INTRODUCTION
The members of the heat shock protein 70 (HSP70) family are among the most conserved and widely studied stress proteins. Due to their high abundance and high conservation of their primary structures and functions, they are also widely used in phylogenetic analysis. For example, sequences of HSP70s have been used to confirm monophyly of Metazoa (Borchiellini et al., Reference Borchiellini, Boury-Esnault, Vacelet and Le Parco1998).
The HSP70 is a large multigene protein family with members residing in a number of subcellular compartments, including the cytoplasm, mitochondria and endoplasmic reticulum (ER). Members of the HSP70 family assist in a large variety of cellular processes to ensure cellular homeostasis during normal and stressful conditions. The functions of HSP70s include the folding and assembly of newly synthesized proteins, refolding of misfolded and aggregated proteins, membrane translocation of organellar and secretory proteins, and control of the activity of regulatory proteins (Mayer & Bukau, Reference Mayer and Bukau2005). Heat shock proteins participate in minimizing the aggregation of non-native proteins and targeting non-native or aggregated proteins for degradation and removal from the cell (Feder & Hofmann, Reference Feder and Hofmann1999). The broad range of functions has been achieved through the amplification and diversification of HSP70 genes in evolution (generating specialized HSP70 chaperones), a special range of co-chaperones that target HSP70s to their substrates, and cooperation with other chaperone systems (Mayer & Bukau, Reference Mayer and Bukau2005).
The need for molecular chaperones is accelerated under stressful conditions that could potentially damage the cellular and molecular structures in the cell (Buchberger et al., Reference Buchberger, Bukau and Sommer2010). Factors of both anthropogenic and natural origin have been shown to induce the synthesis of HSPs in organisms ranging from bacteria and plants to mammals (Sanders, Reference Sanders1993). In marine organisms, factors that have all been linked to stress comprise those of environmental origin, as well as those caused by anthropogenic pollution, introduced species and nutrient enrichment (Lafferty et al., Reference Lafferty, Porter and Ford2004).
The HSP70s are among the most conserved proteins, having 60–78% identity of amino acid sequences among eukaryotic organisms and 40–60% identity among eukaryotic HSP70s and Escherichia coli DnaK sequence (Bardwell & Craig, Reference Bardwell and Craig1984; Kregel, Reference Kregel2002).
Eukaryotic genomes have multiple HSP70 genes including the genes that are solely inducible, those constitutively expressed but inducible by stress, and those solely constitutively expressed (heat shock cognates) (Hendrick & Hartl, Reference Hendrick and Hartl1993). The human genome harbours at least 17 genes coding for proteins of the HSP70 family (Brocchieri et al., Reference Brocchieri, Conway de Macario and Macario2008). Glucose regulated proteins GRP75 and GRP78 that also have different levels of inducibility, form a separate group among the HSP70 protein family. Glucose regulated proteins are stress-inducible chaperones that mostly reside in the ER or mitochondria. They are constantly expressed at basal level and elevated levels of GRP78 are used as indicators for ER stress. Glucose regulated protein 78 takes part in a vast number of physiological processes: the stress response, proliferation, apoptosis, angiogenesis, inflammation and immune response (Lee, Reference Lee2014).
The use of HSPs as biomarkers is most widely used in aquatic toxicology (Feder & Hofmann, Reference Feder and Hofmann1999). The sessile lifestyle of sponges and their constant filtering of surrounding water, with their ability to respond to environmental stress, makes them especially suitable as biomarkers to assess the effects of pollution (Agell et al., Reference Agell, Uriz, Cebrian and Martí2001; Cebrian et al., Reference Cebrian, Agell, Martí and Uriz2006). It has been shown that in addition to heat stress, the increase in HSP70 levels can also be induced by a number of factors including heavy metals (Agell et al., Reference Agell, Uriz, Cebrian and Martí2001), cold (Koziol et al., Reference Koziol, Wagner-Hülsmann, Mikoc, Gamulin, Kruse, Pancer, Schäcke and Müller1996), acoustic stress (Filiciotto et al., Reference Filiciotto, Vazzana, Celi, Maccarrone, Ceraulo, Buffa, Di Stefano, Mazzola and Buscaino2014), and space competition or aggression among sessile marine invertebrates (Rossi & Snyder, Reference Rossi and Snyder2001). Recently HSP70s have reattracted a lot of attention as markers for heat stress in the light of climate warming (Fan et al., Reference Fan, Liu, Simister, Webster and Thomas2013; Webster et al., Reference Webster, Pantile, Botté, Abdo, Andreakis and Whalan2013).
The first documentation of HSP response in sponges goes back to 1993 when elevated immunologically detectable HSP70 levels were seen in cubes of the marine sponge Geodia cydonium that were treated with tributyltin in laboratory conditions (Batel et al., Reference Batel, Bihari, Rinkevich, Dapper, Schacke, Schroder and Müller1993). A significant increase in immunologically detectable HSP70 levels was observed in that sponge in response to temperature and pH changes (Koziol et al., Reference Koziol, Wagner-Hülsmann, Mikoc, Gamulin, Kruse, Pancer, Schäcke and Müller1996). Koziol et al. (Reference Koziol, Wagner-Hülsmann, Mikoc, Gamulin, Kruse, Pancer, Schäcke and Müller1996) cloned the first cytosolic putatively inducible HSP70 cDNA from G. cydonium. In an attempt to solve the deep phylogeny of sponges, cytosolic HSP70s were also cloned from Sycon raphanus, the marine sponge that belongs to the class Calcarea, and Rhabdocalyptus (Acanthascus) dawsoni, the marine sponge that belongs to the class Hexactinellida (Koziol et al., Reference Koziol, Leys, Müller and Müller1997). In 1998, ER protein GRP78 from Suberites domuncula was added to the cloned HSP70 family proteins from sponges (Koziol et al., Reference Koziol, Kobayashi, Müller and Müller1998).
The expression of proteins from the HSP70 superfamily has been studied in several sponge species in different stressful conditions from heat and pollutants to disease-like conditions as well as through developmental processes of a freshwater sponge (Müller et al., Reference Munro and Pelham1995, Reference Müller, Koziol, Dapper, Kurelec, Batel and Rinkevich1998; Schröder et al., Reference Schröder, Batel, Lauenroth, Hassanein, Lacorn, Simat, Steinhart and Müller1999; Agell et al., Reference Agell, Uriz, Cebrian and Martí2001; Efremova et al., Reference Efremova, Margulis, Guzhova, Itskovich, Lauenroth, Müller and Schröder2002; Cebrian et al., Reference Cebrian, Agell, Martí and Uriz2006; Schill et al., Reference Schill, Pfannkuchen, Fritz, Köhler and Brümmer2006; López-Legentil et al., Reference López-Legentil, Song, McMurray and Pawlik2008).
Several studies have been conducted to assess the effects of water-based drill cuttings/sedimentation to benthic communities (Trannum et al., Reference Trannum, Nilsson, Schaanning and Øxnevad2010; Trefry et al., Reference Trefry, Dunton, Trocine, Schonberg, McTigue, Hersh and McDonald2013). Offshore drilling activities discharge large amounts of waste material in the form of drill cuttings that cause increased sedimentation around oil and gas installations, which cause a significant reduction in the number of taxa, abundance, biomass and diversity of macrofauna in benthic communities (Trannum et al., Reference Trannum, Nilsson, Schaanning and Øxnevad2010). Water-based mud causes sedimentation and therefore physical stress to sessile benthic animals up to a distance of 500 m from the actual drilling site (Bakke et al., Reference Bakke, Klungsøyr and Sanni2013).
The marine sponge Thenea muricata (Bowerbank, 1858) (Demospongiae, Astrophorida, Theneidae, Thenea) has adapted to life in the deep sea (60–3000 m), preferably on soft muddy bottoms. The habitats of T. muricata, the most abundant species of the genus Thenea, are widely distributed in the Atlantic Ocean from the waters of North Greenland to the southern Mediterranean Sea (Cárdenas & Rapp, Reference Cárdenas and Rapp2012). The data about molecular biology and biochemistry of this widely distributed soft bottom sponge are scarce: in one study the metabolic activity and oxygen consumption of T. muricata was determined (Witte & Graf, Reference Witte and Graf1996), in another one the nucleotide profile of T. muricata was studied, which revealed the presence of unusual 2′,5′-linked oligonucleotides, the natural metabolites of 2′,5′-oligoadenylate synthetase (Lopp et al., Reference Lopp, Reintamm, Kuusksalu, Tammiste, Pihlak and Kelve2010). Cárdenas et al. (Reference Cárdenas, Xavier, Reveillaud, Schander and Rapp2011) sequenced a fragment of the mitochondrial cytochrome c oxidase I gene and C1-D2 domain of 28S ribosomal gene, which are so far the only genetic data about T. muricata available in the GenBank.
In the framework of the SPONGRAM (Sponge Risk Assessment and Monitoring) project (2008–2009), whose main goal was to assess the potential negative impact on deep water sponge communities caused by elevated sedimentation rates associated with offshore oil and gas drilling activity, T. muricata was chosen to represent deep water sponges. The SPONGRAM project combined the background data of the sponge physiology under in situ conditions and laboratory data to assess cultivation and sedimentation effects on sponge survival, cell physiology and microbial community. The results of the project which suggested stress conditions of the sponges inspired us to study HSP70 expression in different sponge samples to gain more specific information about their stress response.
The aim of the present work was to study the expression of HSP70 during cultivation of T. muricata under laboratory conditions and subsequent sediment exposure experiments. Stress response of sponges was assessed on protein and mRNA levels. Several gene fragments from T. muricata coding for HSP70 family proteins were identified and the observed HSP70 set in the studied sponge samples was described.
MATERIALS AND METHODS
Sponge samples
Sponges were collected from the North Sea on a soft bottom sediment off Korsfjord (close to Bergen) at 300 m depths (60°07′44N 004°49′9E) in the framework of the SPONGRAM project (2008–2009). Sponges were shock frozen in liquid nitrogen directly on board the research vessel or transported to the laboratory for cultivation studies. The sponge samples were transported to the aquarium facility of the University of Bergen in water from the sampling site and maintained in natural running seawater from a depth of 200 m. They were cultivated in tanks with running seawater (seawater culture) or planted in sediment (sediment culture) and flushed continuously with running seawater (the temperature was 9–11°C in the cultivation tanks throughout the year). In sediment exposure experiments, sponges were continuously exposed during a 4-week period to a certain amount of sediment slurry dripping into the cultivation tank. One month after the start of cultivation, the sponges were exposed to 5 mg L−1 of sediments, then the sediment addition was stopped for 4 weeks and the sponges were exposed to an increased load of sediments (10 mg L−1). In two different experiments with the sediment loads of 10 and 30 mg L−1, the addition to sponges started after 2 months of their maintaining in culture. Samples for analyses throughout the experimental period were shock frozen in liquid nitrogen and subsequently stored at −80°C. Every sample corresponded to a different sponge specimen.
From 30 samples of T. muricata chosen for the analysis, four were frozen in situ and 26 were cultivated in seawater or sediment tanks for different time periods. The number of cultivated sponges analysed at the beginning of cultivation studies (5 days) was five (three of them were dead), after 1 month of cultivation – four, after 2 months – six, after 3 months – six, after 4 months – four, and after 7 months of cultivation – one sponge.
Sponge protein extracts, SDS PAGE and Western blotting
Sponges were powdered at liquid nitrogen temperature by using tissue homogenizer TissueLyser (Qiagen, Retsch GmbH, Germany) at 30 Hz for 1 min. 50 µL of lysis buffer (140 mM NaCl, 2 mM EDTA, 1 mM phenylmethylsulphonyl fluoride (PMSF), 20 mM Tris-HCl, pH 7.5) was added to 50–100 mg of a powdered sponge sample. The mixture was vortexed and kept on ice for 20 min followed by centrifugation (13,200 rpm, 4°C) for 15 min. The concentration of proteins remaining in the supernatant was determined by the Bradford method (Bradford, Reference Bradford1976) using bovine serum albumin (BSA) for calibration curve.
Proteins (20 µg of total protein) were separated using electrophoresis in 10% sodium dodecyl sulphate polyacrylamide gel (SDS PAGE) (Laemmli, Reference Laemmli1970). Proteins were visualized by staining with PageBlue™ Protein Staining Solution (Thermo Fisher Scientific, Waltham, MA). For Western blot analysis proteins were blotted to Hybond C Extra membrane (Amersham®, GE Healthcare, Little Chalfont, UK) overnight at 175 mA. Membranes were blocked with 5% powdered skim milk in TBS-Tween 20 (140 mM NaCl, 0.1% Tween 20, 20 mM Tris-HCl, pH 7.5). Monoclonal Anti-Heat shock protein 70 (HSP70) clone BRM-22 (Sigma Aldrich, St. Louis, MO) diluted 1:5000 in TBS-Tween 20 was used as a primary antibody and goat anti-mouse IgG, F(ab’)2-HRP (Santa Cruz Biotechnology, Dallas, TX) in 1:5000 dilution was used as a secondary antibody. Western blot results were visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce®, Thermo Fisher Scientific, Rockford, IL) and ImageQuant™Las4000 equipment with attached software (GE Healthcare, Little Chalfont, UK).
RNA extraction and RT-PCR
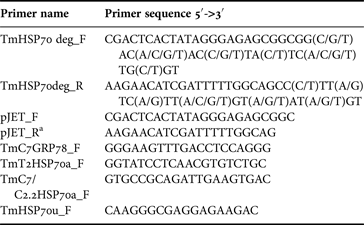
Sponge RNA was extracted using TRIzol reagent (Invitrogen). 1 µg of RNA and 100 pmol of 3′-RACE adapter primer from FirstChoice® RLM-RACE kit (Ambion®, Thermo Fisher Scientific, USA) were used for cDNA synthesis with RevertAid™ H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). HotStarTaq®Plus PCR mixture (Qiagen, Hilden, Germany) with TmHSP70deg_F and TmHSP70deg_R primers (Table 1) were used for the first amplification of T. muricata HSP70 fragments from cDNA. The second PCR was conducted using the amplicon together with primers pJET_F and pJET_R* (Table 1). The first PCR profile was as follows: initial denaturation at 94°C for 5 min, followed by 10 cycles at 94°C for 1 min, 48°C for 1 min, 72°C for 3 min, followed by 25 cycles at 94°C for 1 min, 60°C for 1 min and 72°C for 3 min with final extension at 72°C for 10 min. The second PCR was conducted using the following profile: initial denaturation at 94°C for 5 min, followed by 25 cycles at 94°C for 1 min, 50°C for 1 min, 72°C for 3 min with final extension at 72°C for 10 min.
Table 1. Primers used for amplification reactions and sequencing

*Modified pJET_R.
The PCR products were analysed in 1% agarose gel and DNA was visualized with ethidium bromide. The PCR products of an expected length (1500 bp) were purified from the gel using GeneJET™ PCR Purification Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. Purified PCR products were subjected to the direct sequencing from both ends using pJET_F and pJET_R* primers (Table 1) and subcloned using InsTAclone™ PCR Cloning Kit (Thermo Fisher Scientific, Waltham, MA). Plasmid DNA was extracted by alkaline lysis (Sambrook & Russell, Reference Sambrook, Russell, Argentine and Irwin2001), the presence of an insert of an expected length was determined by restriction analysis. 3–5 plasmids with inserts of expected length were chosen for sequencing with pJET_F and pJET_R* primers (Table 1).
The 3′-RACE procedures were performed using FirstChoice® RLM-RACE kit (Ambion®, Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. To obtain 3′ regions of HSP70a coding mRNAs, the primer TmHSP70u_F (Table 1) was used in combination with the FirstChoice® RLM-RACE kit 3′-adaptor specific outer primer. Subsequently, the nested PCR was conducted with a 3′-adaptor specific inner primer in combination either with the primer TmT2HSP70a_F (designed on the basis of the obtained HSP70a sequence from the specimen T2) or the primer TmC7/C2.2HSP70a_F (designed on the basis of the obtained HSP70a sequences from the specimens C2.2 and C7, Table 1). Primer TmC7GRP78_F was used in combination with FirstChoice® RLM-RACE kit (Ambion®, Thermo Fisher Scientific, Waltham, MA) 3′-adaptor specific outer primer to obtain 3′ regions of GRP78 mRNA from all three specimens analysed. PCR products were analysed in 1% agarose gel and DNA was visualized with ethidium bromide. PCR products of expected length (550–650 bp) were purified from the gel using GeneJET™ PCR Purification Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions.
Sequencing reactions were performed using BigDye® Terminator v3.1 Cycle Sequencing chemistry (Applied Biosystems®, LifeTechnologies®, Thermo Fisher Scientific, Waltham, MA), and Genetic Analyzer 3130 (Applied Biosystems®, LifeTechnologies®, Thermo Fisher Scientific, Waltham, MA) was used for the analysis. Sequence manipulations and illustrations were made using the BioEdit program. The phylogenetic tree was constructed using online software: http://www.phylogeny.fr (Dereeper et al., Reference Dereeper, Guignon, Blanc, Audic, Buffet, Chevenet, Dufayard, Guindon, Lefort, Lescot, Claverie and Gascuel2008).
Sequences of human, bacterial and known sponge HSP70 proteins used for comparison were obtained from the NCBI GenBank (http://www.ncbi.nim.nih.gov/). GenBank accession numbers of the sequences are indicated in the Supplementary Material, Appendix 3.
RESULTS
Immunological detection of T. muricata HSP70s
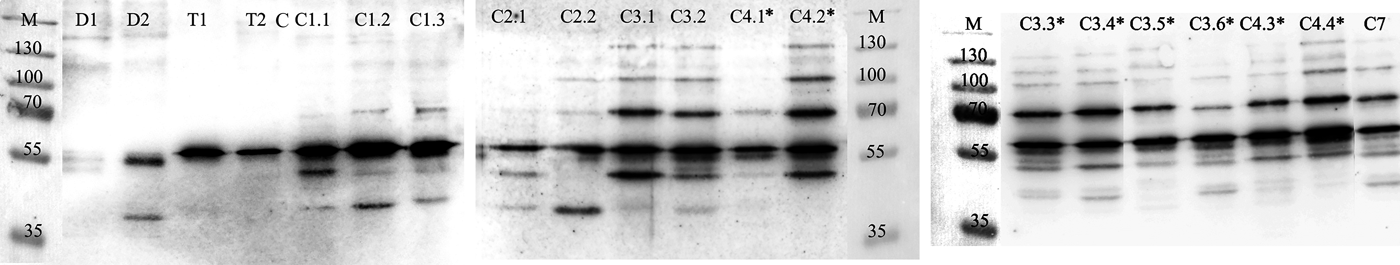
The representative set of the analysed samples is given in Figure 1. 70 kDa proteins were not detected in the sponges frozen directly on the research vessel (Figure 1, lanes T). At the beginning of cultivation and after 1 or 2 months cultivation in seawater or sediment tanks, low levels of HSP70 were detected in some, but not in all sponge samples (Figure 1, lanes C1–C2). Also, HSP70 was not detected in those sponges that died at the beginning of cultivation (Figure 1, lanes D). After longer cultivation (3 or more months), HSP70 was detected in every sample with generally higher expression levels (Figure 1, lanes C3–C7). In addition to the cultivation duration that had an obvious effect on HSP70 expression level, the latter was influenced by differences between sponge individuals subjected to the same treatments (see Figure 1, lanes C1.1 and C1.3; C4.1* and C4.2*). However, the impact on HSP70 expression caused by elevated sediment load did not become evident from the sediment exposure experiments, e.g. see Figure 1 lanes C3.1, C3.3* and C3.5*, which corresponded to sponge specimens cultivated without sediments, exposed to 5 or 30 mg L−1 of sediments, respectively.

Fig. 1. The proteins recognized by bovine HSP70 antibody in the samples of T. muricata. Names above the lanes indicate particular specimens. Symbols in the names are as follows: D, specimens that were dead; T, specimens that were frozen on research vessel; C, cultivated specimens: the first number indicates cultivation time in months and the second number indicates the particular specimen. *, specimens exposed to sediments, in particular C3.3* and C3.4* correspond to 5 mg L−1; C3.5* and C3.6* correspond to 30 mg L−1; C4.1* and C4.2* correspond to 5 mg L−1 +10 mg L−1; C4.3* and C4.4* correspond to 10 mg L−1 of sediment load. Lane M – molecular weight marker (kDa).
Remarkably, in all samples tested (except those that originated from two out of the three dead sponges, see Figure 1, lane D1; D3 not shown) cross-reactivity of bovine HSP70 antibody with proteins (or protein doublets) of about 60 kDa was observed. In some samples, proteins of about 100 and 50 kDa were also recognized. The identity of these proteins is not clear at present.
HSP70 mRNAs and conserved sequence motifs in T. muricata
To elucidate the genes of the HSP70 family proteins, three sponge specimens were chosen for a further analysis of HSP70 expression at mRNA level: specimens T2, C2.2 and C7. Specimen T2 was a representative of samples frozen on the research vessel, C2.2 represented short-term cultivated samples, and C7 – long-term cultivated samples.
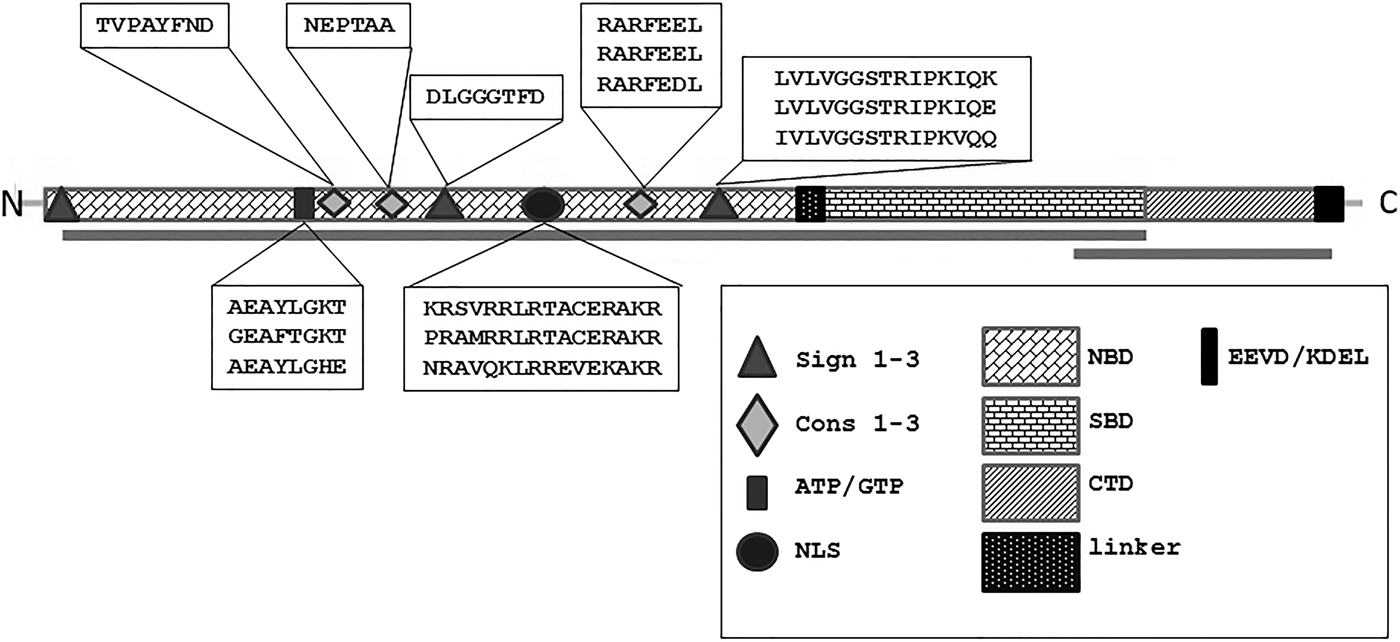
Initially, 1500 bp fragments of HSP70 cDNA were amplified using degenerate primers that were designed on the basis of previously known HSP70 sequences of sponges and other animals. The obtained fragments included the majority of the N-terminal nucleotide (ATP) binding domain (NBD) and most of the substrate (peptide) binding domain (SBD) of HSP70s (Figure 2) – thus covering the most conserved part of HSP70s. Subcloning and sequencing revealed the presence of three types of proteins belonging to the HSP70 superfamily: HSP70a type sequences were detectable in all three specimens, GRP78 was detectable in the cultivated specimens C2.2 and C7, HSP70b was present only in the specimen T2 (frozen in situ). Two HSP70a sequences that were obtained from the specimen T2 had a few single nucleotide differences, but none of those led to amino acid substitution. In addition, one HSP70b type sequence was identified from T2, while GRP78 was not detectable in this specimen. Three HSP70a sequences sharing a 99% sequence identity on amino acid level were obtained for C2.2; GRP78 sequence was detectable on the electropherogram of direct sequencing as minor peaks under major peaks of HSP70a sequence. On the contrary, GRP78 was obtained as the main sequence from the long-term cultivated specimen C7; the HSP70a sequence was detected under major peaks of GRP78 sequence. According to blastp search in non-redundant protein sequences database of NCBI, HSP70a type had best matches with stress-induced or constitutive HSP70 proteins from sponges and other organisms. The GRP78 from T. muricata had best matches with another member of the HSP70 superfamily – glucose-regulated protein of 78 kDa. HSP70b type protein sequence had best BLAST matches mainly with amoeboid HSP70 sequences, indicating that it might originate from a sponge-associated eukaryotic microorganism.

Fig. 2. The topology and conserved motifs of HSP70 proteins from T. muricata. Three domains of HSP70 superfamily proteins are shown: the nucleotide binding domain (NBD), the substrate binding domain (SBD) and the C-terminal domain (CTD). HSP70 specific signature sequences (Sign 1–3), conserved motifs (Cons 1–3), the ATP/GTP binding sequence and the nuclear localization signal (NLS) are located in NBD. Proteins of HSP70 superfamily end with the EEVD/KDEL motif specific for HSP70/GRP78, respectively. The regions amplified from T. muricata are indicated as lines below the topology chart. The sequences of the motifs found in T. muricata HSP70 proteins are written in textboxes (if the motif sequences are not identical they belong to HPS70a, HSP70b or GRP78 proteins, respectively).
As the primer HSP70deg_F was designed on the basis of signature sequence 1, specific to all HSP70 proteins, only the other two HSP70 signatures defined in PROSITE database of protein domains (Bairoch, Reference Bairoch1992) are present in HSP70 sequences of T. muricata: signature 2 DLGGGFD, identical in all sequences (positions 184–191, Supplementary Material, Appendix 1), and signature 3 [LI]VLVGGSTRIPK[IV]Q[KE] having slight differences between HSP70a, HSP70b and GRP78 (positions 319–333, Supplementary Material, Appendix 1). The additional conserved motifs Cons1–Cons3, defined by Rensing & Maier (Reference Rensing and Maier1994), are also detectable in all obtained sequences, including conserved motif 3 characteristic of eukaryotic non-mitochondrial or non-plastid HSP70s (Figure 2; Supplementary Material, Appendix 1).
A potential ATP/GTP binding P-loop motif [AG]x(4)GK[ST] (Saraste et al., Reference Saraste, Sibbald and Wittinghofer1990) is present in all sequences (positions 116–123) with minor differences in C7_GRP78.
Nuclear localization signal NLS (positions 241–257, Supplementary Material, Appendix 1) is present in HSP70a sequences but in GRP78 and HSP70b the first position of the signal has been substituted, suggesting that those proteins do not locate to nucleus. Several specific single amino acid positions have been identified in literature that distinguish between eukaryotic and prokaryotic, ER and cytoplasmic HSP70s (Gupta et al., Reference Gupta, Aitken, Falah and Singh1994). Positions that can be used to distinguish between cytosolic and ER HSP70s are highlighted in the Supplementary Material, Appendix 1; those positions support the specification of T. muricata HSP70 sequences.
3′-RACE was performed to amplify 3′regions of obtained HSP70 mRNAs to further confirm the specification of the obtained sequences. The eukaryotic HSP70s that are located in cytoplasm usually end with regulatory sequence EEVD whereas GRP78 proteins end with ER retention signal KDEL (Munro & Pelham, Reference Müller, Batel, Lacorn, Steinhart, Simat, Lauenroth, Hassanein and Schröder1987). Our 3′-RACE results for HSP70a of the specimen C2.2, for GRP78 of the specimen C7 and for HSP70a of the specimen T2 supported the previous results. Additionally, using primers designed on the basis of GRP78 sequence from C7, a 3′ region was obtained for GRP78 of the specimen C2.2. An additional 3′ region that ends with EEVD signature was obtained for C2.2, which was not compatible with the previously known sequences (Supplementary Material, Appendix 2). No 3′-RACE result was obtained for HSP70b, so the true identity of this protein remains obscure.
The obtained sequences of T. muricata which were deposited to Genbank have the following accession numbers: KP863863 (GRP78_spC7), KP863864 (HSP70a_spC7), KP863865 (HSP70a1_spT2), KP863866 (HSP70a2_spT2), KP863867 (HSP70a1_spC2.2), KP863868 (HSP70a2_spC2.2), KP863869 (HSP70a3_spC2.2) and KP863870 (HSP70b_spT2).
Multiple sequence alignment and phylogenetic analysis
T. muricata HSP70 sequences were compared with HSP70 sequences from other sponge species and from human as well as with bacterial DnaK protein sequence. Only full length sequences of sponge HSP70s were taken for comparison. From the human HSP70 protein sequences HSPA1A was selected as a representative of cytosolic stress-induced HSP70s, HSPA8 as a representative of constitutively expressed cytosolic HSP70s and HSPA5 as a representative of ER located GRP78s.
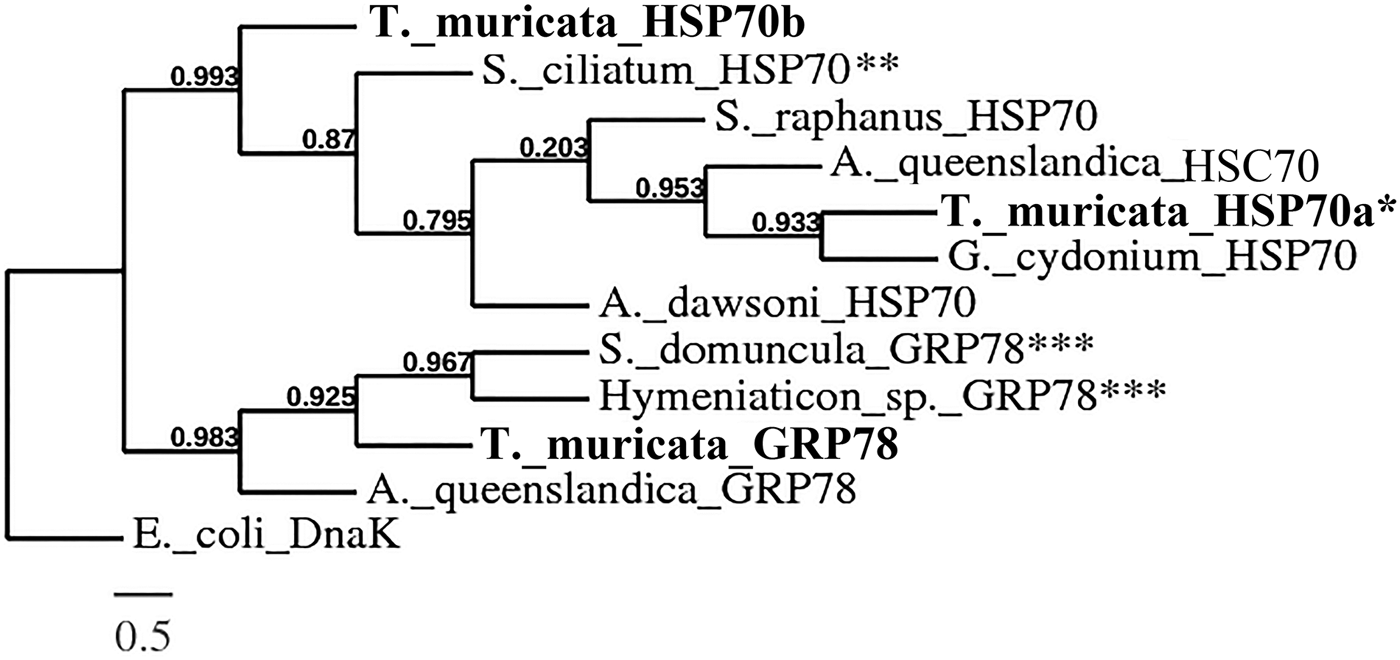
The C-terminal region of HSP70s is weakly conserved (Rensing & Maier, Reference Rensing and Maier1994), thus only the sequences obtained from the degenerate PCR experiment from T. muricata and the corresponding regions of the sequences from other organisms (including the conserved region of HSP70s) were used for the multiple alignment. An identity/similarity matrix (Supplementary Material, Appendix 4) was constructed on the basis of multiple sequence alignment of the above mentioned HSP70 family protein sequences. HSP70a type sequences from T. muricata had sequence identities and similarities with stress-induced or constitutive HSP70s in the range of 75–90% and 86–95%, respectively; identities and similarities with GRP78 protein sequences were lower. The GRP78 sequence of T. muricata (the specimen C7) was more similar to glucose-regulated proteins from sponges and humans, whereas the identities and similarities with HSP70s were lower. The HSP70b sequence was more similar to the HSP70 than the GRP78 protein sequences (Supplementary Material, Appendix 4). A phylogenetic tree was constructed on the basis of known sponge HSP70 protein sequences. All HSP70 proteins of T. muricata branch together with the HSP70 superfamily proteins from other sponges. GRP78 is more closely related to GRP78 from other sponges while HSP70a is grouped together with the stress-induced or constitutive HSP70s (Figure 3).

Fig. 3. A phylogenetic tree based on multiple sequence alignment of amino acid sequences of the HSP70 superfamily proteins from sponges (Supplementary Material, Appendix 3). Branch support values are indicated on branches (the branch length is proportional to the number of substitutions per site). The sequences obtained from T. muricata are shown in bold. *HSP70a1 from specimen C2.2 was chosen for the phylogenetic analysis as a representative of T. muricata HSP70a type sequences. **The protein has been deposited to the database as GRP78, but due to the EEVD motif at the end of the sequence, the protein was renamed as HSP70. ***The protein has been deposited to the database as HSP70, but due to the ER retention signal KDEL at the end of the sequence, the protein was renamed as GRP78.
DISCUSSION
In the course of the SPONGRAM project, cultures of T. muricata were successfully established for ex situ impact studies that enabled us to assess the cultivation effect as well as the impact of elevated sediment load on sponge survival and cell physiology. There was a high mortality of sponges at the beginning of cultivation, however, after an adaptation period of 1–2 months the populations became stable. Also, the metabolic activity estimated by adenylate energy charge (AEC) values indicated a beneficial effect of long-term cultivation (4–7 months) of sponges: AEC values increased after 3 months of cultivation (results published in the final report of the SPONGRAM project). However, the effect of long-term cultivation on the expression of proteins of the HSP70 family in sponges had not been studied before our research project. Long-term cultivation of sponges in laboratory conditions, despite many studies that have been devoted to the subject, is still a difficult task with many questions yet to be answered (Schippers et al., Reference Schippers, Sipkema, Osinga, Smidt, Pomponi, Martens and Wijffels2012).
The results presented in this work indicate that 70 kDa proteins were detected immunologically at different levels in T. muricata samples. In preliminary experiments, two HSP70 antibodies (polyclonal Geodia cydonium HSP70 and monoclonal bovine brain HSP70) were tested for their suitability to recognize 70 kDa proteins in different samples. Interestingly, the marine sponge antibody produced against recombinant HSP70 from G. cydonium was not able to detect 70 kDa proteins in the samples of T. muricata from the adaptation period (1–2 months) of cultivated sponges (data not shown). The proteins of 70 kDa from those samples were detected at low levels using bovine HSP70 antibody (Figure 1); therefore, the latter antibody was used for the subsequent immunological studies. Higher levels of the stress protein HSP70 observed after 3 months of cultivation seemed to be in conflict with the adaptation of sponges to culture conditions and the improved physiological status of sponges (as assessed by AEC values (Witte & Graf, Reference Witte and Graf1996)) after the adaption period. However, these data may suggest that though the mortality of sponges had been reduced to a minimum, the maintenance of the remaining sponges in culture needs a continuous induction of HSP70 proteins. As there was no clear effect of elevated sediment load on the levels of HSP70, possible sedimentation effects (if they were manifested at all) may have been masked by the cultivation effects or by the variations among different sponge individuals. Some tolerance to sedimentation effects may be characteristic of this sponge species, as T. muricata lives on soft muddy bottoms and, interestingly, as revealed from sediment exposure experiments, the sponges can use their basal spicules to ‘crawl’ out of the sediment (H.T. Rapp, unpublished data).
In addition to 70 kDa proteins, bovine HSP70 antibody recognized several other proteins in the extracts of T. muricata: primarily the proteins of about 60 kDa were detected in the cultivated samples as well as in those frozen in situ (Figure 1). Similarly, bovine HSP70 antibody recognized the proteins of about 60 kDa in the case of marine sponges Crambe crambe and Chondrosia reniformis (Agell et al., Reference Agell, Uriz, Cebrian and Martí2001; Cebrian et al., Reference Cebrian, Agell, Martí and Uriz2006). Agell et al. (Reference Agell, Uriz, Cebrian and Martí2001) explained this phenomenon as a cross-reaction of the specific antibody with the proteins from two different HSP (HSP60 and HSP70) families. Yet another possibility may exist according to which the protein(s) of about 60 kDA is (are) splice variant(s) of a member of the proteins belonging to the HSP70 family. Tsukahara et al. (Reference Tsukahara, Yoshioka and Muraki2000) have characterized HSC54, a novel 54 kDA splice-variant of the human heat-shock cognate protein 70. The true nature/origin of the proteins of around 60 kDa as well as that of the other proteins recognized by bovine HSP70 antibody in T. muricata remains to be elucidated in future studies.
The specification of particular 70 kDa proteins (stress-induced, constitutive, GRP) recognized by bovine HSP70 by means of the Western blot analysis is hardly possible. Both identification by mass-spectrometry and quantification by qPCR that would allow advanced measurements of HSP70 expression levels, require pre-known sequence data that have not been available for T. muricata so far. So our next task was to identify which mRNAs coding for HSP70 proteins could be detected in T. muricata samples. For that three specimens were chosen with different levels of immunologically detectable 70 kDA proteins. We identified three types of HSP70 coding mRNAs in T. muricata; at that, different sets of HSP70 mRNAs were obtained for different specimens. Sequence motifs indicate that the proteins coded by those mRNAs are either stress-induced or constitutively expressed members of HSP70 and GRP78 families. One obtained sequence – HSP70b – seems to be a 70 kDa stress protein from a sponge-associated symbiotic organism. Such a result is not surprising since sponges are known to harbour large amounts of bacterial and eukaryotic symbionts (Fan et al., Reference Fan, Reynolds, Liu, Stark, Kjelleberg, Webster and Thomas2012). The finding of different sets of mRNAs coding for proteins of HSP70 family gives the first implication of the distribution of the corresponding proteins in the specimens of T. muricata, which were subjected to different treatments. The data obtained from direct sequencing experiments show that for the specimen T2 (frozen in situ, no immunologically detectable HSP70), the obtained sequence corresponded to HSP70a. For the specimen C2.2 (short-term cultivation, HSP70 detected at low levels), the main sequence corresponded to HSP70a; in addition, GRP78 was identified on the basis of accompanying minor peaks. For the specimen C7 (long-term cultivation, HSP70 detected at higher levels), the main sequence corresponded to GRP78; HSP70a was identified from accompanying minor peaks. Although direct sequencing is not as good as the qPCR method for quantification purposes, it still gives us a hint about the distribution of different HSP70 mRNAs (and the corresponding proteins) in the particular sponges. These data allow us to speculate that the induction of GRP78, which may be the result of the accumulation of misfolded proteins inside the ER, is important for long-term cultivation of T. muricata. However, high levels of any of the stress proteins point to persistent stress conditions of sponges that survived the adaptation period and had been maintained for months under laboratory conditions.
In addition, the repertoire of HSP70s in sponges, including T. muricata, may be much wider than presently known. Six mRNAs sequences from different sponge species, covering the complete coding part of HSP70s, have been deposited to GenBank: four of them were cytosolic stress-induced HSP70s and two belonged to the GRP78 family. Furthermore, a number of mRNA sequences that code for proteins of the HSP70 superfamily can be found in Amphimedon queenslandica (Conaco et al., Reference Conaco, Neveu, Zhou, Arcila, Degnan, Degnan and Kosik2012), the only sponge with a published genome (Srivastava et al., Reference Srivastava, Simakov, Chapman, Fahey, Gauthier, Mitros, Richards, Conaco, Dacre, Hellsten, Larroux, Putnam, Stanke, Adamska, Darling, Degnan, Oakley, Plachetzki, Zhai, Adamski, Calcino, Cummins, Goodstein, Harris, Jackson, Leys, Shu, Woodcroft, Vervoort, Kosik, Manning, Degnan and Rokhsar2010). At least 17 genes coding for proteins belonging to the HSP70 superfamily have been described in humans (Brocchieri et al., Reference Brocchieri, Conway de Macario and Macario2008). Besides, considering that proteins from several different HSP superfamilies may contribute to the stress response of sponges under various conditions, the detailed understanding of the contribution of each stress protein involved calls for future studies.
In conclusion, the results of the present study indicate that HSP70 expression levels in T. muricata were mainly affected by the duration of cultivation of sponges. After a cultivation period of 2–3 months when sponges had adapted to life in culture, persistent higher levels of HSP70 expression were observed. Our data referred to changes of the pattern of induced stress proteins during long-term cultivation of sponges. The involvement of ER stress protein GRP78 was evidently needed to provide supplementary protection for the survival of cultivated sponges. The results obtained in this study provide a basis for future quantitative stress studies in this field.
FINANCIAL SUPPORT
This work was supported through the SPONGRAM project funded by The Norwegian Deep Sea Program (NDP) through contract number 4501533230, and also by the Estonian Ministry of Education and Research (grant No. 0140108) and the Estonian Science Foundation (grant No. 9185).
SUPPLEMENTARY MATERIALS
To view supplementary material for this article, please visit http://dx.doi.org/ 10.1017/S0025315415002234