Introduction
RNA interference (RNAi) is a gene silencing mechanism in which the expression of a gene is specifically inhibited by its cognate double-stranded RNA (dsRNA). The inhibition produced by RNAi closely resembles the loss-of-function or gene knockout phenotype (Hammond et al., Reference Hammond, Bernstein, Beach and Hannon2000). Studies carried out on the mechanism of RNAi have revealed that small interfering RNAs (siRNAs) are generated as intermediates in all the organisms undergoing RNAi; thus, siRNAs serve as molecular markers of the RNAi process.
RNAi has been demonstrated to be functional in several insect cell lines, eggs and in vivo using a DNA expression vector to express and synthesize dsRNA, such as in Drosophila (Lum et al., Reference Lum, Yao, Mozer, Rovescalli, Von Kessler, Nirenberg and Beachy2003). Also, injecting adult termites or larval Tribolium castaneum and Blattella germanica with dsRNA or siRNA caused targeted gene silencing and subsequent phenotypes (Tomoyasu et al., Reference Tomoyasu and Denell2004; Arakane et al., Reference Arakane, Muthukrishnan, Kramer, Specht, Tomoyasu, Lorenzen, Kanost and Beeman2005; Cruz et al., Reference Cruz, Mane-Padros, Belles and Martin2006; Zhou et al., Reference Zhou, Oi and Scharf2006). Recently, RNAi has been successfully demonstrated in a small number of lepidopteran insects, including Spodoptera litura (Rajagopal et al., Reference Rajagopal, Sivakumar, Agrawal, Malhotra and Bhatnagar2002), Hyalophora cecropia (Bettencourt et al., Reference Bettencourt, Terenius and Faye2002), Bombyx mori (Quan et al., Reference Quan, Kanda and Tamura2002), Spodoptera frugiperda (Meyering-Vos et al., Reference Meyering-Vos, Merz, Sertkol and Hoffmann2006) and Manduca sexta (Eleftherianos et al., Reference Eleftherianos, Marokhazi, Millichap, Hodgkinson, Sriboonlert, ffrench-Constant and Reynolds2006a,Reference Eleftherianos, Millichap, ffrench-Constant and Reynoldsb, Reference Eleftherianos, Gokcen, Felfoldi, Millichap, Trenczek, ffrench-Constant and Reynolds2007). Most of this research is aimed at functional analysis of target genes. However, more data on lethal phenotypes, e.g. mortality, are needed to explore RNAi as a potential pest control agent.
Chitin synthases (CHS) are large membrane proteins that catalyze the polymerization of N-acetylglucosamine (GlcNAc) into chitin, which is a major component of the exoskeletons and peritrophic membranes of insects (Coutinho et al., Reference Coutinho and Henrissat1999). Earlier, we have reported cloning the chitin synthase A gene from Spodoptera exigua, an important pest that damages a wide range of economically important crops, and found that expression of this gene was high in cuticle and trachea tissue, indicating that the chitin synthase gene may be very important during insect development (Chen et al., Reference Chen, Yang, Senthil Kumar, Tang, Sun, Qiu, Hu and Zhang2007).
In this study, RNAi was used to study the impact of silencing chitin synthase A in S. exigua (SeCHSA). In vitro RNAi in High 5 cells showed that the expression of transgene (SeCHSA) was inhibited by either dsRNA or siRNA corresponding to the gene. In vivo injection of siRNA or dsRNA into S. exigua larva resulted in developmental arrest and cuticle and trachea defects. These results suggest that the silencing of CHSA can be induced by dsRNA/siRNA injection into lepidopteran larvae to cause their abnormal development.
Materials and methods
Cell culture and insect culture
An insect cell line (Trichoplusia ni High 5) was maintained in Grace's medium (Gibco BRL) supplemented with 10% fetal bovine serum. S. exigua larvae were reared at 26±1°C and a L14:D10 photoperiod using artificial diet as described previously (Chen et al., Reference Chen, Yang, Senthil Kumar, Tang, Sun, Qiu, Hu and Zhang2007).
Selection of sequence and synthesis of dsRNA /siRNA
S. exigua chitin synthase A (SeCHSA) was cloned previously (GenBank accession no. DQ062153). A cDNA fragment containing an SeCHSA partial sequence (2060–2464 bp) was amplified using forward (Up1: 5′-TAA TAC GAC TCA CTA TAG GGT GAA TCG ATT CGT AA-3′) and reverse primers (Down1: 5′-TAA TAC GAC TCA CTA TAG GGA TGA ATA CGG CCG CA-3′). Both the sense and antisense strands of the amplified fragments contained a T7 RNA polymerase binding site. The PCR products were purified with QIA quick columns and were then used for in vitro transcription with T7 RNA polymerase. All the reagents and enzymes used were from Ambion MEGAscript RNAi kit, and the reactions were carried out following the manufacturer's instructions. The dsRNAs were annealed by incubation at 37°C for 3.5 h, followed by slow cooling to room temperature. The annealed dsRNAs were treated with DNase I at 37°C for one hour and then purified. The purified dsRNAs were analyzed on a 1% agarose gel before being stored at −20°C.
Processing dsRNA by ShortCut RNase III (NEB) digestion in the presence of manganese produces a heterogeneous population of short (18–25 bp) siRNAs and completely eliminates the longer dsRNA. Before transfection, the heterogeneous population of short (18–25 bp) siRNAs must be settled and ethanol purified. The reactions were carried out following the manufacturer's instructions. The siRNAs were analyzed on a 4% agarose gel and then stored at −20°C.
Co-transfection of High 5 cells
To make sure that the dsRNA and siRNA are working for silencing the target gene in vitro, High 5 cell and pEGFP-C3 vector, a good expression system as our previous work showed (data not presented), are used to express our target gene even though T. ni is a different species to S. exigua of our interest. A cDNA fragment containing an SeCHSA partial sequence (2112–2963 bp) was amplified by PCR with two primers which contain two sites of restriction endonucleases, BamH I and Hind III, and then inserted into a pEGFP-C3 vector to construct the recombined plasmid pEGFP-C3-CHS. High 5 cells were co-transfected with pEGFP-C3-CHS and dsRNA/siRNA using Cellfection (Invitrogen) according to the manufacturer's protocol. Briefly, cells were seeded and allowed to settle overnight (50% confluence). Plasmid and dsRNA/siRNA were mixed with liposome at a ratio of 1:3 (w/v) in Grace's SFM without antibiotics. The mixture was incubated at room temperature for 30 min. The medium was removed, and the cells were washed with Grace's SFM without antibiotics. Then, the above mixture was added to cells in 0.8 ml Grace's SFM and incubated for 5 h. After that, the transfection mixture was removed; 2 ml of Grace's SFM containing antibiotics were added. The cells were incubated at 27°C for 48 h. Ten microgram plasmid DNA, with 10 μg dsRNA/siRNA, was transfected 1×106 High 5 cells. Buffer treated cells were used as control.
Observation of fluorescence and immuno-blot
After harvesting, the cells were counted by hemocytometer and observed under fluorescence microscope. For immuno-blotting, the cells were collected by centrifugation at 800 g for 5 min and resuspended in PBS. The cells were pelleted by centrifugation at 1000 g for 5 min and the supernatant was discarded. The cells were lysed by adding 400 μl RIPA Lysis Buffer (Clemens et al., Reference Clemens, Worby, Simonson-Leff, Muda, Maehama, Hemmings and Dixon2001), pipetting up and down to mix well. (The amount of protein loaded for each treatment was equal to 0.5×106 cells in 100 μl of sample buffer.) Laysate were boiled for 5 min, and 10 μl total cell extract was loaded onto 15% SDS–PAGE and electrotransferred onto a nitrocellulose membrane. Western blot analysis was carried out as described previously (Chen et al., Reference Chen, Yang, Senthil Kumar, Tang, Sun, Qiu, Hu and Zhang2007). Immunoreactivity was detected by incubating the membrane with anti-SeCHS serum (1:500 dilution) followed by incubation with AP-conjugated goat anti-rabbit IgG (Sigma) (1:2000 dilution). Colour reaction was detected after the addition of NBT/BCIP.
Injection of dsRNA/siRNA into S. exigua larvae
The last day of 4th instar S. exigua larvae were used for injection experiment because larvae at earlier stages of development proved to be too small for satisfactory injections. Three micrograms of siRNA/dsRNA dissolved in 3 μl DEPC water were injected into the side of the thorax of S. exigua larvae using 10 μl microliter syringes (Hamilton), and the injection point was sealed immediately with wax. Control larvae were injected with an equivalent volume of buffer alone or were not injected. Each group had 20 individual larvae with the experiment repeated three times (n=60 larvae per group).
Observation of larval phenotype and data analysis
Larvae were observed at 12 h intervals after injection. Abnormal larvae were identified as individuals with delayed molting, reduced size, slow development or slow melanization of the cuticle, relative to untreated or buffer-injected control larvae. When observed, abnormal larvae were removed and stored at −80°C for subsequent RNA extraction.
To test for an effect of treatment, ANOVAs were performed using the cumulative percentage of abnormal larvae as the dependent variable and group (no injection, buffer injection, dsRNA injection, siRNA injection) as the independent variable. Post-hoc Duncan's tests were used to determine which groups differed when treatment effects were detected. These analyses were repeated for time intervals of 0, 24, 36, 48 and 72 h post-injection. Percentage values were arcsine square root transformed prior to analyses to correct for the non-normal distribution of percentage values.
RT-PCR analysis of gene silencing
After data analysis, all abnormal larvae and control larvae were used for RT-PCR. Total RNA was extracted for individual larvae using reverse transcriptase AMV (Takara). The cDNA was employed as a template for amplification and detection of SeCHSA transcripts. The primers used were: Down2 (5′-GTC GCC TAT GGT GGT TTC GT-3′) and Up2 (5′-TTT GTA TTC TTC TTC GCC TTG A-3′) for SeCHSA, which generated a 636 bp product. The primers Up3 (5′-GGTTGG TAT GGG TCA GAA GGA-3′) and Down3 (5′-GCG GTG GTG GTG AAA GAG TA-3′) were used to amplify S. exigua β-actin gene (GenBank accession no. AY507963), a loading control with a 477 bp PCR product.
PCR was carried out in a 25 μl reaction. The PCR program (28 cycles) comprised a denaturing step at 94°C for 1 min, annealing at 58°C for 1 min and extension at 72°C for 1 min. PCR products were analyzed on 1% agarose gel.
Electron microscopy and tracheal chitin staining
The abnormal and control larvae of S. exigua were dissected under a stereomicroscope, and cuticles were treated with paraformaldehyde-lysine-periodate fixative as described previously (Zimoch & Merzendorfer, Reference Zimoch and Merzendorfer2002). After fixation, the samples were infiltrated with epon resin for 48 h and then polymerized at 60°C for 24–48 h. Ultrathin sections were obtained using an ultramicrotome (Leica Ultracut UCT) mounted onto formvar-coated copper grids and stained with 2% uranyl acetate in water and lead citrate. Then, sections were observed under a JEM-1010 electron microscope (Jeol, Japan) and photographic negatives were scanned (HP scanner).
The trachea tissues were prepared from both abnormal and control larvae. After removing fat body, trachea were treated with paraformaldehyde-lysine-periodate fixative and Triton-X 100 as described previously (Chen et al., Reference Chen, Yang, Senthil Kumar, Tang, Sun, Qiu, Hu and Zhang2007) and then incubated overnight in PBS containing 0.01% (w/v) FITC (NEB). FITC was visualized with an Olympus IX70 fluorescence microscope using a mercury short ARC photo optic lamp (HBO, Osram) and a NUA filter set (Olympus).
Results
dsRNA and siRNA synthesis
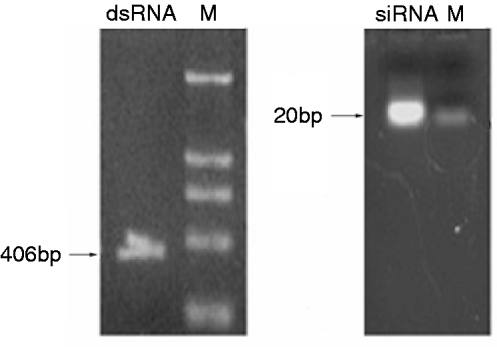
dsRNA was synthesized in vitro and was diluted in nuclease-free water. Agarose gel electrophoresis showed that the dsRNA used in the present study was 406 bp. After synthesis, the dsRNA were digested into siRNAs (18–25 bp) with RNase III. The siRNAs diluted in nuclease-free water were a mixture of differently sized SeCHSA siRNA fragments and were analyzed on 4% agarose gel to ensure that longer dsRNA were completely eliminated (fig. 1).
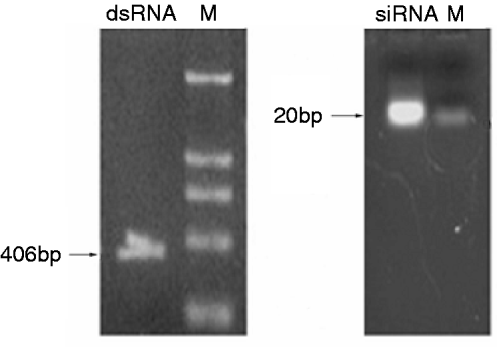
Fig. 1. Generation and analysis of dsRNA and siRNA. dsRNA was synthesized in vitro and then cut into siRNA with RNaseIII. The dsRNA and siRNA were analyzed on 1% and 4% agarose gel, respectively. Lane ‘M’ contains a DNA molecular weight marker.
SeCHSA expression in High 5 cells
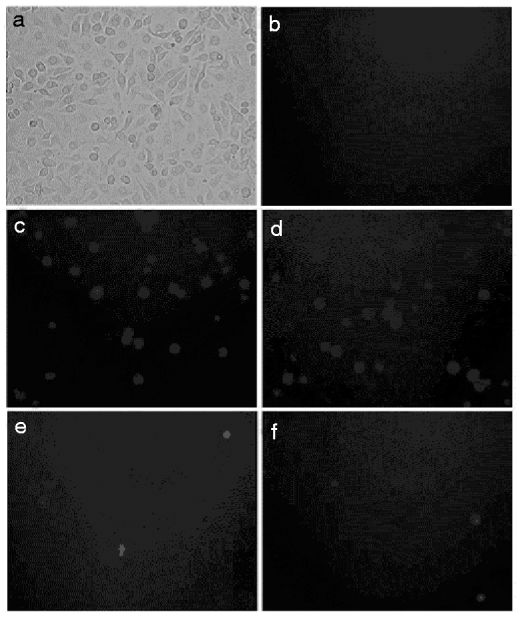
To assess whether dsRNA can inhibit gene expression in S. exigua, we adopted the High 5 cell culture model to determine the effect of RNAi. High 5 cells were transfected with a reporter gene plasmid pEGFP-C3-CHS that expresses enhanced GFP and SeCHSA. GFP expression was assayed 48 h post-transfection. Fluorescence was observed in cells transfected with the reporter gene plasmid pEGFP-C3-CHS but not in cells without transfection (fig. 2). These results indicated that SeCHSA was expressed in High 5 cell.
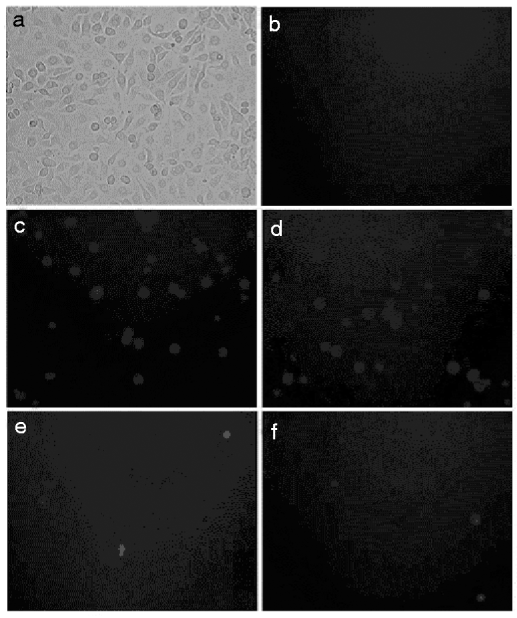
Fig. 2. In vitro SeCHSA RNAi in High 5 cells. Co-transfected dsRNA (e) or siRNA (f) with the reporter plasmid pEGFP-C3-CHS into High 5 cells. (a) bright field views of the cells, (b) uninfected cells and (c and d) buffer treated cells.
RNAi in High 5 cells
SeCHSA was targeted, and cognate dsRNA and siRNA duplexes were examined for their ability to interfere with transient expression of SeCHSA in High 5 cells. High 5 cells were co-transfected with the plasmid pEGFP-C3-CHS and either dsRNA or siRNA. We found significant reduction of the fluorescence intensity of GFP in the cells co-transfected with SeCHSA and dsRNA/siRNA, while no reduction was observed when High 5 cells were transfected with plasmid alone (fig. 2).
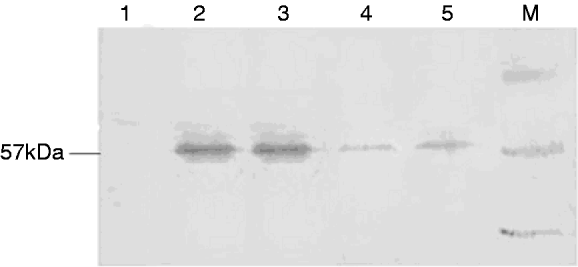
To detect SeCHSA protein in cells, total cell extracts were loaded onto SDS-PAGE and transferred to PVDF membrane. Western blot analysis showed that addition of dsRNA/siRNA to the infected cell culture resulted in specific reduction in the expression of SeCHSA protein. No reduction of SeCHSA protein was observed in cells treated with plasmid alone (fig. 3). Although reduction in the expression of SeCHSA protein was caused by co-transfection of dsRNA/siRNA, complete inhibition of the transcription of the target gene was not observed. The cell RNAi experiment was repeated three times, and all of the cell samples were examined and got the consensus of results.
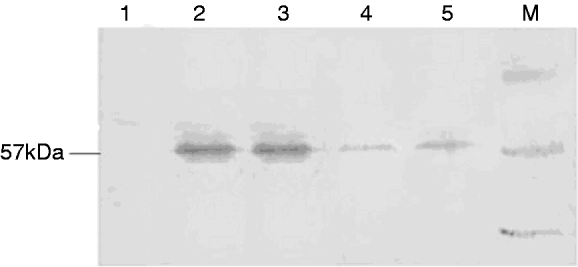
Fig. 3. Immune blot analysis of SeCHSA RNAi in High 5 cells. Co-transfected dsRNA (5) or siRNA (4) with the reporter plasmid pEGFP-C3-CHS into High 5 cells resulted in the specific reduction of expression of SeCHSA. In controls, no reduction was observed when High 5 cells were transfected with plasmid alone (2 and 3). There was no band in cell without transfection (1). Lane ‘M’ contains a protein molecular weight marker.
Abnormal larval phenotype
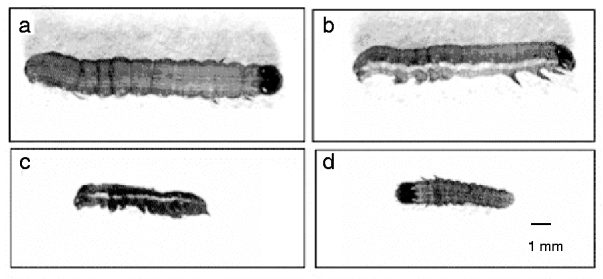
The effect of sequence-specific gene silencing of a chitin synthase gene in S. exigua larvae was determined by observing morphological changes and developmental arrest along with normal phenotypes of the control larvae. Two different lethal phenotypes were observed in abnormal larvae after SeCHSA knockdown. Most of the abnormal larvae were unable to molt to the next larval stage, and only a small number molted to the next instar stage. In the abnormal larvae that were unable to molt, melanization of the larval cuticle was evident, which was possibly due to the weakened new cuticle, and epidermis was torn during separation from the old cuticle. In the abnormal larvae that molted to the next instar, larvae were much smaller in size compared to control larvae; and they all failed to pupate and died without any cuticle splitting (fig. 4).
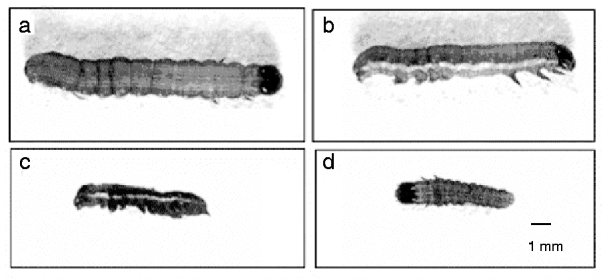
Fig. 4. In vivo SeCHSA RNAi in S. exigua. The phenotypes of larvae injected with siRNA/dsRNA. Most abnormal larvae were unable to molt to the next larval stage (c); a small number of siRNA-injected larvae molted to the next instar (d), but they were smaller in size compared to controls with no injection (a) or buffer injection (b).
The accumulated percentage of abnormal larvae injected with siRNA/dsRNA reached 50.00% and 31.67% 72 h post-injection, respectively, and was significantly higher than that of control larvae (11.67% for buffer injection and 0% for non-injected) (table 1). The abnormal phenotypes were observed 24 h post-injection, and abnormal larvae injected with siRNA/dsRNA accounted for 35.00% and 11.67%, respectively, of all injected larvae.
Table 1. Accumulated percentage of abnormal larvae in the treatments and the controls. The data are mean±SEM. Means labeled with different letters (a, b, c and d) within each column are significantly different by Duncan's test (n=3; p=0.01).
In vivo RNAi
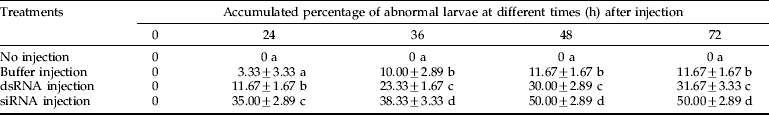
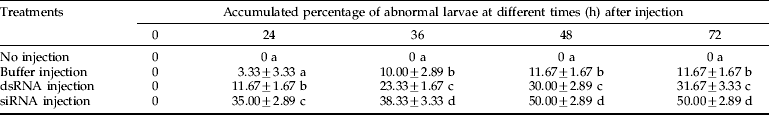
To determine the impact of dsRNA/siRNA mediated knockdown of the CHSA gene in S. exigua, RT-PCR was carried out after dsRNA/siRNA injection (fig. 5). In our previous study on the developmental expression pattern of SeCHSA gene (Chen et al., Reference Chen, Yang, Senthil Kumar, Tang, Sun, Qiu, Hu and Zhang2007), we observed that SeCHSA was expressed maximally on the first and last day of the 5th instar stage. In the present study, SeCHSA-specific dsRNA/siRNA was injected on the last day of the 4th instar stage, when expression of SeCHSA does not reach the highest expression, to achieve the maximum effect of down-regulation. Total RNA was isolated from abnormal insects (collected in the first three days post-injection) for RT-PCR analysis.
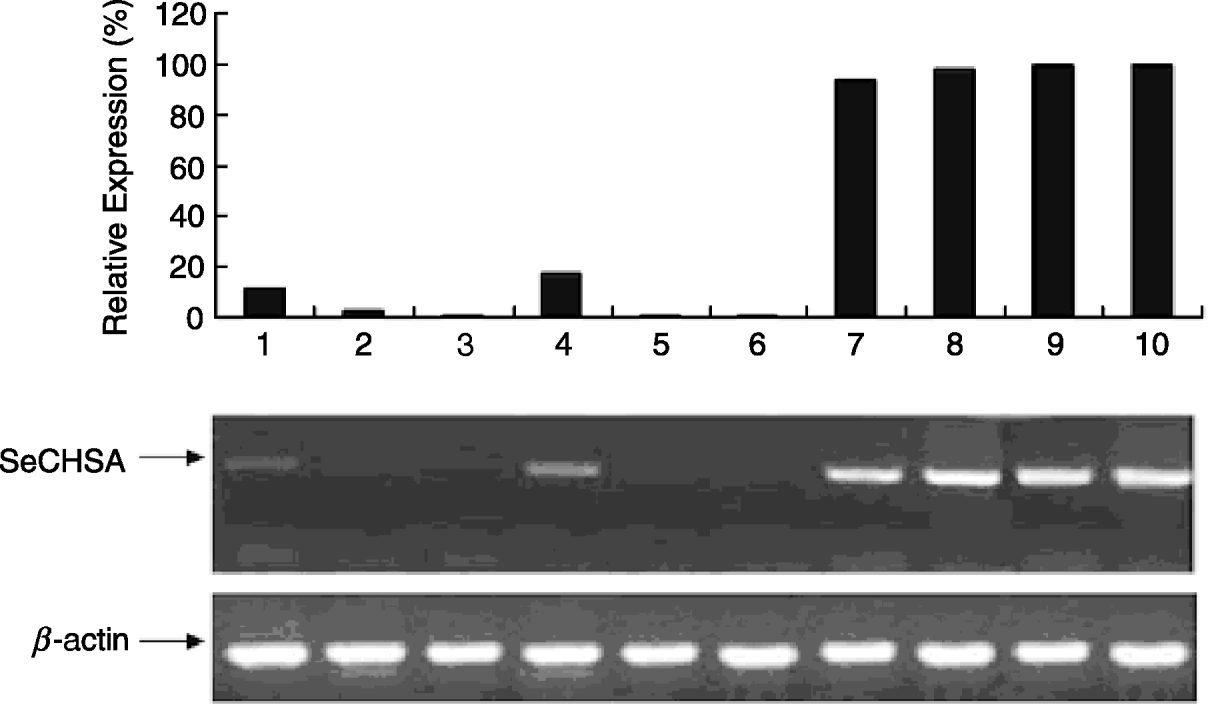
Fig. 5. RT-PCR analysis of SeCHSA mRNA. Three samples from dsRNA-injected abnormal larvae (1, 2, 3) or siRNA-injected abnormal larvae (4, 5, 6) collected in the first three days post-injection, and three samples from buffer-injected (7, 8, 9) and no injection control larvae (10) were selected (■, SeCHSA).
When dsRNA or siRNA specific for SeCHSA was injected into the 4th instar larvae, we observed significantly lower levels of SeCHSA transcript compared to those in the control larvae, and the effect of dsRNA was very similar to that of siRNA (fig. 5).
Impact of gene silencing on cuticle and tracheal formation
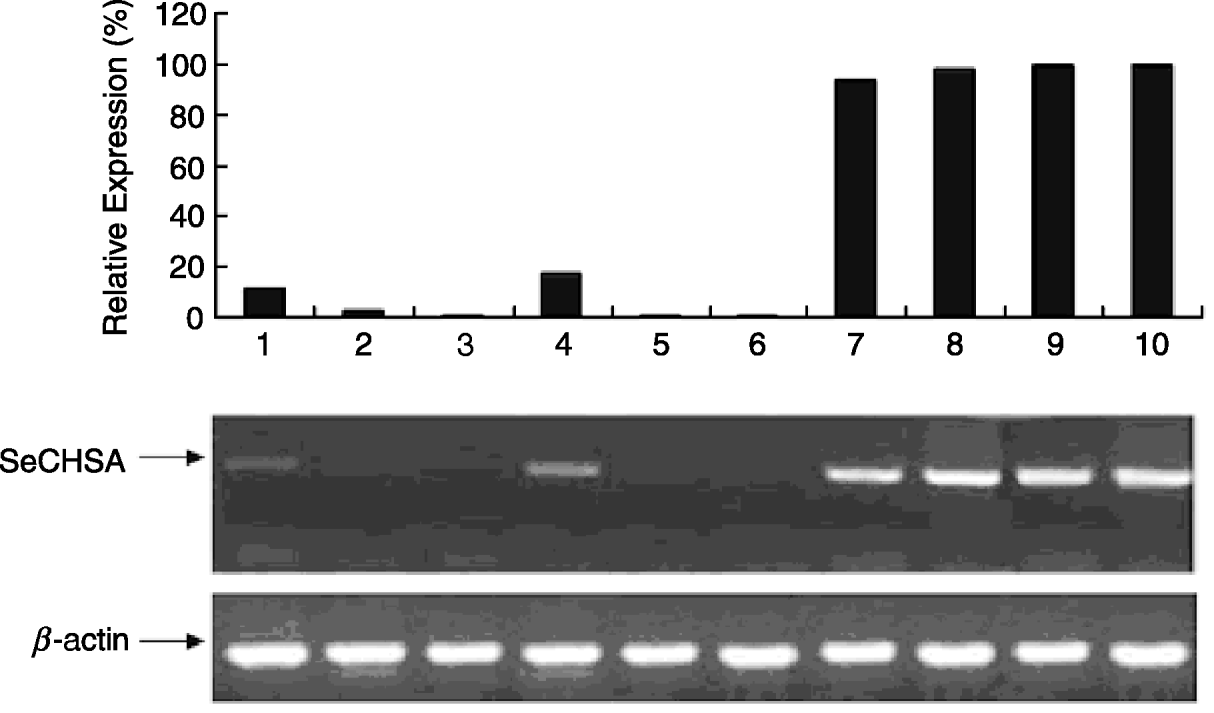
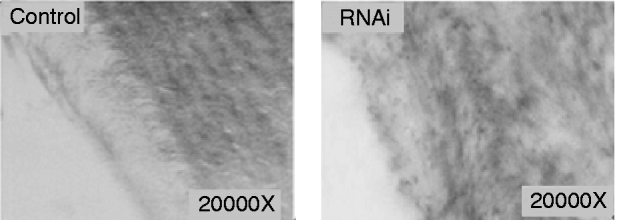
The electron microscope observation of the cuticle from abnormal larvae injected with siRNA or dsRNA indicated that the chitin layer was disordered compared with cuticles from control larvae (fig. 6). In addition, the epithelial walls of larval trachea from siRNA/dsRNA-injected larvae did not expand uniformly because of the reduction of chitin; and the resultant tubes were grossly misshapen, with constricted and distended regions all along their lengths (fig. 7). These results suggest that reduced expression of SeCHSA is associated with defects in cuticle formation, reduced chitin accumulation in trachea and defects in trachea development.
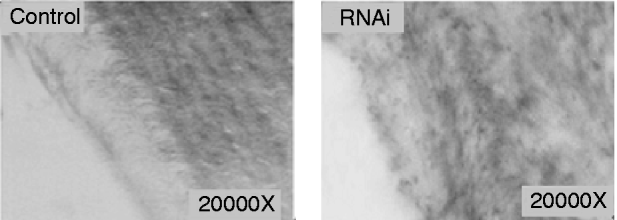
Fig. 6. Cuticle and trachea of larvae injected with siRNA/dsRNA. The super-micro structure of cuticle slices of the abnormal larvae injected with siRNA/dsRNA. The chitin layer of the abnormal larval cuticle from siRNA/dsRNA-injected larvae was in disarray compared to the controls.
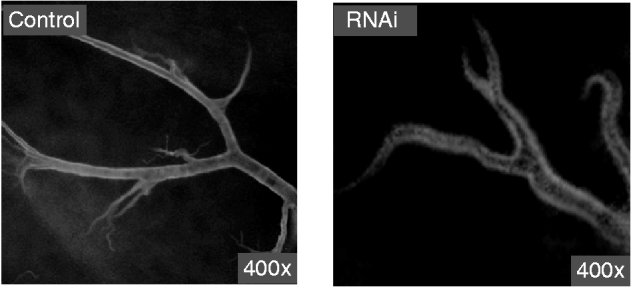
Fig. 7. Tracheal chitin labeled with fluorescein-conjugated chitin-binding probe. The trachea tissues were prepared from abnormal larvae in siRNA/dsRNA injected and control larvae.
Discussion
RNA interference (RNAi) is an evolutionarily conserved gene silencing mechanism. Both dsRNAs and siRNAs have been used successfully to specifically prevent the expression of exogenous and endogenous insect genes. In Lepidoptera, few genes have been studied using RNAi, and little is known about quantitative mortality caused by RNAi.
In the present study, we demonstrated that expression of the transgene SeCHSA in High 5 cells was inhibited by dsRNA/siRNA. The inhibition of SeCHSA expression was observed 36 h after co-transfection. Our result is similar to that of aminopeptidase N in Sf9 cells (Agrawal et al., Reference Agrawal, Malhotra and Bhatnagar2004). However, compared to Drosophila S2 cells, the inhibitory effect in High 5 cells was slightly lower (Caplen et al., Reference Caplen, Parrish, Imani, Fire and Morgan2001). Inability to attain complete inhibition of CHS expression in High 5 cells treated with dsRNA or siRNA suggested that uptake of dsRNA/siRNA is limited. Although High 5 cells have the cellular machinery to initiate RNAi and can generate specific degradation of target mRNAs, the proteins required for amplification and/or the spread of this effect are either less active or absent in these cells (Caplen et al., Reference Caplen, Parrish, Imani, Fire and Morgan2001).
It has been reported that RNAi could occur by injecting dsRNA into several lepidopteran insects, including M. sexta, H. cecropia and S. litura (Vermehren et al., Reference Vermehren, Qazi and Trimmer2001; Bettencourt et al., Reference Bettencourt, Terenius and Faye2002; Rajagopal et al., Reference Rajagopal, Thamilarasi, Venkatesh, Srinivas and Bhatnagar2005). In these cases, the injected dsRNA might be cut into siRNA by DICER in insects, which would then cause the interfering effect. We injected both dsRNA and siRNA in our study. The results indicated that both dsRNA and siRNA could suppress expression of SeCHSA gene in S. exigua, and siRNA is significantly more effective than dsRNA for inducing the abnormal phenotypes (table 1).
RNAi effectiveness depends on the concentration of dsRNA or siRNA and the developmental stage of individual larvae. In our experiments, we chose to inject 3 μl (1 μg μl−1) dsRNA/siRNA per larva, based on preliminary studies that identified this amount as the lowest dosage producing a detectable effect. In the range of 3–5 μl (1 μg μl−1), higher dosage did not make a difference; but we could not find gene silencing with less than 3 μl dosage. Compared with other RNAi experiments in insects, less dsRNA/siRNA was injected in S. exigua as compared to S. litura (4 μg per larva) (Rajagopal et al., Reference Rajagopal, Sivakumar, Agrawal, Malhotra and Bhatnagar2002), but more was injected than in T. castaneum (0.2 μg per larva) (Arakane et al., Reference Arakane, Muthukrishnan, Kramer, Specht, Tomoyasu, Lorenzen, Kanost and Beeman2005). The larvae in the last day of 4th instar stage were selected for injection in this study because expression of SeCHSA reached highest level in the first day and last day of the 5th instar stage (Chen et al., Reference Chen, Yang, Senthil Kumar, Tang, Sun, Qiu, Hu and Zhang2007). When expression of SeCHSA in the 5th instar larvae was inhibited, the RNAi effect may be maximized. In addition, there is little haemolymph in the larvae at that stage, so it is suitable for injection.
The inability to molt and reduce size were two consequences of SeCHSA knockdown in injected larvae (fig. 4). These findings were consistent with previous research on T. castaneum. In this species, TcCHSA-specific RNAi in T. castaneum disrupted all three types of molt (larval-larval, larval-pupal and pupal-adult) (Arakane et al., Reference Arakane, Muthukrishnan, Kramer, Specht, Tomoyasu, Lorenzen, Kanost and Beeman2005). In addition, larval phenotypes of S. exigua were observed every 12 h post-injection, and the accumulated percentage of abnormal larvae was calculated (table 1). This is important for exploring the potential application of RNAi in pest control.
Down-regulated SeCHSA expression also resulted in defects in cuticle formation and tracheal development (figs 6 and 7). Our results showed that the tracheae from abnormal larvae injected with siRNA/dsRNA could not expand uniformly, which was similar to Drosophila (Araujo et al., Reference Araujo, Aslam, Tear and Casanova2005). Because our RNAi was not 100% effective, some SeCHSA transcript was translated to produce chitin. Also, SeCHSA that was already present prior to RNAi could still catalyze chitin synthesis. These might explain why the insects still have new cuticles, and chitin in the cuticle and tracheae could still be observed. However, reduction of chitin has an impact on formation of the cuticle and development of tracheal tubes and may cause the death of S. exigua larvae.
In our study, both in vitro and in vivo RNAi are powerful tools to study the development of related genes in lepidopteran insects, and silencing of the SeCHSA gene could produce morphological defects in S. exigua larvae. Recently, transgenic plants engineered to express insect dsRNAs were reported for entomological research and the field control of insect pests (Baum et al., Reference Baum, Bogaert, Clinton, Heck, Feldmann, Ilagan, Johnson, Plaetinck, Munyikwa, Pleau, Vaughn and Roberts2007; Mao et al., Reference Mao, Cai, Wang, Hong, Tao, Wang, Huang and Chen2007). Since CHSA is an important gene for the growth and development of S. exigua, it could be used as a good target for transgenic dsRNA-producing plants which will be protected by insect RNAi triggered by ingestion of the dsRNA.
Acknowledgements
We thank Prof. Yi Pang and Mrs Miner He (Sun Yat-sen University, China) for their help in cell culture and Prof. Shuanglin Dong (Nanjing Agricultural University, China) for his help in insect culture. We are also grateful to Dr Xiaoqiang Yu (University of Missouri-Kansas City, MO, USA) for critical reading of the manuscript. This work was supported by National Basic Research Program of China (grant IBN-2006CB102001) and National Natural Science Foundation of China (grant IBN-30471143).