Institutional antibiograms are recommended to guide empiric antibiotic therapy,Reference Barlam, Cosgrove and Abbo 1 , Reference Tamma, Robinson and Gerber 2 but they have several limitations. Because traditional antibiograms provide organism-specific susceptibilities, they do not estimate the risk of organism-drug mismatch related to empiric antibiotic selection prior to culture identification and susceptibilities. Second, composite antimicrobial susceptibility data reported in traditional antibiograms may miss differences in resistance across clinical syndromes (eg, respiratory tract vs bloodstream infections). Furthermore, antibiograms in the ambulatory setting do not exclude patients with chronic medical conditions. Thus, antibiograms may overestimate resistance for patients who were previously healthy, which is particularly important in the ambulatory setting because microbiological susceptibility data may be less accessible. The objective of our study was to use the electronic health record (EHR) to generate and validate syndrome-specific antibiograms for urinary tract infections (UTIs) in previously healthy children in the outpatient setting.
Methods
This retrospective descriptive study of antimicrobial susceptibilities was conducted at Ann & Robert H. Lurie Children’s Hospital in Chicago, Illinois, a 288-bed tertiary-care children’s hospital with more than 12,000 admissions and 60,000 emergency department visits each year. Study subjects included all pediatric patients aged ≤21 years with urine isolates positive for gram-negative bacilli (GNB) collected from all outpatient locations, including the emergency department, from October 2, 2016, to May 1, 2017. If a patient had duplicate isolates for the same organism during this period, only the first isolate was included for the study.
We defined a ‘healthy’ outpatient cohort by excluding children who had (1) urine collected from pediatric subspecialty clinics, (2) complex chronic conditions defined by International Classification of Disease, Tenth Revision (ICD-10) codes,Reference Feudtner, Feinstein, Zhong, Hall and Dai 3 (3) prior hospital admissions within the prior 12 months, or (4) antibiotic use within 90 days. Patients not included in this EHR-derived healthy cohort were classified as ‘complex.’ Patient identification and data extraction were automated using Vigilanz software (Vigilanz, Minneapolis, MN), a web-based clinical support tool, using discrete data queries from administrative, pharmacy, and microbiology records.
Weighted-incidence syndromic antibiograms (WISAs) for urinary GNB were created by calculating a weighted average of individual species susceptibilities, similar to the methodology used to create combination antibiograms described previously.Reference Hebert, Ridgway, Vekhter, Brown, Weber and Robicsek 4 Antimicrobial susceptibilities were determined for the following study antibiotics: amoxicillin/clavulanate, ampicillin, cefazolin, ceftriaxone, and trimethoprim/sulfamethoxazole. WISAs were generated for healthy patients using our EHR-derived definition, complex patients, and for all patients combined. The sensitivity and specificity of the EHR-derived definition to distinguish between healthy and complex patients were determined by a sequential manual review of 50% of the patient charts. Susceptibility differences between various antibiograms (all outpatients, healthy cohort, and complex cohort) were analyzed using the Fisher exact test.
Results
Overall, 686 outpatient UTI specimens were included in the study. The most common organisms in the healthy cohort were Escherichia coli (269; 71.5%), Proteus mirabilis (31, 8.2%), and Klebsiella pneumoniae (23; 6.1%). The most common organisms in the complex cohort were E. coli (166, 53.6%) K. pneumoniae (32; 10.3%), and Pseudomonas aeruginosa (21; 6.8%).
Compared to the cohort of complex patients, urinary isolates from the EHR-derived healthy cohort had statistically significantly greater susceptibility rates (P<.05) for all study antibiotics (Table 1). The greatest difference in susceptibility was seen for trimethoprim-sulfamethoxazole (56.9% vs 75.9%; P<.001), and the least was seen for ampicillin (34.4% vs 42.7%; P=.033). Compared to the WISA for all outpatient children, the WISA for the healthy cohort showed higher susceptibility for all antibiotics. This difference was statistically significant for amoxicillin (P=.012), ceftriaxone (P=.038), and trimethoprim/sulfamethoxazole (P=.048).
Table 1 Urinary Tract Infection Antibiograms for Healthy, Complex, and All Children
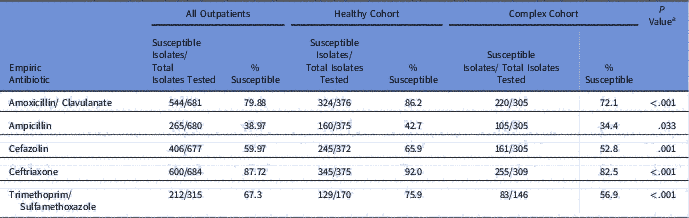
a Comparing healthy vs complex cohorts.
Compared to manual chart review, the EHR-based criteria identified healthy children with a sensitivity of 85%, specificity of 81%, positive predictive value of 76%, and negative predictive value of 89%. Susceptibility rates for healthy children based on the EHR definition were similar to rates for previously healthy children confirmed by manual chart review (Table 2). For all study antibiotics, antibiotic susceptibilities were higher via direct chart review.
Table 2 Urinary Tract Infection Antibiograms for Healthy Children Identified by Chart Review Versus EHR Method
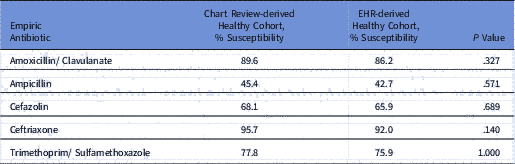
Note. EHR, electronic health record.
Discussion
By defining a healthy cohort and comparing it to the remaining medically complex patients and the ambulatory patient population as a whole, we demonstrated differences in antibiotic susceptibility for multiple antibiotics. While it is expected that healthy patients would have less frequent antibiotic resistance, our methodology utilized discrete data elements, including diagnostic codes, to readily build these customized antibiograms from an EHR. We have previously described that 58% of pediatricians report that overly complex patient populations limit the generalizability of their hospital antibiogram, and 95% of pediatricians would probably or definitely use an antibiogram that compiled antibiotic resistance data from healthy children with common infections.Reference Spiekerman, Patel, Patel and Kociolek 5 Thus, we surmise that using our proposed methodology to develop and disseminate healthy child antibiograms would be well received by general pediatricians.
Dahle et alReference Dahle, Korgenski, Hersh, Srivastava and Gesteland 6 had previously developed a pediatric ambulatory antibiogram for uropathogens and compared it to an existing hospital antibiogram, demonstrating increased susceptibility of E. coli to narrower-spectrum antibiotics. Simpao et alReference Simpao, Ahumada and Larru Martinez 7 embedded an electronic antibiogram into the EHR that can provide customized reports based on inputs for body site, hospital/clinic location, and community versus hospital-acquired infection. Our EHR-derived healthy cohort demonstrated less frequent antibiotic resistance among urine isolates compared to medically complex patients. Furthermore, compared to manual chart review, EHR derivation of the healthy cohort underestimated susceptibility to the study antibiotics. Thus, we were able to define a subset of patients for whom narrower-spectrum empiric antibiotic therapy could be safely administered. Our methodology could be further modified to develop combination antibiograms that estimate the likelihood that combinations of antibiotics will treat a given infectious syndrome, factoring in the local, weighted incidence of pathogens (including polymicrobial infections) causing that syndrome. In the adult population, these antibiograms have been developed for abdominal-biliary infections, urinary tract infections, ventilator-associated pneumonia, and catheter-related bloodstream infections.Reference Hebert, Ridgway, Vekhter, Brown, Weber and Robicsek 4 , Reference Randhawa, Sarwar, Walker, Elligsen, Palmay and Daneman 8
This study has several limitations. Prior antibiotic use prescribed at outside institutions could not be captured by our study methodology. The relative proportion of healthy and complex patients, as well as susceptibility rates for various antibiotics, may differ between hospitals; thus, our criteria may not be generalizable. Risk factors for acquisition of antibiotic-resistant organisms may differ between institutions.
Nonetheless, this study provides proof-of-concept that a syndromic antibiogram for a cohort of healthy children can be accurately identified using ICD-10 codes and other discrete data elements. Our methodology could be automated to inform selection of antibiotic therapy for other patient populations and clinical syndromes. Criteria for a healthy cohort may differ and may be tailored according to each institution’s own risk factors for antibiotic resistant organisms.
Financial support
A medical student research grant to Y.C., through the 2017 Infectious Diseases Society of America (IDSA) Medical Scholars Program, helped fund expenses related to an abstract/poster presentation at the IDWeek 2017 conference; the abstract reported some preliminary data described in this article.
Conflicts of interest
All other authors report no conflicts of interest relevant to this article.
Acknowledgments
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2018.246




