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 30 s initial denaturation at 95°C and 40 cycles of 95°C for 5 s, 60°C for 10 s and 72°C for 34 s, followed by the melting curve analysis (60–95°C). After qPCR, melting curves were evaluated to confirm single peaks and check amplification specificity. Means and standard errors were obtained from the average of three biological replicates with their three respective technical replicates. The fold changes were evaluated using the 2−ΔΔCt method (Pfaffl, Reference Pfaffl2001). Statistical analysis was conducted using GraphPad Prism 5 software (San Diego, CA).
Results
Identification of 20 putative UGTs
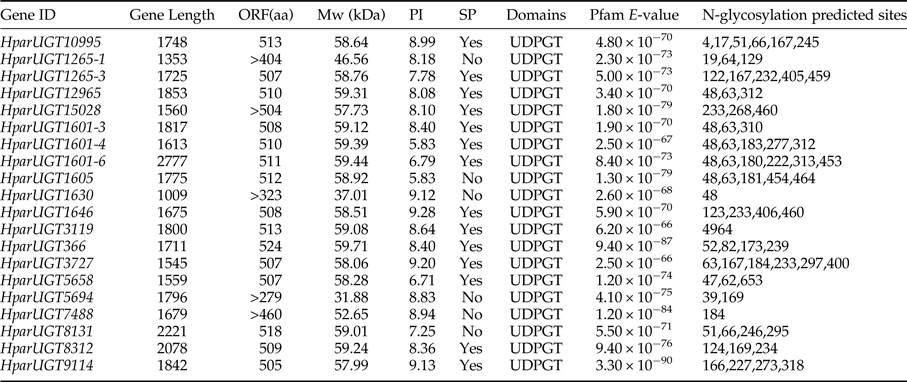
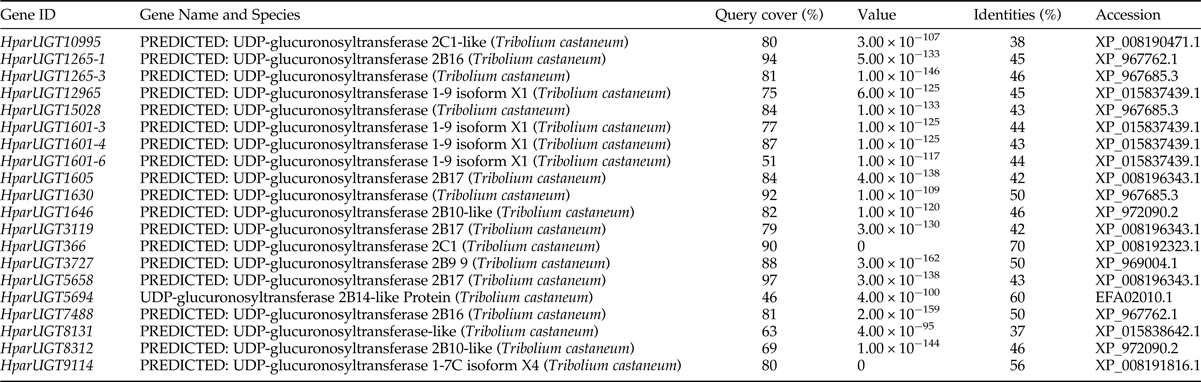
To identify olfaction-related genes and explore the olfactory signal transduction mechanisms in H. parallela, the H. parallela antennal transcriptome sequencing from adult females and males was conducted. Female antennae transcripts were assembled into 71,928 Contigs and 43,624 unigenes, and male antennae transcripts were assembled into 63,485 Contigs and 38,785 unigenes. A total of 19025 unigenes (54.82% of all unigenes) returned the annotation result by searching against non-redundant (NR), Swissprot, KEGG, COG, and GO databases with a cut-off E-value of 10−5. Of them, 47 candidate OR genes and 26 OBP genes were identified in the male and female antennal transcriptomes (unpublished). In this paper, a total of 20 cDNA fragments encoding putative UGTs were identified from the H. parallela antennal transcriptome, with gene lengths between 1009 and 2777 bp (table 1). These 20 cDNAs all have predicted UDPGT domains with low E-values (6.2×10−66–3.3−10−90) based on Pfam. Of these, 15 sequences have complete ORFs, while four sequences (HparUGT1625-1, HparUGT1630, HparUGT5694, and HparUGT7488) are incomplete sequences truncated at 5′-regions, and one sequence (HparUGT15028) contains a truncated 3′-region. The lengths of the 15 deduced full-length UGT proteins range from 505 to 524 amino acids, with predicted isoelectric points ranging from pI 5.83 to 9.28 and calculated molecular masses between 57.99 and 59.71 KDa.
Table 1. Summary of H. parallela antennal UGTs sequences.
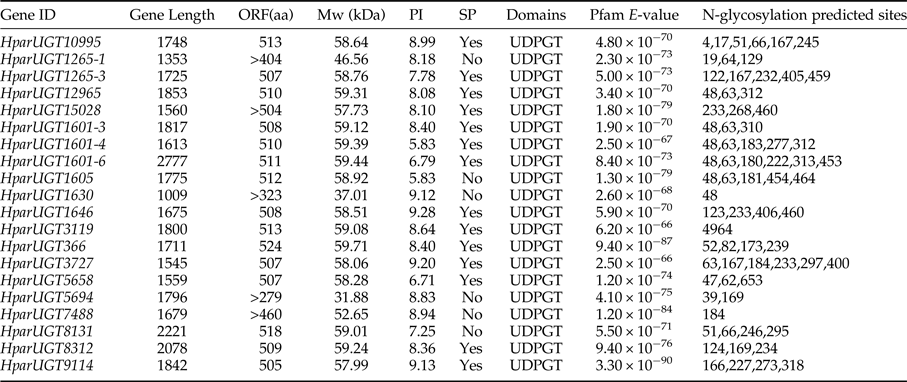
ORF, open reading frame; MW, molecular weight; PI, isoelectric point; SP, signal peptide.
Structural prediction of UGT proteins
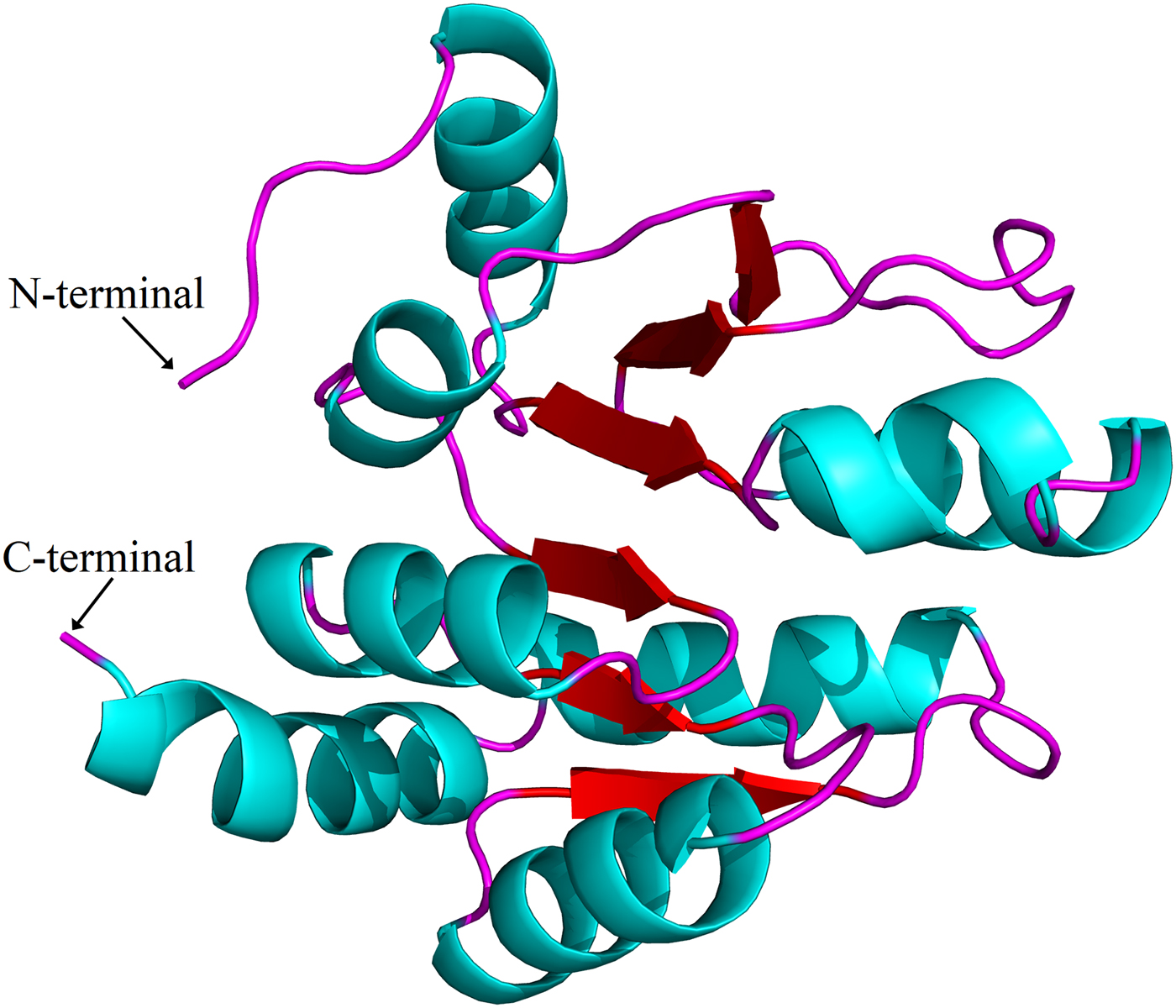
Sequence alignments combined with multiple bioinformatics methods revealed the major structural features of the putative H. parallela UGT proteins (fig. 1). In general, the N-terminal half is highly variable, whereas the C-terminal half is more conserved. All identified H. parallela UGT proteins, including the truncated UGT proteins, displayed the characteristic UGT signature motif in the middle of the C-terminal domain. Signal peptides were detected in 14 H. parallela UGT proteins but not in the four sequences with incomplete N-terminal regions (fig. 1). In addition, transmembrane domains, followed by cytoplasmic tails, were also identified at the C-terminal of all H. parallela UGTs. A highly conserved aspartate residue, which is a negatively charged amino acid residue, is immediately in front of the transmembrane domain on the luminal side. Based on comparison with the crystal structure of human UGT2B7, there are two predicted sugar binding regions (DBR1 and DBR2) observed in the H. parallela UGT proteins, and several important residues that interact with the sugar donor are also conserved (fig. 1). Finally, the two catalytic residues located in the N-terminal substrate binding site are also evolutionarily conserved between two species (fig. 1). In addition, the three-dimensional structure of HparUGT1646, the highest homologous UGT of H. parallela with human UGT2B7, was predicted using human UGT2B7 as a template (fig. 2), confirming these homologous features between two proteins.

Fig. 1. Multiple alignment of 20 H. parallela UGTs with human UGT2B7. The N-terminal signal peptides predicted by SignalP 4.1 are underlined. The transmembrane domain and cytoplasmic tails based on human UGT2B7 (accession number NM_001074) are in white boxes under alignment. The signature motif is indicated by arrows above the alignment. All of the putative β-sheets in the C-terminal half predicted by comparison with human UGT2B7 crystal structure are denoted as Cβ + number with an arrow. Asterisks (![]() ) below the alignment indicate the important catalytic residues (H and D). Two donor binding regions (DBR) are boxed and several important residues interacting with the sugar donor are indicated by letters (a,b and c) below the alignment.
) below the alignment indicate the important catalytic residues (H and D). Two donor binding regions (DBR) are boxed and several important residues interacting with the sugar donor are indicated by letters (a,b and c) below the alignment.
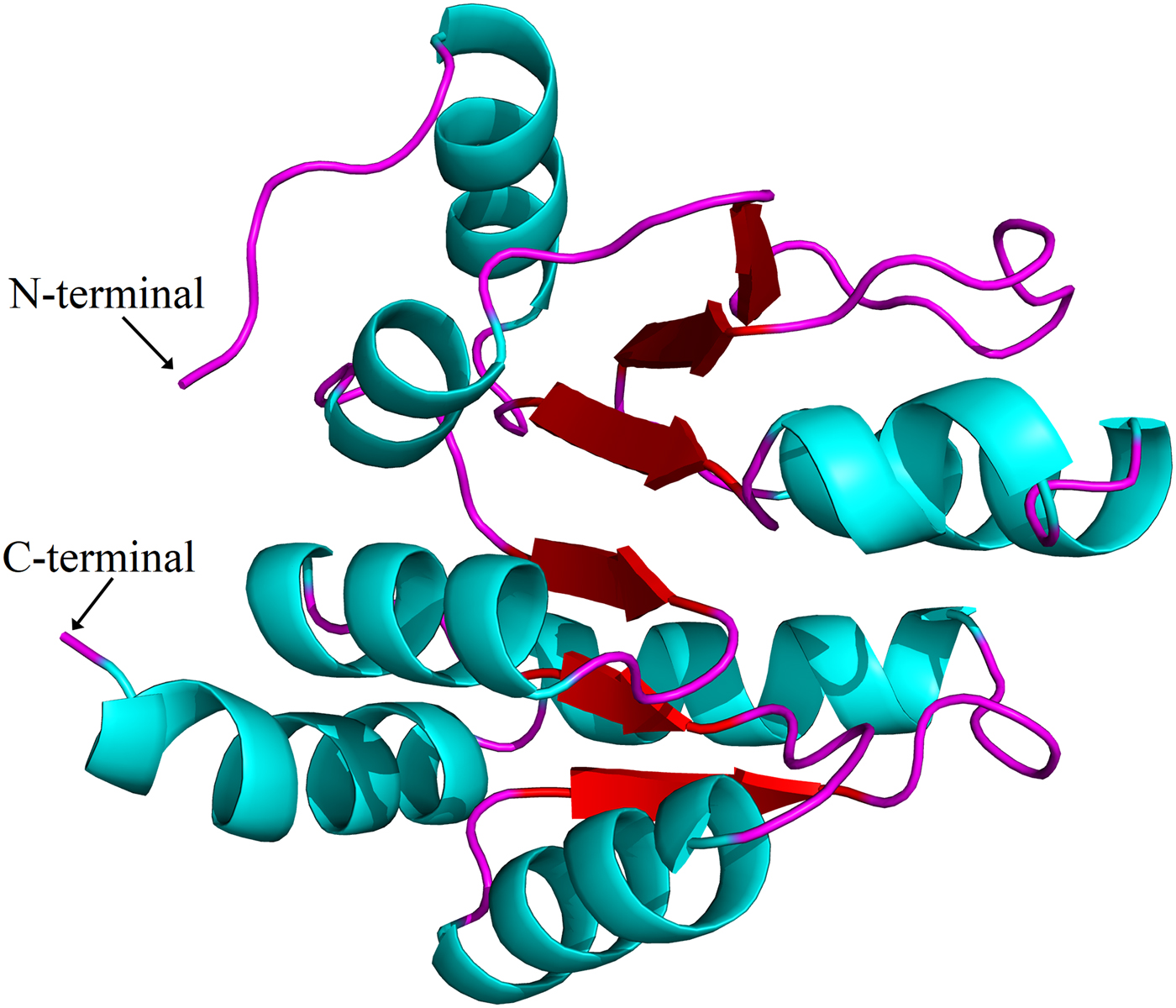
Fig. 2. Putative three-dimensional structure of HparUGT1646, the highest homologous UGT of H. parallela with human UGT2B7. The three-dimensional structure was predicted using human UGT2B7 as the template.
Homology and phylogenetic analysis
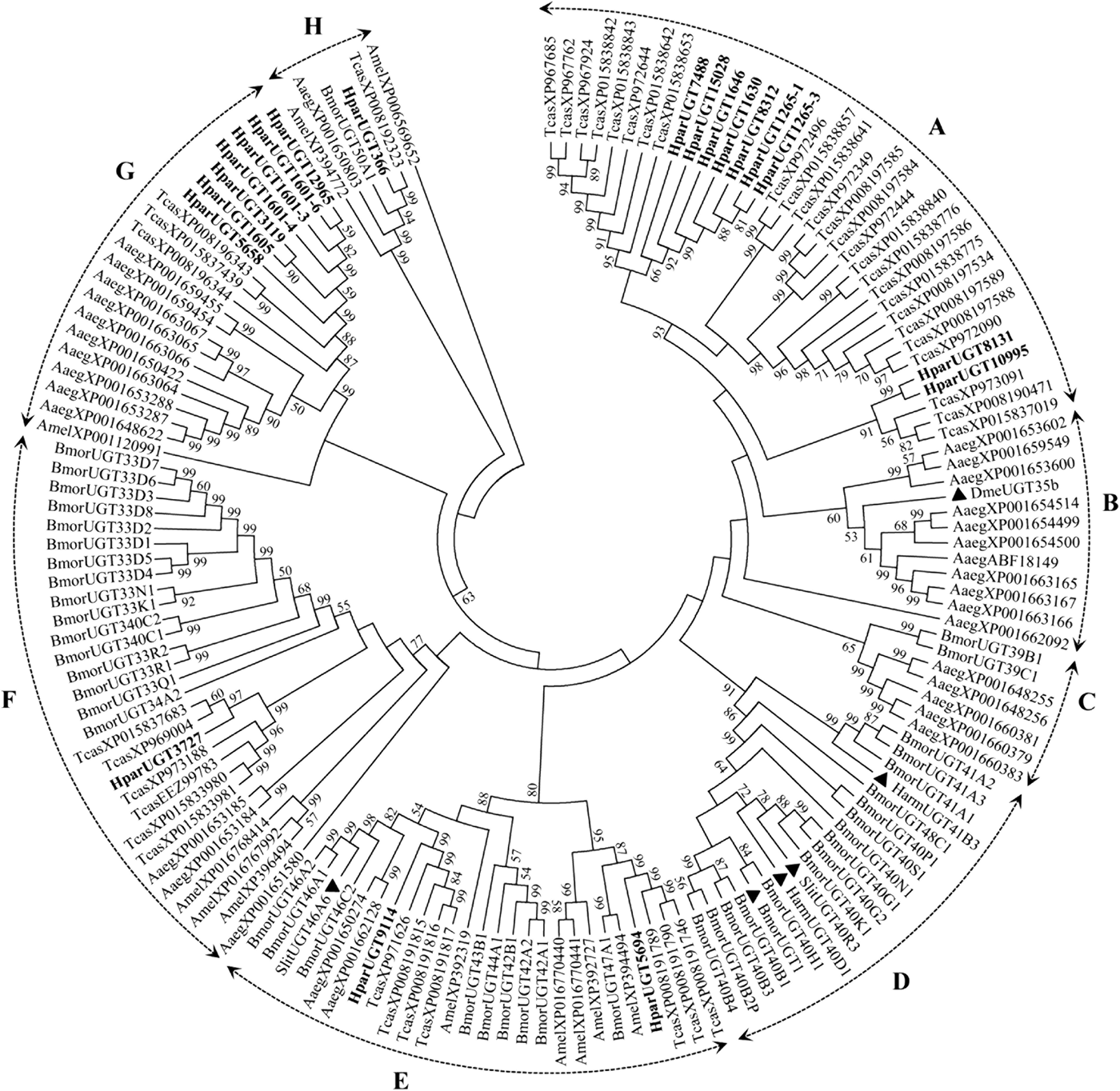
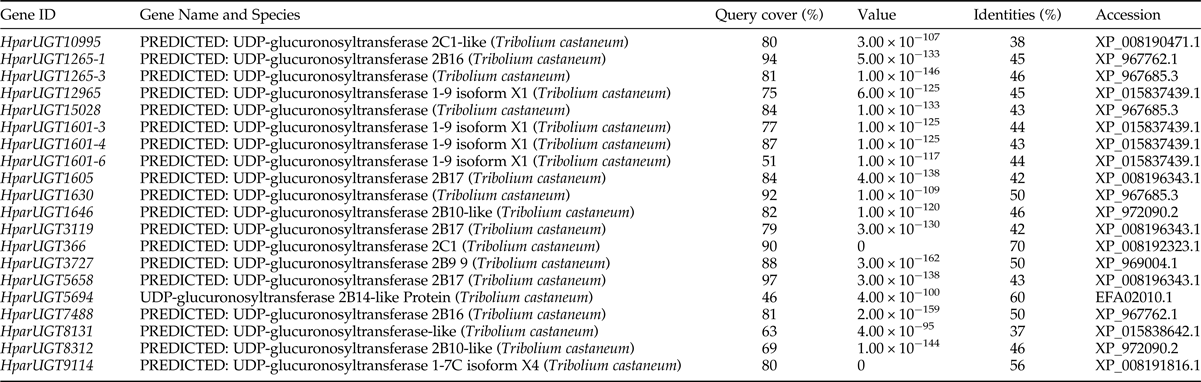
The 20 H. parallela UGTs as well as the UGTs from Tribolium castaneum, B. mori, Aedes aegypti, and Apis mellifera distribute into 8 major groups (group A-H) that localize to distinct branches of the phylogenetic tree (fig. 3). The phylogenetic tree contains some order-specific groups, such as the Lepidoptera- (D), Coleoptera- (A), and Diptera-specific groups (B), and several conserved groups which are common to more than one order (C, E, F, G, and H). Most groups are supported by high bootstrap values. The H. parallela UGTs belong to five groups and fall into a branch together with the T. castaneum UGTs. The largest group, Group A, contains nine H. parallela UGT genes. Group F is the second largest group and contains seven members. The remaining groups, E and F, contain two members and a single member, respectively. The phylogenetic tree is reconstructed by Maximum likelihood methods (Supplementary fig. 1). The cluster pattern of a phylogenetic tree, reconstructed by Maximum likelihood methods, is quite similar to that of the neighbor-joining phylogenetic tree, verifying the reliability of neighbor-joining phylogenetic tree. Consistent with this phylogenetic tree, BLASTX best hits analysis of the H. parallela UGTs showed that their respective orthologs are all sequences from T. castaneum with sequence identities ranging from 37 to 70% and E-values from 0 to 4 × 10−95 (table 2). Of the 15 full-length UGTs, HparUGT366 shared the highest identity (70%) with the T. castaneum UGTs; 12 other H. parallela UGTs have identities ranging from 42 to 56%, and two remaining H. parallela UGTs have less than 40% identity with their respective orthologs.
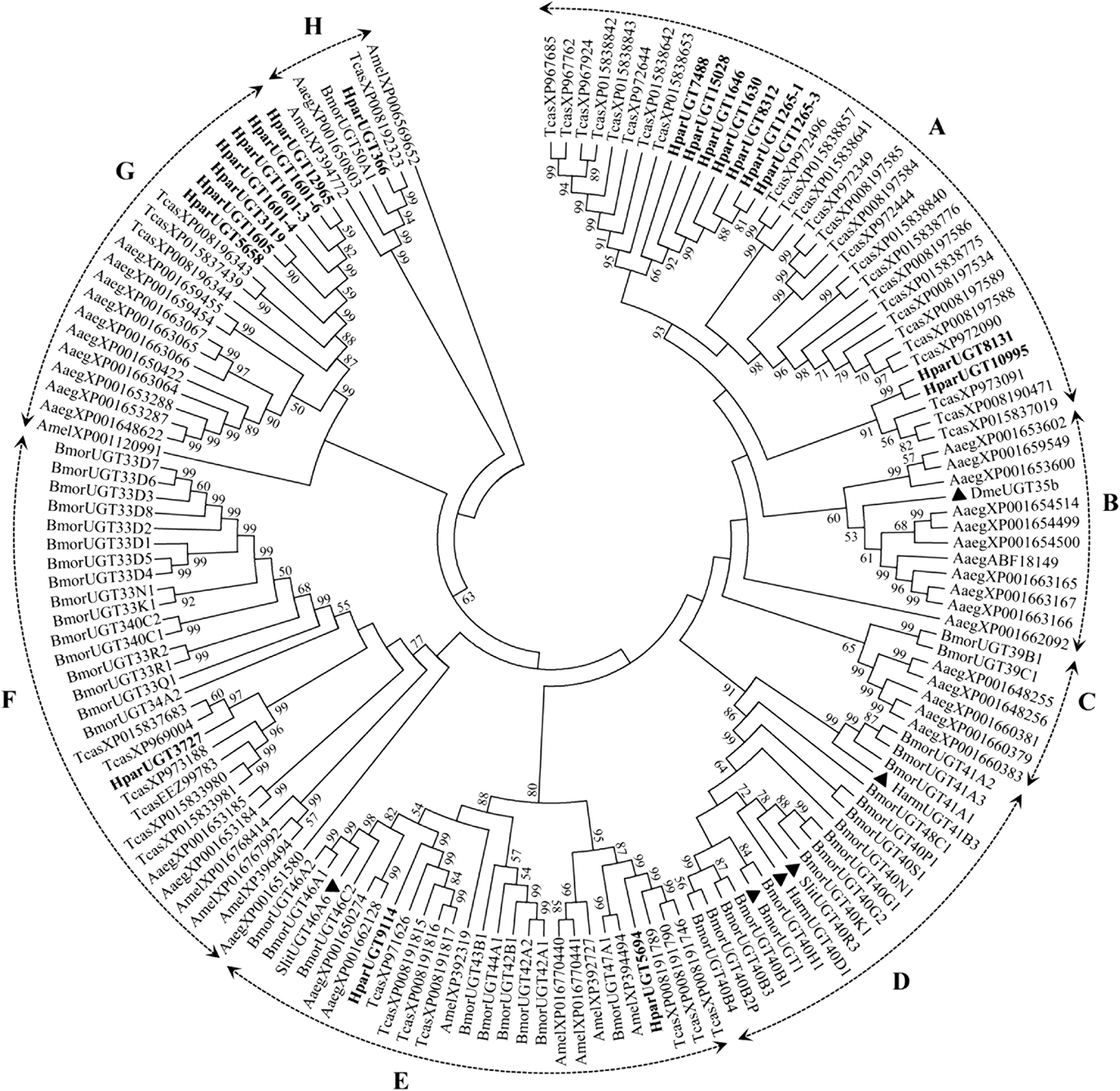
Fig. 3. Phylogenetic tree of H. parallela antennal UGTs with UGTs of other insects. Phylogenetic tree of UGT protein sequences from various insect species, including H. parallela, T. castaneum, B. mori, A. aegypti, A. mellifera, Helicoverpa armigera, S. littoralis, D. melanogaster. The GenBank accession numbers are included in the Supplemental table 2. Phylogenetic tree was constructed by MEGA 6 program using the neighbor-joining method with p-distance model and 1000 bootstrap replicates (Ahn et al., Reference Ahn, Vogel and Heckel2012). Hpar, H. parallela; Tcas, T. castaneum; Bmor, B. mori; Aaeg, A. aegypti; Amel, A. mellifera; Harm, H. armigera; Slit, S. littoralis; Dme, D. melanogaster. The H. parallela UGTs are bold and the insect UGTs biochemically characterized were marked with black triangle. A-H represent eight major groups that localize to distinct branches of the phylogenetic tree, the phylogenetic pattern follows that described previously for the insect UGTs in the literature (Ahn et al., Reference Ahn, Vogel and Heckel2012).
Table 2. The BLASTX best hits summary of H. parallela antennal UGT genes.
Expression analysis of H. parallela UGT genes
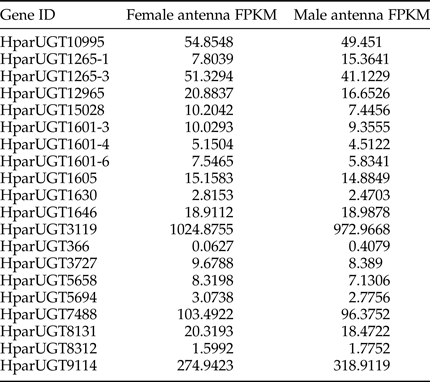
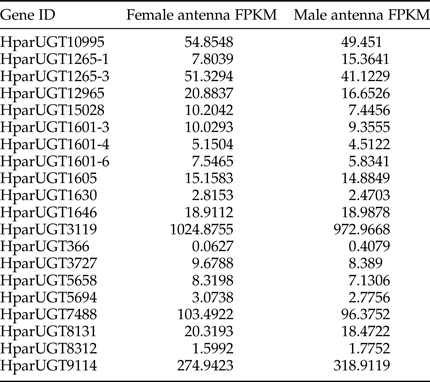
Expression levels of H. parallela UGTs were determined in terms of fragments per kilobase of transcript per million mapped reads (FPKM) estimated from the antennae RNA-seq data (table 3). Except HparUGT366, all UGTs were found to be expressed in antennae at FPKM>1, and 11 of them had an FPKM>10 in male or female antennae. Compared to other UGTs, HparUGT3119 showed the highest expression in both female and male antennae, with FPKM = 1024.8755 and FPKM = 972.9668, respectively (table 3). HparUGT9114 and HparUGT7488 had an FPKM of >100 in at least one of the female or male antennae.
Table 3. Fragments per Kilobase per Million Mapped reads (FPKM) of H. parallela antennal UGTs estimated from antennae RNA-seq data.
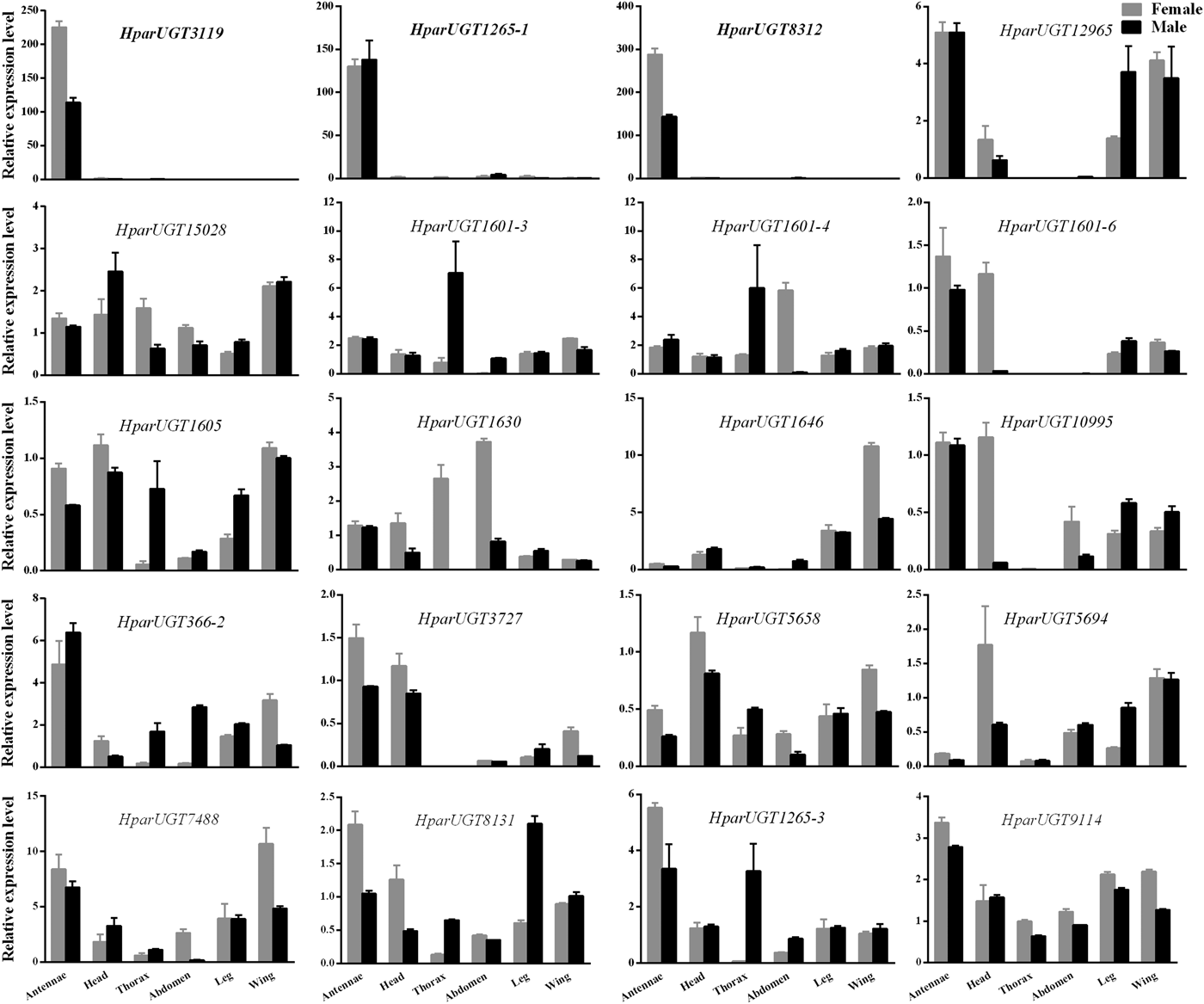
qPCR analysis revealed a wide range of expression patterns of the H. parallela UGTs in different tissues (fig. 4), including antennae and various non-olfactory tissues. Most H. parallela UGTs were ubiquitously expressed in most tissues, although some H. parallela UGTs were preferentially expressed in a specific tissue. Of note, HparUGT1265-1, HparUGT3119, and HparUGT8312 were restricted to the antennae and barely detectable in other tissues. In addition to antennae, HparUGT366, HparUGT1265-3, HparUGT7488, HparUGT1601-6, HparUGT12965, HparUGT10995, HparUGT8131, HparUGT9114, and HparUGT3727 were highly expressed in other tissues. Of the remaining UGTs, HparUGT1601-3 was mainly expressed in the thorax, HparUGT1646 was mainly expressed in the wings and HparUGT5694 in the heads. HparUGT1630 and HparUGT1601-4 were highly expressed in both the abdomen and thorax.
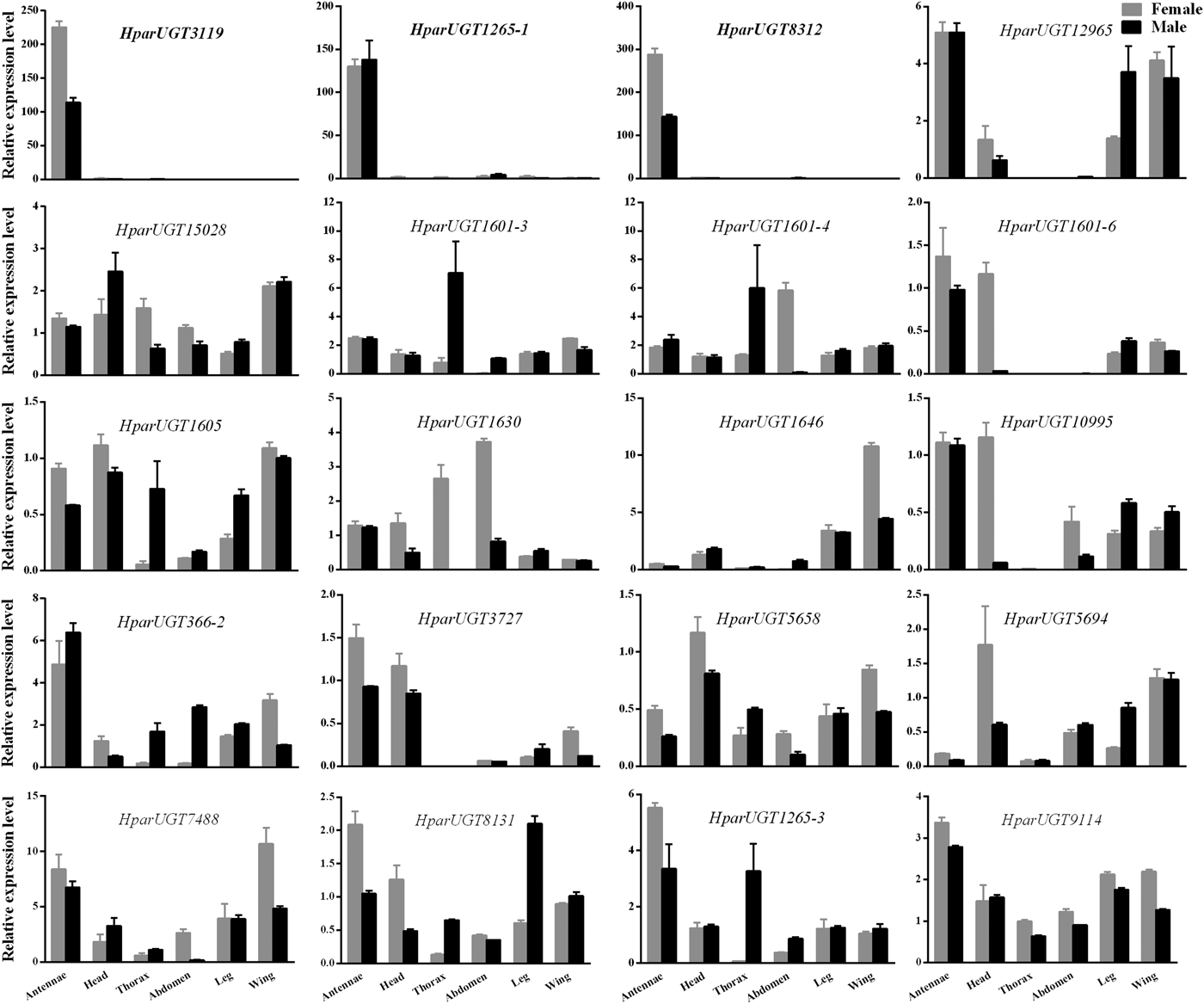
Fig. 4. Expression pattern of H. parallela antennal UGT genes by qPCR across various tissues. Fold changes were calculated using the 2−ΔΔCt method. The gene expression levels in various tissues were normalized relative to that in female head. GADPH gene was used as the reference gene to normalize target gene expression. Data are shown as averages of biological and technical replicates ± SE of the mean. The three UGT genes (HparUGT 3119, HparUGT1265-1 and HparUGT 8312), highlighted in bold in this figure, were highly expressed in antennae.
Discussion
Based on the previous genome-wide analysis, UGT gene numbers ranging from 12 to 58 have been identified in various insect species including B. mori (45), T. castaneum (43), three Diptera species, two Hymenoptera species and Acyrthosiphon pisum (58). In addition, 45 H. armigera UGTs were identified from RT-PCR analysis of a cDNA library (Ahn et al., Reference Ahn, Vogel and Heckel2012). However, knowledge of UGT tissue distribution is limited. In this study, a total of 20 UGT members were identified from the H. parallela antennal transcriptome. This number of antennal UGTs in H. parallela is greater than that in S. littoralis (11 genes, Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014), suggesting that a large proportion of the H. parallela UGT repertoire has been detected by our transcriptome method. However, T. castaneum is known to have 43 UGTs, so other H. parallela UGTs might remain to be found. The high diversity of UGTs identified in H. parallela antennae likely reflects the functional importance of UGTs in antennae olfaction.
UDPGT domains were predicted with low E-values by Pfam software, and the UGT signature motif, a hallmark of prokaryotic and eukaryotic UGTs (Mackenzie et al., Reference Mackenzie, Owens, Burchell, Bock, Bairoch, Bélanger, Fournel-Gigleux, Green, Hum and Iyanagi1997), was found in the H. parallela UGTs, strongly supporting these sequences as genes belonging to the UGT superfamily. As described for other insect UGTs (Ahn et al., Reference Ahn, Vogel and Heckel2012), the H. parallela N-terminal substrate binding domain is less conserved than the C-terminal sugar donor binding domain. In a previous study, critical information regarding donor binding regions, sugar binding residues, and catalytic residues in insect UGTs was determined based on the crystal structures of human UGT2B7 and two plant UGTs (Miley et al., Reference Miley, Zielinska, Keenan, Bratton, Radominska-Pandya and Redinbo2007; Radominska-Pandya et al., Reference Radominska-Pandya, Bratton, Redinbo and Miley2010; Ahn et al., Reference Ahn, Vogel and Heckel2012). These key residues and donor binding regions are also conserved in H. parallela UGTs. In animals, a signal peptide found at the N-terminus of the UGT mediates the integration of the protein precursor into the endoplasmic reticulum (ER), and is subsequently cleaved. Then, the UGT is N-glycosylated and retained in the ER membrane by virtue of its hydrophobic transmembrane domain (Magdalou et al., Reference Magdalou, Fournel-Gigleux and Ouzzine2010). Similarly, we predicted N-glycosylation sites, signal peptides, and transmembrane domains for all of the H. parallela UGT sequences, and these features have also been reported in other insect UGTs (Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). In summary, the structural features of H. parallela UGTs revealed by these data indicated that H. parallela UGTs were probably active enzymes with similar structures and functions to other known insect and animal UGTs.
The phylogenetic tree constructed in this study follows the phylogenetic pattern described previously for the insect UGTs (Ahn et al., Reference Ahn, Vogel and Heckel2012). For example, the UGT50 family comprises one member from each insect species, and includes BmorUGT50A1 and HparUGT366. The genes within each group likely arose from a common ancestor and have a similar function. Phylogenetic analysis of plant UGTs has shown that phylogenetic grouping can be useful for predicting the substrates of a specific enzyme (Lim et al., Reference Lim, Baldauf, Li, Elias, Worrall, Spencer, Jackson, Taguchi, Ross and Bowles2003; Cartwright et al., Reference Cartwright, Lim, Kleanthous and Bowles2008; Osmani et al., Reference Osmani, Bak and Møller2009; Barvkar et al., Reference Barvkar, Pardeshi, Kale, Kadoo and Gupta2012). The predicted positions of the insect UGTs that have been characterized to some extent biochemically and were also superimposed onto the tree. Whereas HarmUGT41B3 and HarmUGT40D1 are capable of glycosylating gossypol (Krempl et al., Reference Krempl, Sporer, Reichelt, Ahn, Heidel-Fischer, Vogel, Heckel and Joußen2016), BmorUGT1 catalyzes the glucosidation of a wide variety of substrates (Luque et al., Reference Luque, Okano and O'Reilly2002), and DmeUGT35b and SlitUGT4040R3 were preferentially expressed in olfactory organs of D. melanogaster and S. littoralis (Wang et al., Reference Wang, Hasan and Pikielny1999; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). However, these UGTs do not provide clues to the function of H. parallela UGTs, as they belong to order-specific groups. Despite high homology with T. castaneum UGTs, H. parallela UGT functions cannot be determined by homology analysis due to limited reports on T. castaneum UGT catalytic activity. However, most H. parallela and T. castaneum UGTs share a common family or subfamily according to the current UGT nomenclature guidelines, which defines families and subfamilies as sharing at least 40 and 60% amino acid sequence identity (aaID), respectively (Mackenzie et al., Reference Mackenzie, Bock, Burchell, Guillemette, Ikushiro, Iyanagi, Miners, Owens and Nebert2005).
The expression patterns of H. parallela UGTs may be useful to predict their functions. In vertebrates, UGT2A1 is highly expressed in the rat olfactory epithelium, can glucuronidate odorants, and glucuronidation products abolish the ability of odorants to induce an olfactory response, suggesting that this olfactory-specific UGT participates in terminating odorant signals (Lazard et al., Reference Lazard, Zupko, Poria, Nef, Lazarovits, Horn, Khen and Lancet1991; Heydel et al., Reference Heydel, Leclerc, Bernard, Pelczar, Gradinaru, Magdalou, Minn, Artur and Goudonnet2001; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). The olfactory-restricted expression has been established as a useful criterion for identifying specific olfactory genes (Durand et al., Reference Durand, Carot-Sans, Chertemps, Montagné, Jacquin-Joly, Debernard and Maïbèche-Coisne2010; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014) such as odorant-binding proteins, olfactory receptors, and some odorant-degrading enzymes (Benton et al., Reference Benton, Vannice and Vosshall2007; Durand et al., Reference Durand, Carot-Sans, Chertemps, Montagné, Jacquin-Joly, Debernard and Maïbèche-Coisne2010). In this study, three genes (HparUGT8312, HparUGT1265-1, and HparUGT3119) were identified as being mainly expressed in antennae and are therefore implicated in a specific olfactory function. Indeed, several UGT genes that may be involved in specific roles in olfactory organs have also been found in other insects. For example, DmeUgt35b is preferentially expressed in the third antennal segment of D. melanogaster, and UGT40R3 and UGT46A6 are specifically expressed or overexpressed in antennae (Wang et al., Reference Wang, Hasan and Pikielny1999; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). An increasing body of literature reported a possible involvement of UGTs in insect olfaction (Ahn et al., Reference Ahn, Vogel and Heckel2012; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014; Younus et al., Reference Younus, Chertemps, Pearce, Pandey, Bozzolan, Coppin, Russell, Maïbèche-Coisne and Oakeshott2014). For example, preferential expression of UGT genes were also found in the antennae of B. mori (Huang et al., Reference Huang, Chai, Zhang, Liu, Dai, Lu and Xiang2008) and M. sexta (Robertson et al., Reference Robertson, Martos, Sears, Todres, Walden and Nardi1999). The powerful functions of UGTs in the metabolism of endogenous and exogenous compounds, resulting in the elimination and inactivation of their substrates, have also been shown in various insects (Luque et al., Reference Luque, Okano and O'Reilly2002; Sasai et al., Reference Sasai, Ishida, Murakami, Tadokoro, Ishihara, Nishida and Mori2009; Ahn et al., Reference Ahn, Vogel and Heckel2012; Bock, Reference Bock2016) . Furthermore, it is worthy of note that the substrates of a UGT from B. mori, named BmUGT1, include a number of odorants, such as vanillin, eugenol, β-citronellol, isomenthol, P-hydroxybiphenyl, and guaiacol (Luque et al., Reference Luque, Okano and O'Reilly2002). All these findings suggest that insect UGTs play possible roles in deactivation of olfactory signal, as already shown in vertebrate (Lazard et al., Reference Lazard, Zupko, Poria, Nef, Lazarovits, Horn, Khen and Lancet1991; Ahn et al., Reference Ahn, Vogel and Heckel2012; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014; Younus et al., Reference Younus, Chertemps, Pearce, Pandey, Bozzolan, Coppin, Russell, Maïbèche-Coisne and Oakeshott2014). Interestingly, SlitUGT46A6, HparUGT9114, and HparUGT5694 cluster in a conserved group (E) with a high bootstrap (80%). The antennal-specific SliUGT46A6 exhibited upregulation or downregulation after insecticide or odorant exposure, respectively, suggesting its specific function in olfaction (Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). The expression of UGTs in olfactory tissues was also reported in vertebrate (Heydel et al., Reference Heydel, Holsztynska, Legendre, Thiebaud, Artur and Le Bon2010; Olender et al., Reference Olender, Keydar, Pinto, Tatarskyy, Alkelai, Chien, Fishilevich, Restrepo, Matsunami, Gilad and Lancet2016; Hanser et al., Reference Hanser, Faure, Robert-Hazotte, Artur, Duchamp-Viret, Coureaud and Heydel2017). For example, UGT1A6 expressed in rat bulb, UGT2A1 in the epithelium of bovine, mouse, and human (Heydel et al., Reference Heydel, Holsztynska, Legendre, Thiebaud, Artur and Le Bon2010). Furthermore, UGT2A1 and UGT2A2 have been detected in the rat olfactory sensory cilia, the important tissue in olfactory process (Lazard et al., Reference Lazard, Zupko, Poria, Nef, Lazarovits, Horn, Khen and Lancet1991; Mayer et al., Reference Mayer, Ungerer, Klimmeck, Warnken, Schnölzer, Frings and Möhrlen2008; Heydel et al., Reference Heydel, Holsztynska, Legendre, Thiebaud, Artur and Le Bon2010, Reference Heydel, Coelho, Thiebaud, Legendre, Le Bon, Faure, Neiers, Artur, Golebiowski and Briand2013).
Although we detected some H. parallela UGT genes in various non-olfactory tissues, suggesting that these genes may have other functions, they might still have olfactory functions, similar to how SexiCXE13 and SlituCXE13, which are not antennae-specific genes, can also degrade sex pheromones and plant volatiles (He et al., Reference He, Li, Liu, Liu and Dong2014). A UGT from B. mori exhibited a wide substrate specificity towards plant allelochemicals (Luque et al., Reference Luque, Okano and O'Reilly2002). It is also reported that the glucuronidated small hydrophilic molecules disabled the ability of odorants to elicit an olfactory response (Lazard et al., Reference Lazard, Zupko, Poria, Nef, Lazarovits, Horn, Khen and Lancet1991; Leclerc et al., Reference Leclerc, Heydel, Amossé, Gradinaru, Cattarelli, Artur, Goudonnet, Magdalou, Netter, Pelczar and Minn2002; Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014). Thus, the precise roles of H. parallela antennal UGTs in olfactory function awaits further investigation.
Conclusions
In summary, a total of 20 UGT genes were identified from the H. parallela antennal transcriptome. Bioinformatics analysis supported the classification of these genes as members of the UGT superfamily, and sequence alignments enabled the prediction of structural features of the H. parallela antennal UGT proteins. Phylogenetic mapping revealed that H. parallela antennal UGTs are divided into eight groups. Specifically, HparUGT9114 clusters in a conserved group with SliUGT46A6 (Bozzolan et al., Reference Bozzolan, Siaussat, Maria, Durand, Pottier, Chertemps and Maïbèche-Coisne2014), suggesting these UGTs might have similar functions in olfactory organs. In addition, qPCR analysis revealed that HparUGT8312, HparUGT1265-1, and HparUGT3119 are highly expressed in antennae, indicating that their gene products may participate in specific olfactory functions. The data presented in this study provide an overview of dark black chafer UGTs, which will contribute to future functional studies of these enzymes.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485318000068
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2017YFD0200600) and the Natural Science Foundation of Jilin Province, P.R. China (20150309006NY). JJZ received research funding from BBSRC UK-China Partnering Awards (BB/J020281/1) and China-UK Cooperation Programme in Global Priorities (BB/L001683/1).