Introduction
FMRFamide-like peptides (FLPs) are involved with nematode growth and development and are broadly associated with neuromuscular regulation, affecting essential components of behaviour (Liu et al., Reference Liu, Kim, Li and Barr2007), feeding (Rogers et al., Reference Rogers, Reale, Kim, Chatwin, Li, Evans and de Bono2003; Papaioannou et al., Reference Papaioannou, Marsden, Franks, Walker and Holden-Dye2005), reproduction (Moffett et al., Reference Moffett, Beckett, Mousley, Geary, Marks, Halton, Thompson and Maule2003), and other basic functions (Maule et al., Reference Maule, Mousley, Marks, Day, Thompson, Geary and Halton2002; Kimber & Fleming, Reference Kimber and Fleming2005; Cohen et al., Reference Cohen, Reale, Olofsson, Knights, Evans and de Bono2009). Nematode FLPs comprise a large, complex family with a characteristic C-terminal -RFamide motif, and extensive variations within the N-terminal region, providing structural variety and functional complexity. Numerous homologues are distributed across all nematode genera (Maule et al., Reference Maule, Mousley, Marks, Day, Thompson, Geary and Halton2002; Li, Reference Li2005; McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005, Reference McVeigh, Geary, Marks and Maule2006), and FLPs are pursued as targets for novel parasite control strategies (Greenwood et al., Reference Greenwood, Williams and Geary2005; Kimber & Fleming, Reference Kimber and Fleming2005; Kimber et al., Reference Kimber, McKinney, McMaster, Day, Fleming and Maule2007). The metabolism of FLPs and attenuation of their signals by endogenous proteases, and the biological consequences, are also major considerations (Husson et al., Reference Husson, Mertens, Janssen, Lindemans and Schoofs2007; Masler, Reference Masler2007, Reference Masler2008). Taken together, FLP sequence variety and related catabolic mechanisms offer a rich source of targets for which to design species-specific control agents. Accordingly, discovery of possible differences in the proteolytic digestion of FLPs by different nematode species is crucial. Differences among protease complements should reside in both substrate preference and susceptibility to specific inhibitors. The present work examined proteolytic activities from two of the most important pathogens of agricultural crops, the plant-parasitic nematodes Heterodera glycines and Meloidogyne incognita, and a free-living nematode, Panagrellus redivivus, for specificity in the degradation of selected FLP substrates and the response of these different nematode systems to a variety of protease inhibitors. Results revealed FLP substrate preferences among species and, for the first time, unique responses to inhibitors by M. incognita proteases.
Materials and methods
Nematode rearing and extraction
Panagrellus redivivus was reared in sterile liquid culture at 22°C (Chitwood et al., Reference Chitwood, Lusby, Thompson, Kochansky and Howarth1995), and harvested after 1 week as a mixed-age population. Worms were washed with autoclaved, distilled and de-ionized (D/D) water to remove culture medium, most water was removed by centrifugation, and the worms were stored at − 20°C. Meloidogyne incognita was raised on pepper (Capsicum annuum ‘PA-136’) and H. glycines was raised on soybean (Glycine max ‘Kent’), using a constant moisture system (Sardanelli & Kenworthy, Reference Sardanelli and Kenworthy1997), at 27°C and 16 h light:8 h dark photoperiod. Plants were inoculated near the rhizosphere with five M. incognita egg masses or 2000 H. glycines eggs per plant, and 5 weeks after inoculation plants were harvested. Egg masses were collected from infected C. annuum roots under a dissecting microscope and placed on modified Baermann funnels kept at the above temperature and photoperiod for infective M. incognita juvenile hatching. Cysts were harvested from infected G. max roots, broken open in water to release eggs, and the freed eggs placed on funnels for collection of infective H. glycines juveniles as above. Freshly collected worms were centrifuged to remove most water, and stored at − 20°C.
Worms were thawed and suspended in ice-cold, autoclaved D/D water (~10 volumes of water/worm volume) and mechanically disrupted in 1.5 ml polypropylene tubes containing 0.5 mm zirconia/silica beads shaken in a Bead Beater (BioSpec Products, Bartlesville, Oklahoma, USA). Processing consisted of six rounds of shaking (30 s per round) alternated with six periods of cooling on ice (3 min each). The processed homogenates were centrifuged at 16,000 × g for 30 s at room temperature and the supernatants (S1) were transferred to glass high-speed centrifuge tubes on ice. The 16,000 × g pellets were washed using three rounds of shaking and cooling, and the resulting wash supernatant (S2) was added to S1. Washed pellets were microscopically examined at 40 × and 100 × to ensure that all worms were disrupted. The pooled S1+S2 supernatant was centrifuged at 40,000 × g, 20 min, 5°C, and the 40,000 × g supernatant was dried as aliquots by vacuum centrifugation and stored at − 20°C. Total protein was determined using the microBCA assay (Pierce Chemical Co., Rockford, Illinois, USA).
Peptide substrates and protease inhibitors
Substrates were chosen to represent a variety of FLPs. Some sequences are encoded by M. incognita or H. glycines genes (KHEFVRFa, KHEYLRFa, KSAYMRFa, SAPYDPNFLRFa) or known to be present in P. redivivus (KHEYLRFa, KSAYMRFa) (Maule et al., Reference Maule, Mousley, Marks, Day, Thompson, Geary and Halton2002; Li, Reference Li2005; McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005). Products from seven flp genes (-1, -3, -6, -8, -9, -12, -14) were included and an eighth was represented by RPKPKFIRFa, a variant of APKPKFIRFa (flp-5) with the Arg1-for-Ala1 replacement providing an additional trypsin cleavage site and providing a comparison with the RNK- N-terminus of RNKFEFIRFa. Additional FLPs used are predicted from expressed sequence tag (EST) analyses (McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005) to be present in plant-parasitic nematodes.
Substrates were synthesized as FRET-modified sequences representing nine different nematode FLPs. QXL520-KHEYLRF-K(5-FAM)a, QXL520-KSAYMRF-K(5-FAM)a, QXL520-RNKFEFIRF-K(5-FAM)a and QXL520-SAPYDPNFLRF-K(5-FAM)a were from AnaSpec (Fremont California, USA). Abz-KHEFVRF-Y(3-NO2)a, Abz-KNEFIRF-Y(3-NO2)a, Abz-KPSFVRF-Y(3-NO2)a, Abz-RPKPKFIRF-Y(3-NO2)a and Abz-SAEPFGTMRF-Y(3-NO2)a were from Biomatik USA (Wilmington, Delaware, USA). Each substrate consisted of a peptide sequence (FLP) flanked by a fluorescent donor (5-FAM or Abz) and a fluorescence quencher (QXL520 or 3-NO2). While the peptide is intact, fluorescence is quenched. Upon peptide cleavage, donor and quencher become separated, and fluorescence increases.
All substrates were dissolved in either 25% acetonitrile (CH3CN) (ABZ/3-NO2 peptides) or 50% CH3CN (QXL520/5-FAM peptides) and dried as aliquots by vacuum centrifugation, then stored at − 20°C. Inhibitors were chosen to cover a wide range of protease specificities, and all were from G-Biosciences (St. Louis, Missouri, USA). 4-(2-Aminoethyl) benzenesulfonyl fluoride (AEBSF), acetyl-leucyl-leucyl-norleucinal (ALLN), antipain-HCl, aprotinin, bestatin, chymostatin, disodium EDTA, E-64, leupeptin, pepstatin, phosphoramidon and phenylmethanesulphonylfluoride (PMSF) were supplied as stock solutions with concentrations provided by the manufacturer. Aliquots of stock solutions were diluted into assay buffer as needed to obtain the indicated concentrations (see below) for individual experiments.
Protease assays
Substrate screens
Reactions were carried out at 27°C in 384-well assay plates (flat bottom, non-treated, Corning, Inc. Corning NY, USA) in 100 mm Tris, pH 7.8 assay buffer, 25 μl reaction volume. Extract aliquots were dissolved directly into assay buffer. Peptide substrate aliquots were first dissolved in dimethylsulphoxide (DMSO) prior to addition to the assay mixture (final concentration of DMSO in all reactions was 2%). Reactions containing 0.08–0.1 μg/μl extract protein and 4 pmol/μl peptide substrate were initiated by the addition of substrate, and monitored over 1 h for increasing fluorescence (SpectraMAX EM fluorescent plate reader; Molecular Devices, Sunnyvale, California, USA). For the Abz/3-NO2 peptides, excitation and emission wavelengths were set to 320 nm and 420 nm, respectively. For the QXL520/5-FAM peptides excitation was at 493 nm and emission was 520 nm. Individual substrates were screened from 4–8 times for each species over 6–8 different experiments. Substrate controls (no extract) were used to correct for background, and extract was incubated for 10 min at 100°C to confirm that activity was heat labile.
The reaction rate was quantified using SoftMax Pro instrument control and data collection software (Molecular Devices). The rate is defined as the increase in relative fluorescence units (RFU) over time and expressed as slope (Vmax) of the linear portion of the reaction. Rate data were then converted to Vmax/min/μg total protein.
Inhibitor screens
Assays were conducted as above, except that inhibitors were added 15 min prior to the addition of substrate. Aliquots of inhibitor stock solutions were first diluted into assay buffer and then added to reactions to provide the following concentrations for inhibitor screening: AEBSF (1.67 mm), ALLN (41.7 μm), antipain (118 μm), aprotinin (0.49 μm), bestatin (208 μm), chymostatin (159 μm), E-64 (44.8 μm), EDTA-Na2 (8 mm), leupeptin (18.8 μm), pepstatin (16.3 μm), phosphoramidon (29.4 μm), PMSF (1.60 mm). Inhibitor screens were conducted at least twice for all inhibitors across all species. Selected substrates and inhibitors were examined further to quantify inhibitor potency (IC50) using the same assay design as above, except that dilution series of inhibitors were prepared in assay buffer and added to reaction mixtures 15 min prior to adding substrate.
Inhibitor data are presented as the percentage reduction in Vmax/min/μg in test versus control reactions: ((Vmax/min/μgcontrol − Vmax/min/μginhibitor) /Vmax/min/μgcontrol) × 100.
Data analysis and information resources
Individual data means were compared using Mann–Whitney non-parametric t-test with P indicated for each comparison. Relationships among data means were analysed by one-way ANOVA with Tukey's multiple comparison using GraphPad Prism (GraphPad Software, La Jolla, California, USA). Inhibitor and substrate attributes (specificities, effective concentrations, primary structure features) were evaluated with online resources provided by MEROPS Peptidase Database (http://www.merops.sanger.ac.uk) and Swiss Institute of Bioinformatics (http://www.expasy.org/). Nucleotide and amino acid sequences were examined using Nematode.net (http://www.nematode.net) and NCBI (http://www.ncbi.nlm.nih.gov) resources.
Results
Differential digestions of FLPs
All nine FLP-FRET substrates were digested to varying degrees by extracts from all three species (fig. 1a and b). Of the nine substrates, seven of them (KHEYLRFa, KSAYMRFa, RNKFEFIRFa, SAPYDPNFLRFa, KNEFIRFa, RPKPKFIRFa and SAEPFGTMRFa) were digested more extensively by the soluble fraction of P. redivivus extract (P < 0.05) than by those fractions from the two plant parasites. KPSFVRFa was digested equally (P>0.1) by extracts of all three species (fig. 1b). KHEFVRFa was digested significantly more extensively (P < 0.05) by M. incognita extract than by extracts of either P. redivivus or H. glycines (fig. 1b). The most highly degraded substrates among the three species were KSAYMRFa and KPSFVRFa (fig. 1a and b). Comparing digestion rate ratios among all three species across all substrates, P. redivivus extract showed a mean digestion rate four- to fivefold greater than either plant parasite (table 1) while M. incognita extract was approximately twofold more potent than that from H. glycines.

Fig. 1 Protease activities in Panagrellus redivivus, Heterodera glycines and Meloidogyne incognita extracts detected using FLP-FRET substrates. Extracts and peptide substrates were obtained as described, and digestion reactions prepared in 25 μl 100 mm Tris, pH 7.8 with 0.08–0.1 μg/μl extract protein, 4 pmol/μl peptide substrate and 2% DMSO. Reactions were initiated by substrate addition and monitored for 1 h at 27°C for increasing fluorescence. Activities are expressed as mean Vmax/min/μg ± SEM (n = 4–8). Means were compared using one-way ANOVA and Tukey's multiple comparison test. For each substrate, means were compared across species and those followed by different letters were significantly different (P < 0.05). For each species, means were compared across substrates and those followed by different numbers were significantly different (P < 0.05). White bars, P. redivivus; grey bars, H. glycines; black bars, M. incognita. Substrates tested were: (a) KHEYLRF [QXL520-KHEYLRF-K(5-FAM)a], KSAYMRF [QXL520-KSAYMRF-K(5-FAM)a], RNKFEFIRF [QXL520-RNKFEFIRF-K(5-FAM)a], and SAPYDPNFLRF [QXL520-SAPYDPNFLRF-K(5-FAM)a]; (b) KHEFVRF [Abz-KHEFVRF-Y(3-NO2)a], KNEFIRF [Abz-KNEFIRF-Y(3-NO2)a], KPSFVRF [Abz-KPSFVRF-Y(3-NO2)a], RPKPKFIRF [Abz-RPKPKFIRF-Y(3-NO2)a], SAEPFGTMRF [Abz-SAEPFGTMRF-Y(3-NO2)a].
Table 1 Comparison of overall protease activities in extracts prepared from three nematode species, Heterodera glycines (Hg), Meloidogyne incognita (Mi) and Panagrellus redivivus (Pr) for nine different FLP substrates.
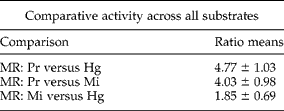
Average activity (Vmax/min/μg) of each species for each substrate was calculated over 6–8 separate experiments. Each of these average activity values was used to calculate an activity ratio between species (activityspecies-x/activityspecies-y) for each substrate. These nine ratios were averaged across all substrates to generate a mean ratio (MR) for each species and used to compare overall activity between species.
Responses to protease inhibitors
To examine the effects of protease inhibitors on the ability of extracts from different species to digest nematode FLPs, two substrates were selected: KPSFVRFa, the only FLP substrate digested nearly equally by extracts of all three species, and KHEFVRFa because of its rapid digestion by M. incognita extract. The inhibitor screen with Abz-KPSFVRF-Y(3-NO2)a as substrate revealed a wide range of responses (fig. 2). All inhibitors had some effect on the P. redivivus and H. glycines preparations, but seven inhibitors (AEBSF, aprotinin, bestatin, EDTA, pepstatin, phosphoramidon and PMSF) did not inhibit M. incognita extract activity (fig. 2). Only two inhibitors reduced activity in all three species by at least 25% (ALLN, chymostatin), and chymostatin was the most consistent single inhibitor, at 62–69% for all three species (fig. 2). In addition to chymostatin, antipain (72%) and E-64 (70%) were the most effective inhibitors of M. incognita activity. In comparison, antipain inhibited H. glycines activity by 53% and P. redivivus activity by only 21%. E-64 was a weak inhibitor with these species at 23% (H. glycines) and 20% (M. incognita).

Fig. 2 Effect of 12 protease inhibitors on the digestion of KPSFVRF [Abz-KPSFVRF-Y(3-NO2)a] by extracts from Panagrellus redivivus, Heterodera glycines and Meloidogyne incognita. Assays were conducted as described, except that inhibitors were added 15 min prior to the addition of substrate. Aliquots of inhibitor stock solutions were first diluted into assay buffer and then added to reactions to provide the following concentrations: AEBSF (1.67 mm), ALLN (41.7 μm), antipain (118 μm), aprotinin (0.49 μm), bestatin (208 μm), chymostatin (159 μm), E-64 (44.8 μm), EDTA-Na2 (8 mm), leupeptin (18.8 μm), pepstatin (16.3 μm), phosphoramidon (29.4 μm), PMSF (1.60 mm). Results are expressed as the average of 2–3 separate assays ± range. Percentage inhibition was calculated as: ((Vmax/min/μgcontrol − Vmax/min/μginhibitor)/Vmax/min/μgcontrol) × 100. White bars, P. redivivus; grey bars, H. glycines; black bars, M. incognita. Dashed lines represent 25% and 50% inhibition levels.
The inhibitor screen with Abz-KHEFVRF-Y(3-NO2)a (fig. 3) was done using P. redivivus and M. incognita extracts only, because of the very low activity with this substrate in H. glycines extracts (fig. 1b). Of the seven inhibitors that did not suppress digestion of Abz-KPSFVRF-Y(3-NO2)a by M. incognita extract, three of them (aprotinin, EDTA and pepstatin) also did not inhibit M. incognita digestion of Abz-KHEFVRF-Y(3-NO2)a, and three others (bestatin, phosphoramidon and PMSF) were only weak inhibitors. Phosphoramidon completely inhibited P. redivivus activity. AEBSF inhibited both M. incognita (60%) and P. redivivus (70%) activities, unlike the result with Abz-KPSFVRF-Y(3-NO2)a (fig. 2). Chymostatin inhibited activities of both species more than 75%, just as it had done with Abz-KPSFVRF-Y(3-NO2)a. Antipain (17%) and E-64 (22%) were very poor inhibitors of P. redivivus activity, yet antipain completely inhibited M. incognita activity and E-64 was over 95% effective (fig. 3). These results were similar to those observed with Abz-KPSFVRF-Y(3-NO2)a (fig. 2).

Fig. 3 Effect of 12 protease inhibitors on the digestion of KHEFVRF [Abz-KHEFVRF-Y(3-NO2)a] by extracts from Panagrellus redivivus and Meloidogyne incognita. Assays were conducted as described, except that inhibitors were added 15 min prior to the addition of substrate. Aliquots of inhibitor stock solutions were first diluted into assay buffer and then added to reactions to provide the following concentrations: AEBSF (1.67 mm), ALLN (41.7 μm), antipain (118 μm), aprotinin (0.49 μm), bestatin (208 μm), chymostatin (159 μm), E-64 (44.8 μm), EDTA-Na2 (8 mm), leupeptin (18.8 μm), pepstatin (16.3 μm), phosphoramidon (29.4 μm), PMSF (1.60 mm). Results are expressed as the average of 2–3 separate assays ± range. Percent inhibition was calculated as: ((Vmax/min/μgcontrol − Vmax/min/μginhibitor)/Vmax/min/ μgcontrol) × 100. White bars, P. redivivus; black bars, M. incognita. Dashed lines represent 25% and 50% inhibition levels.
Since Abz-KHEFVRF-Y(3-NO2)a was the only substrate digested at a higher rate by a plant-parasitic nematode preparation than by P. redivivus extract, and given the effectiveness of antipain and E-64 inhibition of M. incognita activity, these substrate and inhibitor combinations were examined in more detail (fig. 4). E-64 and antipain were equally potent inhibitors of M. incognita protease digestion of Abz-KHEFVRF-Y(3-NO2)a, with IC50 values of 222 nm and 198 nm, respectively. IC90 for E-64 was 1.64 μm and 3.39 μm for antipain.

Fig. 4 Inhibition of KHEFVRF [Abz-KHEFVRF-Y(3-NO2)a] digestion by extracts of Meloidogyne incognita by the cysteine protease inhibitors antipain and E-64. Dilution series of inhibitors were prepared in 100 mm Tris and added to reactions 15 min prior to the addition of substrate. Reaction conditions: 0.09 μg/μl M. incognita extract protein, 2.45–0.03 μm inhibitor, 2% DMSO in 25 μl 100 mm Tris pH 7.8, 27°C. Data are expressed as means ± SD of percent inhibition (3–4 independent assays for each inhibitor) versus log inhibitor concentration. Percent inhibition was calculated as: ((Vmax/min/μgcontrol − Vmax/min/μginhibitor)/ Vmax/min/μgcontrol) × 100. Filled circles, E-64; open circles, antipain.
Discussion
Soluble fractions from the extracts of H. glycines, M. incognita and P. redivivus were capable of digesting a number of FMRFamide-like peptides, which were chosen to represent products of a number of nematode flp genes and provide a variety of amino acid sequences. The significantly higher overall protease activity in P. redivivus extracts, compared with those of the plant parasites, might be attributed in part to the different physiological and developmental states of the populations used to prepare the extract samples. The H. glycines and M. incognita extracts were prepared from newly hatched juveniles, whereas P. redivivus samples were from mixed populations comprising various stages of development, including adults. The protease complement of the mixed population may well have been more diverse than that of the juvenile-only preparations. This phenomenon was observed previously in comparisons of proteolytic activities between a free-living and a plant-parasitic nematode (Masler et al., Reference Masler, Kovaleva and Sardanelli2001). Aminopeptidase activity in Caenorhabditis elegans preparations was fivefold more potent than that from H. glycines, an activity ratio strikingly similar to those reported here. It is interesting to note that these similar results were obtained using assays designed to detect the activity of a specific protease (aminopeptidase) or general protease activity (this report). Whether these differences between free-living and plant-parasitic nematodes were trivial (different population qualities) or fundamental needs further examination.
The overall digestion rates for P. redivivus were markedly higher than those of H. glycines and M. incognita with most of the substrates, and the proteolytic activities of the two plant parasites were similar, except for one substrate. The notable exception was KHEFVRFa, which was digested 7.2-fold more rapidly by M. incognita extract than by H. glycines extract and, even more remarkably, 2.1-fold more than P. redivivus. KHEFVRFa was reported as a novel FLP encoded by M. incognita flp-14 (McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005; Johnston et al., Reference Johnston, McVeigh, McMaster, Fleming and Maule2010) in tandem with the highly abundant KHEYLRFa (Marks et al., Reference Marks, Shaw, Maule, Davis, Halton, Verhaert, Geary and Thompson1995). An EST search did not detect the KHEFVRF motif in any Heterodera species. The motif was only observed in the Meloidogyne species previously reported (McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005; Johnston et al., Reference Johnston, McVeigh, McMaster, Fleming and Maule2010). Thus, KHEFVRFa appears to be primarily confined to Meloidogyne, although it has also been reported in some Pratylenchus species (McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005).
The minor amino acid differences between KHEFVRFa and KHEYLRFa suggest that the two FLPs may have similar physiological effects (Johnston et al., Reference Johnston, McVeigh, McMaster, Fleming and Maule2010). However, these differences had considerable effect upon the relative abilities of M. incognita and P. redivivus extracts to digest these peptides. That KHEFVRFa was digested so efficiently by M. incognita extracts was made all the more remarkable in light of the overall differences between M. incognita and P. redivivus extract activities. The only other FLP that was digested by M. incognita preparations at least as well as by P. redivivus preparations, KPSFVRFa, shared the C-terminal -FVRFa motif. In addition to its presence in Mi-flp-14, this motif was reported in four other flp genes, flp-9, -11, -16 and -17 (Maule et al., Reference Maule, Mousley, Marks, Day, Thompson, Geary and Halton2002; Li, Reference Li2005; McVeigh et al., Reference McVeigh, Leech, Mair, Marks, Geary and Maule2005) with sequences from H. glycines and M. incognita contained in flp-16. FLPs encoded by flp-9 and flp-16, including KPSFVRFa, inhibited pharyngeal action potentials in C. elegans (Li, Reference Li2005), and KPSFVRFa, GQTFVRFa (flp-16) and NGAPQPFVRFa (flp-11) caused flaccid paralysis and reduced muscle contractility in Ascaris suum oviduct (Moffett et al., Reference Moffett, Beckett, Mousley, Geary, Marks, Halton, Thompson and Maule2003). The possibility that M. incognita has a proteolytic component possessing a metabolic association with –FVRFa FLPs is intriguing with regard to the potential for regulating physiological and behavioural processes in these plant parasites.
Response to protease inhibitors revealed both general and specific characteristics among the three enzyme sources. Aspartic proteases and aminopeptidases appeared not to have significant roles in the digestion of either Abz-KHEFVRF-Y(3-NO2)a or Abz-KPSFVRF-Y(3-NO2)a since pepstatin and bestatin were generally ineffective inhibitors, with the exception of a modest inhibition by bestatin of H. glycines activity. The variable results with PMSF and especially AEBSF may have been due in part to the instability of these inhibitors at pH greater than 7 (Lunn & Sansone, Reference Lunn and Sansone1994). While aprotinin was ineffective, the generally strong inhibition by the low molecular weight serine protease inhibitor chymostatin with all three species and both substrates indicated a universal importance for chymotrypsin-like proteases. Metalloprotease inhibition was curious. Although EDTA had some marginal inhibition of P. redivivus and H. glycines activities, it consistently failed to inhibit M. incognita activity. Results were even more striking with phosphoramidon and its markedly divergent effects on M. incognita and P. redivivus preparations. A more detailed examination of these metalloprotease differences, including additional inhibitors and kinetics of isolated enzymes, should be revealing.
Calpain/cysteine protease inhibitors caused at least a 25% decrease in protease activity in all M. incognita assays. Moreover, the potent inhibition of Abz-KHEFVRF-Y(3-NO2)a and Abz-KPSFVRF-Y(3-NO2)a digestions by antipain and E-64 suggests a particularly important role for cysteine proteases in M. incognita, at least with regard to these two FLPs. Antipain (and leupeptin, which had lower but significant inhibitory activity in M. incognita) inhibits cysteine proteases, but some serine proteases as well (Kirschke, Reference Kirschke, Barrett, Rawlings and Woessner2004), while E-64 specifically inhibits cysteine proteases (Barrett et al., Reference Barrett, Kembhavi, Brown, Kirschke, Knight, Tamai and Hanada1982). These include cathepsin L, which is expressed in intestinal tissue in M. incognita and was found to be associated with parasitism (Neveu et al., Reference Neveu, Abad and Castagnone-Sereno2003; Shingles et al., Reference Shingles, Lilley, Atkinson and Urwin2007). An examination of nematode cathepsin L and cathepsin L-like sequences, based upon published reports (Urwin et al., Reference Urwin, Lilley, McPherson and Atkinson1997; Neveu et al., Reference Neveu, Abad and Castagnone-Sereno2003) and EST resources, showed greater that 65% identities among H. glycines and M. incognita amino acid sequences and more than 90% identities in the catalytic core (data not shown). Thus, structural differences among cathepsin L-like proteases cannot fully explain the differences observed in the digestion of certain FLP-substrates.
The distinctly varied abilities of extracts from different nematode species to digest nematode FLP substrate peptides, and the preferential digestion of a novel M. incognita FLP by M. incognita proteases, raise issues regarding peptide signal attenuation, and species-specific metabolic regulation. Expanding the survey of -FVRFa variants should reveal whether KHEFVRFa digestion represents a unique case, or if it represents a more important, fundamental difference among species. Similarly, a closer examination of cysteine and other proteases in plant-parasitic nematodes, including more extensive biochemical characterizations and an increased species coverage, is necessary to further address the use of these enzymes as targets for nematode control.







