INTRODUCTION
Trypanosoma vivax, T. congolense and T. brucei are responsible for livestock mortality and morbidity due to trypanosomiasis in Africa. T. vivax is widespread in Africa, Central and South America, and is predominantly a ruminant parasite infecting cattle, buffalo, goats, sheep and wild bovids (Hoare, Reference Hoare1972; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). Wild ungulates, especially buffalo and antelope, and trypanotolerant cattle are symptomless carriers, despite presenting high T. vivax and other pathogenic trypanosome infection rates (Moloo et al. Reference Moloo, Gettinby, Olubayo, Kabata and Okumu1993, Reference Moloo, Orinda, Sabwa, Minja and Masake1999; Njiokou et al. Reference Njiokou, Simo, Nkinin, Laveissière and Herder2004). In Africa, transmission of T. vivax is both cyclical by tsetse flies and mechanical by tabanid flies, which allows T. vivax to spread in tsetse-free and tsetse-infested areas. T. vivax is the only tsetse-borne trypanosome that has established itself outside Africa, where mechanical transmission by several biting flies, mainly tabanids, is the only mode of transmission (Gardiner and Mahmoud, Reference Gardiner, Mahmoud, Kreier and Baker1992; Jones and Davila, Reference Jones and Davila2001).
T. vivax infecting both African cattle and goat exhibits variable virulence and pathogenicity, ranging from completely asymptomatic chronic infections to wasting disease with severe haematological alterations and death. Although it is commonly alleged that West African isolates are more virulent than East African isolates, disseminated haemorrhagic syndrome caused by T. vivax was reported in Kenya (Gardiner, Reference Gardiner1989). Clinical manifestation of infections caused by T. vivax throughout its geographical distribution depends largely on host immunity, with disease arising when enzootic stability is missing (Gardiner and Mahmoud, Reference Gardiner, Mahmoud, Kreier and Baker1992; Batista et al. Reference Batista, Riet-Correa, Teixeira, Madruga, Simões and Maia2007). In South America, cattle infected with T. vivax are mostly asymptomatic, with rare haematological and nervous system symptoms, abortions and death (Jones and Davila, Reference Jones and Davila2001; Batista et al. Reference Batista, Riet-Correa, Teixeira, Madruga, Simões and Maia2007). We showed that South American T. vivax isolates from either asymptomatic cattle or those exhibiting distinct pathological profiles are closely related to isolate Y486 from Nigeria, West Africa (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006).
Despite the prevalence of T. vivax in buffalo, antelope and other wild ruminants in Africa (Moloo et al. Reference Moloo, Gettinby, Olubayo, Kabata and Okumu1993, Reference Moloo, Orinda, Sabwa, Minja and Masake1999), isolates from these reservoirs were never compared at the molecular level with livestock isolates. Wild reservoirs of T. vivax have been identified using morphology and developmental progression in tsetse flies (Moloo et al. Reference Moloo, Gettinby, Olubayo, Kabata and Okumu1993). Furthermore, sensitive detection of T. vivax has been achieved via PCR using DNA from blood samples of wild ungulates and tsetse flies (Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003; Njiokou et al. Reference Njiokou, Simo, Nkinin, Laveissière and Herder2004).
The phylogenetic position of T. vivax on the edge of the clade constituted by all African trypanosomes agreed with distinct biological and molecular features of T. vivax, including its unique development in tsetse flies (Stevens et al. Reference Stevens, Noyes, Schofield and Gibson2001; Stevens and Rambaut, Reference Stevens and Rambaut2001; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). In our previous phylogenetic study of T. vivax, an Eastern African (Kenya) isolate from cattle (IL3905) was distinct from South American and West African (Nigeria) isolates (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). This result suggested the existence of a divergent T. vivax isolate in East Africa, where other distinct genetic variants (T. vivax-like) have been reported in Tanzania (Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003). All molecular data generated to date were congruent with the differentiation between West and East African T. vivax. These molecular results were supported by morphological, immunological and behavioural data in tsetse and vertebrate hosts (Murray and Clarkson, Reference Murray and Clarkson1982; Moloo et al. Reference Moloo, Kutuza and Desai1987; Gardiner and Wilson, Reference Gardiner and Wilson1987; Vos and Gardiner, Reference Vos and Gardiner1990; Fasogbon et al. Reference Fasogbon, Knowles and Gardiner1990; Dirie et al. Reference Dirie, Murphy and Gardiner1993a, Reference Dirie, Otte, Thatthi and Gardinerb).
The subgenus Duttonella has been considered monotypic, represented only by T. vivax. However, T. uniforme, a species represented by the smallest trypanosome forms and occurring mainly in Central Africa, was placed in this subgenus according to traditional taxonomic traits (Hoare, Reference Hoare1972). Three subspecies of T. vivax have been recognized, each exhibiting differences in geographical origin and in several aspects of their biology, morphology and pathology: T. vivax vivax and T. vivax ellipsiprymni in Africa, and T. vivax viennei in Central and South America (Shaw and Lainson, Reference Shaw and Lainson1972; Hoare, Reference Hoare1972). T. v. vivax and T. v. viennei represent morphologically indistinguishable African and South American isolates but are considered distinct subspecies due to their respective cyclical or mechanical transmission. However, the validity of this subspecies is controversial because T. v. vivax can be mechanically transmitted in areas free of tsetse in Africa (Sinshaw et al. Reference Sinshaw, Abebe, Desquesnes and Yoni2006; Delafosse et al. Reference Delafosse, Thébaud, Desquesnes and Michaux2006). T. v. ellipsiprymni is distinguishable by morphological peculiarities and has been reported in goats and antelopes in Tanzania and Malawi; in water-bucks (Kobus ellipsiprymnus) in Zambia; and in Glossina morsitans in Rwanda-Burundi (Keymer, Reference Keymer1969; Hoare, Reference Hoare1972). The taxonomy of T. v. ellipsiprymni and T. uniforme has not been assessed by molecular approaches. In this study, we adopted T. vivax-like for all trypanosomes positioned within the subgenus Duttonella separated by relevant genetic distance from T. vivax.
Although T. vivax infecting cattle from South America and West Africa have different modes of transmission (cyclical or mechanical), phylogenetic data suggested a very close relationship among all isolates from these regions (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). However, these isolates were distinguished from East African T. vivax by morphological, pathological, differential behaviours in ruminants and tsetse, and molecular features (Keymer, Reference Keymer1969; Hoare, Reference Hoare1972; Fasogbon et al. Reference Fasogbon, Knowles and Gardiner1990; Dirie et al. Reference Dirie, Murphy and Gardiner1993a, Reference Dirie, Otte, Thatthi and Gardinerb; Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). Our previous study justified the recognition of one Kenyan isolate (IL3905) (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006), now referred to as a T. vivax-like trypanosome, as a new subspecies/species if this isolate could be characterized by a combination of molecular, morphological and biological traits as recommended by Gibson et al. (Reference Gibson2007).
Therefore, the current taxonomy of T. vivax and the subgenus Duttonella deserve to be revised. The recognition of new trypanosomes at the subgeneric level, and specific and subspecific classifications should be confirmed in phylogenetic affiliation and in the levels of genetic divergence, taking into account a set of data, including morphology, mode of transmission, vertebrate hosts and vector range, geographical origin and distribution, and pathology (Gibson, Reference Gibson2007). This study was conducted integrating phylogenetic, morphological, biological and geographical data to address phylogenetic relationships within the subgenus Duttonella. The results of this study supported the description of a T. vivax-like trypanosome from a nyala antelope (Tragelaphus angasi) wild-caught in Mozambique (East Africa) classified as a new genotype by biological, morphological and molecular parameters. Analysis of phylogenetic relationships and geographical pattern of T. vivax populations were done using SSU and ITS rDNA sequences from T. vivax and T. vivax-like trypanosomes from South America, West and East Africa.
MATERIALS AND METHODS
Origin, growth, and morphological and molecular diagnoses of Trypanosoma vivax isolates
A new trypanosome isolate (TviMzNy) was recovered from blood of a wild-caught nyala antelope (Tragelaphus angasi) captured in the Province of Sofala, Central region of Mozambique, East Africa in 2006. Blood (~3·0 ml) from the naturally infected nyala was inoculated (via endovenous) into a young goat (~5 months). The goat was maintained in an insect-proof housing and provided water and nutritional concentrate ad libitum. Blood samples of the infected goat were cryopreserved in liquid nitrogen or mixed (v/v) with ethanol for further DNA extraction. Diagnosis of T. vivax was initially done by microhaematocrit (MH) and blood samples from nyala smeared on glass slides. Blood samples from cattle, goats and other antelope from Mozambique (East Africa) were also examined by MH. Giemsa-stained blood smears were photographed for morphological identification. Morphological features and measures of trypomastigotes were compared with morphotypes described by Hoare (Reference Hoare1972) for T. vivax subspecies and allied species.
To confirm the morphological classification, blood samples from naturally and experimentally infected animals, which were either spotted on filter paper or preserved in ethanol, were tested using 2 different PCR assays. Amplification of ITS1 rDNA employed primers and reaction conditions previously described by Rodrigues et al. (Reference Rodrigues, Paiva, Campaner, Stevens, Noyes and Teixeira2006), and TviSL-PCR amplifications according to the method of Ventura et al. (Reference Ventura, Paiva, Silva, Takeda, Buck and Teixeira2001).
DNA preparations, PCR amplification and sequencing of SSU and ITS rDNA sequences
Crude DNA preparations were obtained from naturally infected nyala antelope, cattle and goat blood samples, dropped on filter paper and processed following the protocol of Ventura et al. (Reference Ventura, Takata, Silva, Nunes, Takeda and Teixeira2000). Blood samples of the experimentally infected goat preserved in ethanol (v/v), were incubated in a lysis buffer (1% SDS, 100 mm EDTA pH 8·0, 20 mm Tris-HCl, pH 8·0 and 350 μg/ml of proteinase K) at 37°C for 18 h, centrifuged at 14 000 g for 5 min and DNA purified using Wizard Purification Systems (Promega), and used as template for whole SSU rDNA PCR amplifications as before (Viola et al. Reference Viola, Campaner, Takata, Ferreira, Rodrigues, Freitas, Duarte, Grego, Barrett, Camargo and Teixeira2008). Crude DNA templates were obtained directly from blood collected in filter paper and submitted to PCR amplification of SSU rDNA V7–V8 region and the entire ITS rDNA (ITS1-5·8S-ITS2) using primers and reaction conditions previously described (Rodrigues et al. Reference Rodrigues, Paiva, Campaner, Stevens, Noyes and Teixeira2006; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). The amplified DNA fragments from all sequences were cloned and 3–5 clones of each gene for each isolate were sequenced.
Phylogenetic inferences and polymorphism evaluation among Trypanosoma vivax isolates
Phylogenetic analyses were conducted to infer the relationships of the nyala trypanosome with other trypanosomes using an alignment consisting of conserved sequences from whole SSU rDNA of T. vivax isolate Y486 (clone ILDat1.2) from Nigeria, West Africa (Table 1). SSU rDNA sequences from 23 species of mammalian and 13 non-mammalian trypanosomes were included in the alignment. Data were obtained from GenBank (Accession number) and included the following taxa: T. rotatorium (AJ009161); T. mega (AJ009157); T. boissoni (U39580); T. triglae (U39584); T. avium SIM3 (AF416563); T. avium APO1 (AF416561); T. grayi (AJ005278); T. bennetti (AJ223562); T. grayi-like-F4 (AJ620547); T. sp. H25 (AJ009168); T. rangeli (AJ009160); T. dionisii (AJ009151); T. cruzi (AJ009149); T. cyclops (AJ131958); T. theileri K127 (AJ009164); T. sp. D30 (AJ009165); T. varani (AJ005279); T. sp. H26 (AJ009169); T. pestanai (AJ009159); T. sp. Msubugwe-2006 (AM503350); T. brucei rhodesiense (AJ009142); T. b. brucei (M12676); T. b. gambiense (AJ009141); T. equiperdum (AJ223564); T. evansi (AJ009154); T. simiae (AJ009162); T. simiae Tsavo (U22318); T. sp. Fly9 (AJ563915); T. godfreyi (AJ009155); T. congolense kilifi K45.1 (U22317); T. congolense kilifi WG5 (AJ009144); T. congolense savannah IL1180 (U22315); T. congolense savannah 68Q (AJ223563); T. congolense savannah WG81 (AJ009146); T. congolense forest IL3900; and T. congolense forest Cam22b (AJ009145). Additional SSU and ITS rDNA sequences were retrieved from T. vivax Y486 genome data banks (http://www.genedb.org). Trypanosomatids of the genera Leishmania, Crithidia, Leptomonas, Herpetomonas and Phytomonas were selected as outgroup. Of the 2844 characters in the alignment, only 1797 (343 parsimony informative) were considered well aligned and included in the analysis. This alignment was accomplished using as guidance a published alignment of SSU rRNA gene sequences from a large set of taxa (Hamilton et al. Reference Hamilton, Stevens, Gidley, Holz and Gibson2005) (ALIGN_000791). Alignments are available from authors upon request.
Table 1. Geographical origin of African and South American Trypanosoma vivax isolates used in this study

a Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003; bCortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006; cLeeflang et al. Reference Leeflang, Buys and Blotkamp1976; dSanger Center Genome project; eclone derived from the isolate Y486; fPaiva et al. Reference Paiva, De Lemos, Nakazato, Mori, Brum and Bernardo2000; gSilva et al. Reference Silva, Ramirez, Souza, Ortiz, Pereira and Dávila1999; hBatista et al. Reference Batista, Riet-Correa, Teixeira, Madruga, Simões and Maia2007.
In addition to alignment based on conserved regions of whole SSU rDNA used to place the nyala isolate in the general phylogeny of trypanosomes, to assess the degree of polymorphism and phylogenetic relationships among the isolates nested in the T. vivax clade, we employed different data sets as follows (Table 1). (a) Alignment of the variable V7–V8 region of SSU rDNA of T. vivax isolates (East African isolate IL3905 from Kenya; West African T. vivax Y486 from Nigeria (clone ILDat 1.2) plus other salivarian trypanosomes (~1024 characters, 462 parsimony informative). (b) Alignment of all available T. vivax and T. vivax-like sequences corresponding only to the SSU rDNA V7 region (~289 characters, 59 parsimony informative), including sequence AJ563916 from a putative Tanzanian T. vivax-like trypanosome (Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003). (c) Alignment of the entire ITS rDNA (ITS1-5·8S-ITS2) sequences of T. vivax and T. vivax-like isolates plus other salivarian trypanosome species (550 characters, 313 parsimony informative). Sequences were aligned using ClustalX and manually refined. Parsimony and bootstrap analyses were carried out using PAUP* 4.0b10 (Swofford, Reference Swofford2002) with 100 replicates of random addition sequence followed by branch swapping (RAS-TBR) as previously described (Ferreira et al. Reference Ferreira, Campaner, Viola, Takata, Takeda and Teixeira2007). ML analyses were performed using RAxML v.2.2.3 (Stamatakis, Reference Stamatakis2006). Tree searches employed GTRGAMMA with 500 maximum parsimony-starting trees. Model parameters were estimated in RAxML over the duration of the tree search. Nodal support was estimated with 100 bootstrap replicates in RAxML using GTRGAMMA and maximum parsimony starting trees.
RESULTS
Behaviour in experimentally infected goat and morphology of the nyala trypanosome
A goat infected by inoculation of blood (3·0 ml) from a wild-caught nyala antelope infected with the T. vivax-like trypanosome developed high parasitaemia, severe anaemia, fever and progressive weight loss, with accentuated cachexia 20 days after inoculation, when blood was collected from the goat. Inoculation in mice of blood from infected nyala did not result in infection.
Trypomastigotes in Giemsa-stained blood smears from both the wild-caught nyala antelope and the experimentally infected goat exhibited very similar morphological features, consistent with those described for trypanosomes of the subgenus Duttonella (Hoare, Reference Hoare1972). The flagellates displayed a voluminous and circular kinetoplast with most forms showing a rounded swollen posterior end of the body, and a distinctive free flagellum (Fig. 1). Only trypomastigotes resembling T. vivax were found in the nyala blood, which presented high parasitaemia detected directly from a fresh drop of blood. In addition, in Mozambique we identified 2 goats and 2 cows with trypanosomes morphologically similar to those found in the nyala antelope. However, in this country, cattle were commonly infected by T. congolense and not T. vivax (data not shown).

Fig. 1. Micrographs of Giemsa-stained blood smears of Trypanosoma vivax-like from nyala (A) and T. vivax from cattle (B). Trypomastigote forms detected in blood of naturally infected nyala antelope from Mozambique (a, b); trypomastigotes found on blood from a goat experimentally-infected with the isolate TviMzNy (c–f); (B) blood forms observed in a sheep experimentally infected with the Brazilian isolate TviBrPo originally from cattle. Arrows indicate the (n) nucleus, (f) flagellum, (um) undulant membrane and (k) kinetoplast.
Comparison of blood samples revealed differences in T. vivax trypomastigote size and shape from South America compared with the nyala isolate (TviMzNy). The trypanosome from the nyala antelope exhibited larger trypomastigotes, with a more conspicuous undulating membrane, and subterminal and marginal kinetoplast. Most forms appeared as a heavier structure that gradually attenuated towards the anterior end, and a few presented narrow ends (Fig. 1A). The TviMzNy isolate blood trypomastigotes (mean from 30 flagellates) ranged from 21 to 33 μm (mean 27 μm) in length and from 2·0 to 4·0 μm (mean 3·0 μm) in width. In contrast, South American isolates exhibited blood trypomastigotes slightly reduced in length exhibiting a round and swollen posterior end that tapered rapidly to the anterior extremity and were largely monomorphic. The terminal kinetoplast was typical of most trypomastigotes, as illustrated by forms from a Brazilian cattle isolate (TviBrPo) (Fig. 1B), previously reported (Ventura et al. Reference Ventura, Paiva, Silva, Takeda, Buck and Teixeira2001). These forms and corresponding measurements are typical of T. v. vivax, with length varying from 19 to 31 μm (mean 24 μm) and width ranging from 2·0 to 4·0 μm (mean 3·0 μm) (Hoare, Reference Hoare1972).
Comparison of our data with the morphological features determined by Hoare (Reference Hoare1972) showed that blood trypomastigotes from the nyala isolate (TviMzNy) were more similar to the large forms described for East African T. vivax. The isolate also presented characteristics similar to those described for T. v. ellipsiprymni. Brazilian and Venezuelan T. vivax isolates were morphologically indistinguishable from T. vivax reported in West Africa (Nigeria, Cameroon and Senegal) (Hoare, Reference Hoare1972; Ventura et al. Reference Ventura, Paiva, Silva, Takeda, Buck and Teixeira2001; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006).
Phylogenetic position and taxonomy of the nyala trypanosome based on SSU rDNA sequences
In this study, we sequenced the entire SSU rDNA of the trypanosome isolated from a nyala antelope wild-caught in Mozambique, East Africa. The SSU rDNA sequence generated for the nyala isolate was aligned with the T. vivax Y486 (clone ILDat1.2) sequence from Nigeria, West Africa, and from a range of salivarian and other trypanosome species aiming the placement of the isolate in the general phylogeny of Trypanosoma. Regardless of the analytical approach, all analyses yielded similar topologies and the same relationships for all major clades of trypanosomes, as represented in the ML tree (Fig. 2). According to tree topologies, the nyala isolate clustered with T. vivax Y486, the only T. vivax isolate with a complete SSU rDNA sequence previously determined. These two isolates always formed a clade positioned at the edge of a major clade formed by all other salivarian trypanosomes, a placement congruent to previous studies (Stevens et al. Reference Stevens, Noyes, Schofield and Gibson2001; Hamilton et al. Reference Hamilton, Stevens, Gaunt, Gidley and Gibson2004; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). The isolate TviMzNy was closest to T. vivax Y486 (3·4% of SSU rDNA sequence divergence) than to any other trypanosome, forming a strongly supported clade (100% boostrap) that warrants its classification within the subgenus Duttonella.
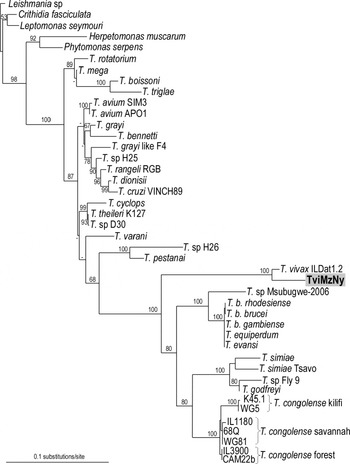
Fig. 2. Phylogenetic tree of trypanosomes inferred by ML analysis using SSU rDNA sequences from Trypanosoma vivax (ILDat1.2) and T. vivax-like (TviMzNy) isolates from a nyala antelope plus 36 additional trypanosome species. Trypanosomatids from 5 other genera were selected as outgroups for Trypanosoma. −lnL=8284·585960. Bootstrap support from 100 replicates is indicated at the nodes.
To assess the genetic relatedness of the nyala trypanosome with several T. vivax and T. vivax-like isolates, we compared the polymorphic V7–V8 or only V7 region of SSU rDNA from African and South American isolates. Only partial sequences of SSU rDNA were obtained for most T. vivax isolates due to difficulties in PCR amplifications using as DNA templates crude preparations of field-collected blood samples, generally showing low parasitaemias and mixed infections. However, we previously demonstrated that V7–V8 SSU rDNA sequences are useful markers to resolve the relationships between T. vivax and other African trypanosomes (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). Furthermore, V7–V8 sequences were adequate to evaluate the degrees of polymorphism and genetic relatedness and to distinguish all species and lineages of trypanosomes that we have examined to date (Maia da Silva et al. Reference Maia da Silva, Noyes, Campaner, Junqueira, Coura, Anez, Shaw, Stevens and Teixeira2004; Rodrigues et al. Reference Rodrigues, Paiva, Campaner, Stevens, Noyes and Teixeira2006; Ferreira et al. Reference Ferreira, Campaner, Viola, Takata, Takeda and Teixeira2007; Viola et al. Reference Viola, Campaner, Takata, Ferreira, Rodrigues, Freitas, Duarte, Grego, Barrett, Camargo and Teixeira2008).
Sequences of V7–V8 SSU rDNA obtained directly from the nyala and infected goat blood samples were identical, indicating that the trypanosome TviMzNy isolated from the experimentally infected goat is the same as that of the field-caught nyala antelope (Fig. 3B). These sequences were aligned with previously determined sequences from other African isolates, including IL3905 (Kenya), ILDat1.2 (Nigeria), 2 cattle isolates from Brazil (TviBrMi and TviBrCa) and 1 from Venezuela (TviVeMe). Despite the geographical distance separating T. vivax-like from East Africa and all other T. vivax isolates, phylogenetic analysis formed a well-supported lineage (100% of bootstrap) comprised of all these isolates that corresponds to the subgenus Duttonella (Figs 2 and 3B). In the phylogenetic tree based on V7–V8 SSU rDNA sequences, the TviMzNy isolate was positioned closest to the isolate IL3905 from Kenya (Fig. 3B). Sequences from V7–V8 SSU rDNA of the nyala isolate TviMzNy showed a GC content of 65%, similar to South American/West African isolates (63%) and T. vivax-like IL3905 from Kenya (64%), corroborating high GC content in all members of the subgenus Duttonella (Stevens and Rambaut, Reference Stevens and Rambaut2001; Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006).
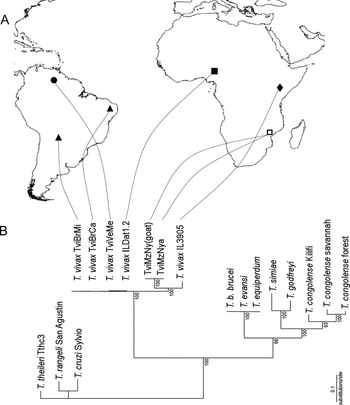
Fig. 3. (A) Map indicating the geographical origin of isolates from East Africa (□ Mozambique, ◆ Kenya), West Africa (■ Nigeria) and South America (▲ Brazil, • Venezuela). (B) Phylogenetic tree based on ML analysis of SSU (V7–V8) rDNA sequences of Trypanosoma vivax isolates and other salivarian trypanosome species. T. theileri, T. cruzi and T. rangeli were used as outgroups representing non-salivarian trypanosomes. Bootstrap support from 100 replicates is indicated at the nodes.
A comparison of the V7 region of SSU rDNA sequences from isolates included in this study with the unique DNA sequence amplified directly from a tsetse fly collected in East Africa (Tanzania) and attributed to a T. vivax-like trypanosome (Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003) was conducted. The Tanzanian T. vivax-like trypanosome was most divergent within the subgenus Duttonella, equally distant from isolates IL3905 (Kenya) (~10·3%) and TviMzNy (~11%). The same branching pattern of T. vivax and T. vivax-like sequences was generated by V7–V8 (Fig. 3B) or only V7 SSU rDNA (Fig. 4) analyses, with trypanosomes from East Africa consistently positioned in distant branches separated from the South American/West African lineage.

Fig. 4. Dendrogram based on partial V7 SSU rDNA sequences of Trypanosoma vivax and T. vivax-like isolates inferred using parsimony analysis. Bootstrap support from 100 replicates is indicated at the nodes.
Genetic polymorphism and relatedness among Trypanosoma vivax isolates based on whole ITS rDNA sequences
To better assess the degree of polymorphisms and divergence pattern within clade T. vivax we compared length and sequence of PCR-amplified entire ITS rDNA (ITS1, 5·8S and ITS2). The whole ITS rDNA (wITS) sequence of TviMzNy was aligned with sequences from South American isolates (3 Brazilian and 1 Venezuelan) plus African isolates IL3905 from Kenya and Y486 from West Africa (clone ILDat1.2). All South American/West African isolates had both wITS and ITS1 sequences of similar length, 490 bp and 143 bp, respectively. However, the East African isolates displayed length polymorphisms. TviMzNy differed slightly in length from South American/West African isolates for both wITS (486 to 488 bp) and ITS1 (145 bp), whereas the isolate IL3905 presented wITS varying from 525 to 534 bp, and ITS1 from 151 to 157 bp.
Analysis of T. vivax ITS rDNA sequence polymorphisms disclosed small divergences among South American isolates (~0·5%) and between these isolates and the West African isolate (~0·8%). However, high divergences separated these isolates from East African isolates. Divergences varied from ~36·4% for TviMzNy to ~44% in IL3905, whereas ~48% of ITS sequence divergence separated TviMzNy from IL3905. Data from TviMzNy were consistent with the heterogeneity observed among ITS sequences within the African isolates, with intra-isolate divergence varying from 0·8% for Y486, 1·0% for TviMzNy to 7·6% for IL3905. We further selected from 2–5 of the most polymorphic cloned wITS sequences from each isolate to infer phylogenetic relationships among T. vivax isolates. The dendrogram branching pattern was similar to that generated by V7–V8 SSU rDNA sequences. ITS sequences from the same isolate always grouped together. Sequences from South American and West African isolates also always clustered together in a very homogeneous clade (~0·8% maximum divergence), as we previously demonstrated (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006), while sequences from Kenya and Mozambique segregated into 2 divergent lineages (Fig. 5).
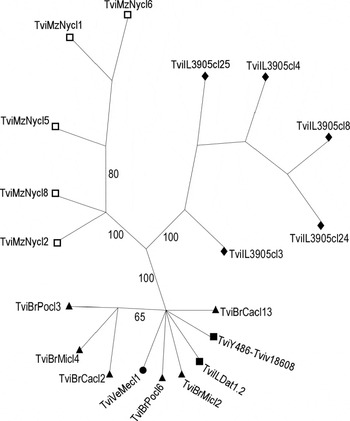
Fig. 5. Dendrogram based on parsimony analysis of the entire ribosomal ITS sequences (ITS1-5·8S-ITS2) from Trypanosoma vivax and T. vivax-like isolates. Symbols correspond to geographical origin of isolates from East Africa (□ Mozambique, ◆ Kenya), West Africa (■ Nigeria) and South America (▲ Brazil, • Venezuela). Bootstrap support from 100 replicates is indicated at the nodes.
Molecular diagnosis and genotyping of Trypanosoma vivax and T. vivax-like isolates
Nyala TviMzNy and all other isolates morphologically identified as T. vivax were tested using a PCR assay based on ITS1 rDNA sequences (Rodrigues et al. Reference Rodrigues, Paiva, Campaner, Stevens, Noyes and Teixeira2006). Methods based on ITS1 rDNA have been used for distinguishing all livestock-infecting trypanosome species according to different sizes of amplified DNA fragments (Desquesnes et al. Reference Desquesnes, McLaughlin, Zoungrana and Davila2001; Adams et al. Reference Adams, Hamilton, Malele and Gibson2007). According to results from the present study, TviMzNy presented ITS1 of similar length compared to isolates from West Africa and South America and were consequently identified as T. vivax (Fig. 6). Polymorphism of ITS1 sequences showed utility as markers for analysis of T. vivax populations by distinguishing individual genotypes. TviSL-PCR based on the spliced leader sequence of T. vivax (Ventura et al. Reference Ventura, Paiva, Silva, Takeda, Buck and Teixeira2001) yielded negative results for TviMzNy indicating that this method is useful only to detect closely related isolates from West Africa and South America (Fig. 6).

Fig. 6. Molecular diagnosis of Trypanosoma vivax and T. vivax-like isolates. Agarose gels (2%) stained with ethidium bromide of amplified ITS1 rDNA (ITS1-PCR) and spliced leader (TviSL-PCR) DNA fragments from East African T. vivax-like isolates IL3905 (Kenya) and TviMzNy (Mozambique). The isolate TviBrCa was selected to illustrate results obtained for all South American/West African T. vivax isolates tested in these assays.
DISCUSSION
A detailed understanding of genetic diversity and phylogeography of T. vivax is crucial for appraisal of taxonomic and phylogenetic relationships among currently recognized T. vivax genotypes, subspecies and allied species, as well as their relationships with other species of Trypanosoma. The complexity and evolutionary history of the trypanosomes classified in the subgenus Duttonella is far from understood and only recently studies have been initiated comparing isolates from South America and Africa. We previously demonstrated that isolates from South America and an isolate from West Africa (Y486) formed a homogeneous lineage. However, only 1 isolate from East Africa has been included in phylogenetic analyses and was positioned distant from the clade comprising South American and West African isolates (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). To further investigate these results, a survey of trypanosomes in domestic and wild ruminants was carried out in Mozambique, East Africa in this work, and a trypanosome from a wild-caught nyala antelope (Tragelaphus angasi) was isolated and compared with cattle isolates from Brazil and Venezuela (South America), Nigeria (West Africa) and Kenya (East Africa). The nyala isolate was diagnosed at both the morphological and molecular levels as a T. vivax-like trypanosome, and its behaviour in an experimentally infected goat was compatible with this species (Hoare, Reference Hoare1972; Masake, Reference Masake1980; Gardiner, Reference Gardiner1989; Gardiner and Mahmoud, Reference Gardiner, Mahmoud, Kreier and Baker1992).
Our current research represents the first study to combine field surveys, biological, morphological and molecular analyses, in addition to phylogenetic inferences and geographical analysis to characterize a new genotype of T. vivax-like, aiming to clarify the phylogenetic and taxonomic relationships between these trypanosomes and T. vivax. The T. vivax-like trypanosome from the nyala antelope clustered together with T. vivax Y486, although separated by a significant genetic distance, supporting its placement within the subgenus Duttonella.
Although relatively few T. vivax isolates have been compared, zymodeme, satellite DNA, kDNA minicircle and karyotype patterns grouped West African and South American (Colombia) isolates together, separated from East African (Kenya) isolates (Fasogbon et al. Reference Fasogbon, Knowles and Gardiner1990; Dirie et al. Reference Dirie, Murphy and Gardiner1993a, Reference Dirie, Otte, Thatthi and Gardinerb). Besides variable pathogenicity and virulence, T. vivax from West and East Africa differ in morphological features. Short trypomastigote forms were associated with acute disease in West African cattle and long forms with chronic infection in cattle, goats and wild ruminants (Hoare, Reference Hoare1972; Gardiner and Mahmoud, Reference Gardiner, Mahmoud, Kreier and Baker1992). T. v. vivax and T. v. viennei are morphologically indistinguishable West African and South American subspecies separated respectively by their cyclical and mechanical mode of transmission. T. v. ellipsiprymni is separated from T. v. vivax by morphological peculiarities (Hoare, Reference Hoare1972). The lack of molecular taxonomic markers and the small number of isolates have hampered progress in generating more accurate diversity levels within the subgenus Duttonella (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006).
In the present study, we addressed the taxonomy of the subgenus Duttonella by analysing morphological and molecular characteristics of African and American trypanosomes. Morphological analysis of a trypanosome from the nyala antelope revealed large trypomastigotes with morphological features previously reported for East African T. v. vivax and T. v. ellipsiprymni. We could not distinguish blood trypomastigotes (described by Hoare, Reference Hoare1972) of these subspecies from those found in blood smears of the nyala antelope. Smaller trypomastigotes of Brazilian T. vivax were similar to those reported in other South American countries (Colombia, Bolivia and Venezuela) and West Africa (Nigeria and Senegal) (Hoare, Reference Hoare1972; Ventura et al. Reference Ventura, Paiva, Silva, Takeda, Buck and Teixeira2001).
Length and sequence polymorphism analyses of the nyala isolate showed a significant degree of variability among ITS gene copies, which appear to be a peculiarity of T. vivax-like trypanosomes in Africa, particularly East Africa as we previously showed for the Kenyan isolate IL3095 (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). The West African isolate Y486 used in most studies of T. vivax has been for a long time maintained in mice, which could have selected a homogeneous clone. Analyses of more isolates field-collected in areas with or without tsetse are needed to clarify if polymorphisms of the African isolate are related to cyclical transmission by tsetse flies, thus justifying the high homogeneity of South American isolates, which are all mechanically transmitted.
The nyala isolate was not detected by PCR based on SL sequences (Ventura et al. Reference Ventura, Paiva, Silva, Takeda, Buck and Teixeira2001), in agreement with the negative result previously shown for IL3905 from Kenya (Cortez et al. Reference Cortez, Ventura, Rodrigues, Batista, Paiva, Añez, Machado, Gibson and Teixeira2006). In addition, attempts to detect T. vivax in Kenya and Tanzania using other PCR assays revealed the existence of distinct populations, indicating that the use of general primers for detection of all populations is crucial in T. vivax surveys (Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003; Njiru et al. Reference Njiru, Makumi, Okoth, Ndungu and Gibson2004).
Data from this and previous studies indicated that T. vivax and the whole subgenus Duttonella are more complex than previously considered. The isolate from the nyala antelope (TviMzNy) was closer to T. vivax than to any other trypanosome species in all phylogenetic analyses, congruent with its morphological and molecular diagnosis. Morphological, biological and phylogenetic data suggested that the nyala trypanosome is a new genotype closely related to T. vivax of the subgenus Duttonella. Phylogenetic affiliation and the relevant genetic distance separating the isolate from Mozambique from South American, Nigerian, Kenyan and Tanzanian T. vivax and T. vivax-like suggests that the nyala trypanosome can be recognized as a new subspecies or even a new species when molecular taxonomic parameters have been defined for the subgenus Dutonella (Gibson et al. Reference Gibson2007). According to morphological features and geographical origin, this isolate could correspond to an ‘atypical’ T. vivax or T. v. ellipsiprymni from East Africa (Hoare, Reference Hoare1972). However, T. v. ellipsiprymni was described in different antelope hosts and to our knowledge this subspecies is not available for comparative studies and has not recently been reported. Consequently, we designated TviMzNy as a trypanosome closely phylogenetically related to T. vivax (T. vivax-like) until additional data are gathered regarding the heterogeneity within T. vivax. Furthermore, sequence data from different genes/gene regions should be pursued to allow a general appraisal of the taxonomy of the subgenus Duttonella.
The recognition of new salivarian trypanosome species is increasing due the availability of new molecular methods. Surveys of trypanosomes in wild tsetse have verified that levels of diversity in these trypanosomes are far greater than previously recognized. Recently, a new species related to T. brucei was described in the subgenus Trypanozoon (Adams et al. Reference Adams, Hamilton, Malele and Gibson2008; Hamilton et al. Reference Hamilton, Adams, Malele and Gibson2008). A molecular survey of trypanosomes in tsetse proboscids reported 1 new T. vivax genotype from Tanzania (Malele et al. Reference Malele, Craske, Knight, Ferris, Njiru, Hamilton, Lehane, Lehane and Gibson2003). Considering the high diversity already observed in limited surveys, future studies of the subgenus Duttonella may reveal even higher levels of diversity leading to the recognition of additional new subspecies and species. Several new genotypes have been ascribed within the subgenus Nannomonas but only 3 species are recognized: T. congolense, T. simiae and T. godfreyi. Genetic divergences in SSU rDNA sequences among T. congolense savannah, forest and kilifi are far greater (4·5%) than that within the whole subgenus Trypanozoon (0·08%), and even larger than that found within the subgenus Duttonella (3·4%). Nevertheless, genotypes of T. congolense have not been so far recognized as separate species or subspecies because they lack obvious morphological, biological or clinical differences (Gibson, Reference Gibson2007).
The trypanosome survey carried out in Mozambique for this study revealed T. vivax in cattle and goats morphologically similar to the T. vivax-like isolate from a nyala antelope. Unfortunately, we were unable to amplify trypanosome ribosomal sequences from the blood of these animals to compare with the nyala trypanosome. The TviMzNy trypanosome is probably transmitted by tsetse flies (Glossina pallidipes), which are abundant in the area where the nyala antelope was captured (data not shown). Experimental infection of a goat with the nyala T. vivax-like indicated that this trypanosome can be infective and virulent for domestic animals. Identification of any new trypanosome genotype, subspecies or species of trypanosome circulating in wild ruminants that can be transmitted to livestock is a matter of considerable veterinary importance in Africa.
We are indebted to people from Mozambique for their generosity and inestimable help in the fieldwork. We specially thank Carmen and José Martins from Chupanga, Mozambique, for their friendly hospitality and invaluable support. We are grateful to Drs E. P. Camargo and W. C. Gibson for their continued criticism and encouragement. This work was supported by grants from the Brazilian agency CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) within the PROAFRICA program. A. C. Rodrigues is a post-doctoral fellow of PROTAX-CNPq; L. B. Viola and A. Marcili are recipients of scholarships from CNPq, and Maia da Silva, F. is a post-doctoral fellow sponsored by PRODOC-CAPES.









