Published online by Cambridge University Press: 24 March 2005
The major surface protease (msp or gp63) of Leishmania plays a major role in the host–parasite interaction. We analysed here the structure of the msp gene locus in Leishmania (Viannia) braziliensis and compared it to results obtained in other species. Physical mapping of cosmid contigs revealed a minimum of 37 genes per haploid genome and at least 8 different msp gene families. Within the same organism, these genes showed a nucleotide sequence varying in certain stretches from 3 to 34%, and a mosaic structure. From an evolutionary point of view, major differences were observed between subgenera Viannia and Leishmania, both in terms of msp gene number and sequence. Within subgenus Viannia, phenetic analysis revealed three clusters in which sequence variants of L. (Viannia) braziliensis and L. (Viannia) guyanensis were interspersed. Functional implications of our results were explored from predicted L. (Viannia) braziliensis protein sequences: regions encoding the msp catalytic site showed a conserved sequence, while regions encoding surface domains possibly involved in the host–parasite interaction (macrophage adhesion sites and immunodominant B-cell and T-cell epitopes) were variable. We speculate that this would be an adaptive strategy of the parasite.
Leishmania are parasitic protozoa responsible for a wide spectrum of clinical forms ranging from self-healing cutaneous lesions to disfiguring mucosal metastasis and life-threatening visceral infection. The parasites are transmitted to their mammalian hosts under a promastigote flagellar form, by the bite of an infected sand fly vector. By receptor-mediated events they will infect the host's macrophages and change into a non-motile amastigote form, which will replicate in the acidic phagolysosomal vacuoles. The ability of a parasite to evade the complement system, attach to the macrophages, invade and finally survive in them likely involves a series of factors. The major surface protease (msp) of Leishmania, is thought to be one of them (for a recent review, see Yao, Donelson & Wilson, 2003) and homologous genes have also been described in other digenetic Trypanosomatidae such as Trypanosoma brucei (El-Sayed & Donelson, 1997) and Trypanosoma cruzi (Grandgennet et al. 2000). Noteworthy is that homologues were also reported in monogenetic parasites as Crithidia fasciculata, suggesting a primary role of the glycoprotein for the parasite survival in the insect gut (Etges, 1992; Inverso et al. 1993). However, this role was not supported in digenetic parasites, at least in L. (Leishmania) major (Joshi et al. 2002).
The genes encoding msp are arranged in multicopy tandem arrays in all Leishmania species studied so far (Medina-Acosta, Beverley & Russell, 1993; Steinkraus et al. 1993; Voth et al. 1998). Earlier studies showed that species of subgenus Viannia generally have more msp genes than species of subgenus Leishmania (Medina-Acosta et al. 1993; Steinkraus et al. 1993; Victoir et al. 1998; Voth et al. 1998). In organisms like Trypanosomatids which essentially regulate their gene expression at post-transcription level (i.e. not at initiation, Stiles et al. 1999), gene repetition is an essential feature to generate a high number of transcripts for proteins needed in large amount. It might also allow the parasite to generate sequence diversity (isogenes) within a same gene array (Victoir & Dujardin, 2002). Accordingly, species of subgenus Viannia should have a potential for a higher number of different msp isogenes than subgenus Leishmania. This hypothesis was supported by data from L. (Viannia) guyanensis (Steinkraus et al. 1993).
In the present work, we further explored this hypothesis in L. (Viannia) braziliensis. Indeed, this is the most pathogenic species of subgenus Viannia and previous studies suggested that this species might harbour the highest number of msp genes (Victoir et al. 1998). We performed a fine mapping of the msp gene locus of that species and sequenced most of the genes encountered. Results were placed in an evolutionary context by comparing them with all msp sequences described so far, and potential functional consequences of the described polymorphism were studied. Particular attention was paid to major functional domains, such as interaction motifs, B and T-cell epitopes.
Genomic cosmid clones containing msp genes were isolated from an L. (Viannia) braziliensis library (MHOM/BR/75/M2903, cloned in pWE15, gift from Professor D. C. Barker, University of Cambridge, UK) by (i) hybridization with a L. (Viannia) braziliensis msp probe (pLb134Sp, Dujardin et al. 1993) and (ii) their differential restriction pattern. Isolated clones were purified at least twice, and their genomic stability was checked over time by several complete digestions. In addition, to minimize the possibility that a cosmid clone would contain artefactual re-arrangements, we only considered those in which msp-hybridizing restriction fragments co-migrated with an msp-hybridizing restriction fragment from total genomic DNA of L. (Viannia) braziliensis. The fine mapping of 7 selected clones (G3411, C4121, C1211, C711, D9311, A811 and B9211) was done by the T3/T7 method. In brief, inserts were taken out of the cloning site by a complete Not I digestion. Then, incomplete digestions were performed with BglI, EcoRI and SalI (serial dilutions of restriction enzyme starting from 1·5 u concentration) and stopped with 0·5 M EDTA after incubation for 15 min. In order to have an optimal resolution in different size ranges, the different restriction fragments were resolved electrophoretically on a 0·5% agarose gel DNA (conventional electrophoresis) or 1% agarose in a field inversion gel electrophoresis configuration (60 V, 2·1 sec forward-0·7 sec backward). DNA was transferred after depurination and denaturation to a Nylon membrane (Hybond-N) according to the manufacturer's instructions (Amersham). Blots where hybridized at 42 °C with T3 and T7 oligonucleotides end-labelled by a kinase reaction (Sambrook, Fritsch & Maniatis, 1989). Post-hybridization washings were performed in 6× SSC, 0·1% SDS, the first 2×5 min at room temperature followed by 1 min a 42 °C. The msp genes were localized by (i) complete digestion of the cosmid clones and hybridization with an msp probe, and (ii) comparison of restriction patterns of msp genes PCR-amplified from the corresponding cosmid clones (see below).
Fragments (1·3 kb) resulting from an amplification of the central portion of the msp genes from different cosmid clones (A 8.1.1., C 7.1.1 and C 4.1.2·1.) were generated by 2 specific msp primers respectively situated at positions 410–422 and 1721–1741 in the lg63c1 cDNA msp sequence of L. (Viannia) guyanensis according to Steinkraus et al. 1993. Amplicons were cloned in the PCR™ 2.1 vector using the TA-cloning kit from Invitrogen. Clones were selected according to their restriction pattern with EcoRI, SalI and BglI (also used for cosmid clone mapping) and sequenced by PCR using the ‘ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit’ from Perkin Elmer. The automatic sequencer used was the model 373 of PE Applied Biosystems using the Sanger enzymatic method. Eleven different sequences were obtained (AJ851007–17). The sequence of a cDNA fragment isolated from an msp-specific L. (Viannia) braziliensis library (Victoir, 2001) was also included (AJ863117). Sequence treatment was done with Seaview (Galtier, Gouy & Gautier, 1996), alignment was done by ClustalW (Thompson, Higgins & Gibson, 1994) and phenetic analysis done by using DAMBE 4.0.26 (http://web.hku.hk/~xxia/software/software.htm). The tree-building method used was the neighbor-joining method (Saitou & Nei, 1987). The genetic distance used for nucleotide sequence analysis is the Poisson correction or the Kimura 80. The following sequences were retrieved from Gene Bank and used for phenetical analysis: L. guyanensis c1,7–10 (M85203, L16776, L16777, L16778, L16779), L. panamensis c1, g2, g3 (AF038028, AF037166, AF037167), L. amazonensis (L46798), L. mexicana c1 (X64394), L. donovani S2, S4, PAA (L19563, L19562, M60048), L. major C (AF039721), L. major (Y00647), L. chagasi stat., C, log. (M80669, M80671, M80672), L. infantum Gen (Z83677), Crithidia fasciculata (M94364), T. brucei (U86345) and T. cruzi g1–4 (AF267161, AF267162, AF267163, AF267505).
The crystallographic coordinates of L. (Leishmania) major msp (Schlagenhauf, Etges & Metcalf, 1998; id. Code: 1lml) were retrieved from the Protein Data Bank. Specific residues or regions of the gene were localized on the 3-dimensional structure of the protein using the computer program Cn3D 3.0, a 3-D viewer for NCBI Databases, Version 3.0.
Seven cosmid clones containing msp genes were mapped and 2 were found to be identical (C711 and D9311); overlapping clones were identified by their restriction pattern (Fig. 1). This allowed identification of 2 different contigs, containing a total of 4 msp tandem arrays (numbered 1 to 4) and 1 single gene (number 5). Arrays were separated by regions in which no msp gene could be detected and of variable length: 7·5 kb (arrays 1 and 2), 18 kb (arrays 3 and 4) and 12·5 kb (array 4 and gene 5). The two contigs would contain 21 and 16 genes respectively. A minimum of 8 different msp isogenes were identified based on the presence and order of restriction sites (Fig. 1). Previous analysis of natural populations of Viannia parasites evidenced a L. (Viannia) braziliensis-specific msp isogene, characterized by an internal SalI site (Victoir et al. 1998). This isogene was localized (star, Fig. 1) and was found to be surrounded by highly heterogeneous msp repeat units. The nature and distribution of isogenes was totally different between the 2 contigs, so that it seems unlikely that the 2 contigs constitute different alleles of the same locus. Consequently, this strain of L. (Viannia) braziliensis would contain a minimum of 37 genes per haploid genome.

Fig. 1. Physical map of 6 cosmid clones of Leishmania (Viannia) braziliensis, containing tandemly repeated msp genes; 8 different msp isogenes were identified, based on the presence and order of restriction sites (B, BglI; S, SalI and E, EcoRI): SEBS, BB, SEEBS, BBBB, SES, SS, SBSS and SESS, (represented by different shading patterns). The 2 copies of the L. (Viannia) braziliensis-specific msp isogene (Victoir et al. 1998) are marked with a star. Numbers under the maps correspond to the arrays mentioned in the text.
Msp genes from cosmid clones C 7.1.1, C 4.1.2.1 and A 8.1.1 (containing the most representative and divergent msp isogenes) were sequenced. The obtained sequence lengths varied between 1325 bp and 1334 bp, with differences ranging from 5 to 21% over the whole sequence and to 34% over some stretches. A closer look at the 11 aligned L. (Viannia) braziliensis sequences (named L. braz. cl1, 2, 4, 5, 9–11, 13–15, 17) revealed a mosaic structure as well at the level of short stretches (8–15 bp, not shown) as well as longer ones (100 to 500 bp) (Fig. 2). From one variant to the other, sequence stretches were represented in different shading if their nucleotide sequence difference was greater than 5%. A clear example of such a structure is shown in L. (Viannia) braziliensis sequences L. braz. cl9, L. braz. cl10 and L. braz. cl11. Indeed, from nucleotide 1 to 580, L. braz. cl9 is identical to L. braz. cl11 and both clones differ by 122 nucleotides (21%) from L. braz. cl10. For the last 780 nucleotides, however, L. braz. cl11 differs only by 2 nucleotides from L. braz. cl10 and both clones differ from L. braz. cl9 by 165 (21·2%) and 163 (20·9%) nucleotides, respectively. Another peculiar mosaic structure was observed in a cDNA msp sequence (Victoir, 2001) here included: this 1·3 kb fragment consisted in a 310-bp msp stretch at its 5′ terminal end (corresponding to position 1–310 in genomic sequence of L. (Viannia) braziliensis cl2), followed by a sequence presenting 95% identity with a subtelomeric L. (Viannia) braziliensis sequence (telo10, Fu & Barker, 1998). This sequence was not a reverse transcription artefact because PCR of genomic DNA with primers targeting each of the two segments generated a fragment of the expected size.

Fig. 2. Representation of the mosaic structure of the different msp genes in a Leishmania (Viannia) braziliensis strain (M2903); msp classes 1–3 refer to the 3 major clusters individualized in Fig. 3. Sequence stretches which share a homology between different clones, equal or higher than 95% are represented by the same shading pattern (unrelated to Fig. 1). The corresponding nucleotide positions are given in bp for the different L. (Viannia) braziliensis sequences.
In order to situate the msp gene polymorphism of L. (Viannia) braziliensis in a broader evolutionary context, we performed a phenetic analysis of all known msp sequences of Leishmania and outgroups (C. fasciculata, T. cruzi and T. brucei) over a length corresponding to positions 412–1743 of the L. (Viannia) guyanensis c1 sequence. Leishmania and C. fasciculata clustered together, separately from Trypanosoma species. Leishmania appeared to be monophyletic, and sequences from subgenera Leishmania and Viannia clustered separately (Fig. 3). Within subgenus Viannia, 3 clusters were observed in which L. (Viannia) braziliensis and L. (Viannia) guyanensis sequences were interspersed (here called classes 1 to 3 respectively). The first cluster, containing L. (Viannia) braziliensis sequences of class 1 was the most divergent (32%) from sequences of subgenus Leishmania. Interestingly, all these L. (V.) braziliensis sequences originated from the same region of the msp gene locus, present in clone C.7.1.1.

Fig. 3. Phenetical analysis of msp gene heterogeneity: sequences of Leishmania (Viannia) braziliensis and other species (accessible in GenBank). Numbers near nodes are bootstrap values (1000 pseudo-replicates). Corresponding GenBank locus names of the sequences are listed in the Materials and Methods section. C (upper case), constitutively expressed, S or stat. (expressed in the promastigote stationary phase), log. (expressed in the promastigote logarithmic phase).
Our genomic sequences allowed the prediction of L. (Viannia) braziliensis proteins from amino acid 15 to 556 of the L. (L.) major msp-6 gene (AF039721). By comparison with the latter species, our predicted L. (Viannia) braziliensis sequences presented differences in identity ranging from 6 to 34% (L. (Viannia) braziliensis class 1). The heterogeneity between the predicted proteins increased towards the C-terminal end (not shown), and was essentially localized at the exterior part of the molecule, as inferred from the crystal structure of L. (Leishmania) major (Schlagenhauf et al. 1998). We searched potential implications of this polymorphism at the level of major functional domains of the protein: catalytic site, attachment motifs and reported epitopes.
Analysing the positions of the 16 essential Cys residues assessed the general conformation of the protein. Positions were conserved in all predicted L. (Viannia) braziliensis proteins, except in L. braz. cl1, which showed a sequence truncated from amino acids 7 to 36 (including one of the Cys residues), which might lead to a difference in conformation. The predicted HexxH motif of the catalytic/zinc-binding site was conserved in all sequences, as well as the 62 aa downstream histidine residue, which is important for the formation of the metallo-protease active groove. Potential N-glycosylation sites, important for the protein structure were conserved in all of the L. (Viannia) braziliensis sequences except those of class 1.
Regions reported to play a role in the internalization of the parasites into the macrophages (aa 215–264 and 290–339 of L. (Viannia) guyanensis L16779 sequence, Puentes et al. 1999) showed heterogeneity among the L. (Viannia) braziliensis sequences: from 3 to 19 aa (6 to 38%) in the first region and from 2 to 18 aa (4 to 36%) in the second one. Again, sequences from class 1 were divergent. Within these regions, there are 2 important adhesion motifs (SRYD and EYLEV, Brittingham et al. 1999). With respect to SRYD, all sequences of class 1 and L. braz. cl10 (class 3) showed a 240-Arg/Ala transition (relative amino acid position in L. (Viannia) guyanensis msp sequence, L16779), i.e. from a large and basic amino acid to a small and hydrophobic one. Also in the EYLEV motif, sequences of class 1 were distinct from all other L. (Viannia) braziliensis sequences, as they showed a T/SFLEL motif. Glu and Tyr, the two first amino acids of the EYLEV motif, are prone to interact and especially a change from Glu to Thr or Ser would have considerable consequences because Thr and Ser are much smaller than Glu and lack its negative charge.
A last point concerned heterogeneity in major reported epitopes. With respect to potential T-cell epitopes determined in former studies by Russo et al. (1993), we found in our L. (Viannia) braziliensis sequences a polymorphism ranging from 1/16 (6%) amino acids (peptide 10 in position 116–130 from the L. (Leishmania) major protein) up to 13/16 (81%) (peptide 11 in position 171–187 from the L. (Leishmania) major protein). This result seems especially noteworthy since this last peptide is especially divergent at the beginning and ending positions of the peptide, which are especially important for T-cell recognition (Janeway & Travers, 1996). With respect to B epitopes, immunodominant determinants of L. (Leishmania) infantum msp were shown to be localized in the C-terminal region (Morales et al. 1997). L. (Viannia) braziliensis msp showed a heterogeneity for these peptides ranging from 4/20 (25%) to 10/20 a.a. (50%). Also within the same organism, considerable heterogeneity (50% (11/20)) could be observed between sequences from two different L. (Viannia) braziliensis phenetic msp classes for an identified immunodominant peptide (position 453 to 475, for the predicted msp amino acid sequence of L. (Leishmania) infantum).
In this study, we analysed the genomic organization of msp genes of L. (Viannia) braziliensis, and distinguished a minimum of 37 genes by haploid genome, distributed in 8 isogenes. This might represent a portion only of the msp gene locus, when considering the gene copy number (70) estimated by densitometry in that species (Victoir et al. 1998). In L. (Viannia) guyanensis, another species of subgenus Viannia, 50 genes constituted by 8 isogenes were reported (Steinkraus et al. 1993). The msp gene organization of Viannia species thus contrasts with that reported in subgenus Leishmania and more particularly in L. (Leishmania) major (7 genes, 2 isogenes; Voth et al. 1998), the species chosen for the Leishmania genome project. Gene amplification, a common phenomenon in Leishmania (Iovannisci & Beverley, 1989; Inga et al. 1998; Kebede et al. 1999) is likely responsible for the difference in the structure of the msp gene locus between the two subgenera. We here demonstrate that it is accompanied by an increase in sequence diversity among genes of the same organism.
The msp gene polymorphism here reported might be generated by synonymous and non-synonymous substitutions (Alvarez-Valin, Tort & Bernardi, 2000). However, another contributing mechanism appeared to be intra-genic recombination as suggested by the mosaic structure observed among L. (Viannia) braziliensis msp genes. This phenomenon is generally due to reciprocal crossing over and segmental gene conversion, as reported in Trypanosoma brucei (Pays & Nolan, 1998). Loci more prone to this type of recombination are generally localized in the subtelomeres, regions known to be especially ‘turbulent’ by comparison with the generally stable central core of the chromosomes (Inga et al. 1998; Wickstead, Ersfeld & Gull, 2003). This might also be the case in msp genes of L. (Viannia) braziliensis, as suggested by the presence of a DNA fragment combining an msp stretch with a subtelomeric sequence. Mosaic patterns have been reported for surface antigens of Theileria annulata (major merozoite piroplasm surface antigen, Gubbels et al. 2000) and Plasmodium falciparum (merozoite surface protein 1, Conway et al. 1999). Altogether, the msp gene polymorphism here reported might constitute an adaptive strategy of the parasite (Guerbouj et al. 2001). Analysing functionally important domains in our predicted proteins allowed exploration of this hypothesis.
The conservation of catalytic regions in all L. (Viannia) braziliensis predicted msp and also among other species, contrasted with the polymorphism in regions involved in the host–parasite interaction. First, structurally important changes were noted in major macrophage adhesion motifs of a series of L. (Viannia) braziliensis isogenes. How this might effectively affect the binding capacities of that species cannot be determined from our data. However, it has been reported elsewhere that inhibition of the internalization of L. (Viannia) braziliensis parasites was only partial when blocking well-defined amino acid stretches based on a L. (Viannia) guyanensis msp sequence (Puentes et al. 1999). Secondly, considerable polymorphism was observed for major epitopes, concerning the primary sequence of T epitopes as well as structural features of B-epitopes. Further work is necessary to understand the functional consequences of the diversity here reported. One should address the hypothesis that this diversity is positively selected because it allows evasion of the host immune response, like in other pathogens: African trypanosomes (VSG, Pays & Nolan, 1998), HIV (gp120, Lewis et al. 1998) or Plasmodium (pfempI, Conway et al. 1999). In addition, reminding that leishmaniasis is an immunopathology, the link between polymorphism of msp (or other immunogens) and clinical pleomorphism should be further explored.
Finally, our results also gave an insight on the evolution of msp genes within the genus Leishmania. In terms of genomic structure of the msp gene locus (and particularly the gene copy number) and considering Crithidia fasciculata (7 msp genes, Inverso et al. 1993) as an outgroup, our results indicate that the Leishmania subgenus shows an ancestral character and that the Viannia subgenus diverged dramatically. This was not supported by phylogenies based on msp amino terminal sequences (our study and Medina-Acosta et al. 1993), but was well supported by those based on msp carboxy terminal sequences (Medina-Acosta et al. 1993). Interestingly, phylogenies based on the sequence of another surface membrane glycoprotein, gp46/M-2, also indicated an ancestral character of Leishmania subgenus (Kerr et al. 2000). Within the Viannia subgenus, msp sequences of L. (Viannia) braziliensis and L. (Viannia) guyanensis were interspersed and clustered in 3 major groups. This could be explained by 2 mechanisms. First, diversification of the corresponding 3 major msp gene classes could have preceded speciation. Secondly, horizontal transfer could have occurred. Indeed, sexual recombination between L. (Viannia) braziliensis and L. (Viannia) guyanensis has been observed in natural conditions, albeit rarely (Bañuls et al. 1997). However, in such a case, the 2 studied strains would be the descendants of a hybridization event, which was not supported for the L. (Viannia) braziliensis strain here studied, by the analysis of other genetic markers.
In conclusion, L. (Viannia) braziliensis and L. (Viannia) guyanensis show the most complex organization of msp genes in the whole genus Leishmania. Why such a structure was selected during evolution and what is the function of the 3 msp classes remain unanswered. We showed that polymorphism in the predicted protein might have fundamental implications for host–parasite interactions. However, this should be further explored, on the one hand by following the expression of the different genes along the parasite life-cycle, and on the other hand, by focusing on the interaction of the most polymorphic domains with the host immune system or with the insect vector.
We gratefully acknowledge financial support from the European Commission (Contracts TS3-CT92-0129, IC18-CT96-0123 and IC18-CT98-0256), WHO-TDR (Contract A00476), Belgian Co-operation (Directorate General for Cooperation and Development) and FWO (Nationale Loterij, Grants 346/1990 and 9.0024.90, 1.5.047.02).
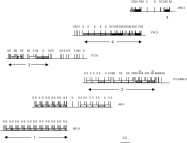
Fig. 1. Physical map of 6 cosmid clones of Leishmania (Viannia) braziliensis, containing tandemly repeated msp genes; 8 different msp isogenes were identified, based on the presence and order of restriction sites (B, BglI; S, SalI and E, EcoRI): SEBS, BB, SEEBS, BBBB, SES, SS, SBSS and SESS, (represented by different shading patterns). The 2 copies of the L. (Viannia) braziliensis-specific msp isogene (Victoir et al. 1998) are marked with a star. Numbers under the maps correspond to the arrays mentioned in the text.

Fig. 2. Representation of the mosaic structure of the different msp genes in a Leishmania (Viannia) braziliensis strain (M2903); msp classes 1–3 refer to the 3 major clusters individualized in Fig. 3. Sequence stretches which share a homology between different clones, equal or higher than 95% are represented by the same shading pattern (unrelated to Fig. 1). The corresponding nucleotide positions are given in bp for the different L. (Viannia) braziliensis sequences.
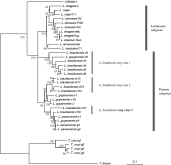
Fig. 3. Phenetical analysis of msp gene heterogeneity: sequences of Leishmania (Viannia) braziliensis and other species (accessible in GenBank). Numbers near nodes are bootstrap values (1000 pseudo-replicates). Corresponding GenBank locus names of the sequences are listed in the Materials and Methods section. C (upper case), constitutively expressed, S or stat. (expressed in the promastigote stationary phase), log. (expressed in the promastigote logarithmic phase).