Introduction
Sweet cherry cultivars grown commercially are either grafted or budded onto the rootstocks from different Prunus species. Different types of rootstocks are used for Prunus species scions on a worldwide basis (Rom, Reference Rom1982), depending on the different attributes each rootstock possesses including adaptation to different geographic regions (Turkoglu et al., Reference Turkoglu, Koc, Ercisli, Bilgener, Akbulut, Yildirim, Gercekcioglu, Esitken and Gunes2012). The choice of the ideal rootstock is very important for the cultivation of sweet cherry. For the identification and characterization of germplasm, DNA-based methods have been favoured by the development of high-resolution melting (HRM) analysis. HRM is an analytical method that uses DNA-intercalating fluorescent dyes in order to measure the rate of the DNA dissociation from a double strand to a single strand (Reed and Wittwer, Reference Reed and Wittwer2004).
Experimental
The main sweet cherry rootstocks used, covering more than 90%, in Greece are: Cab-6P (Prunus cerasus; allotetraploid, AAFF, 2n= 4x= 32); Maxma-14 (P. mahaleb× P. avium; diploid, 2n= 16); Gisela-5 (P. cerasus (var. Schattenmorelle) × P. canescens; triploid, 2n= 3x= 24); Gisela-6 (P. cerasus (var. Schattenmorelle) × P. canescens; triploid, 2n= 3x= 24); Piku-1 (P. avium× (P. canescens× P. tomentosa); triploid, 2n= 3x= 24); PHL-6 (P. avium× P. cerasus; triploid, 2n= 3x= 24). These rootstocks were obtained from the ex situ collection of the Pomology Institute of the Hellenic Agricultural Organisation – ‘DEMETER’ (Chatzicharisis and Kazantzis, Reference Chatzicharisis and Kazantzis2011).
Genomic DNA was isolated with the NucleoSpin Plant II DNA extraction kit (Macherey and Nagel, Germany) following the manufacturer's instructions. For simple sequence repeat (SSR) analysis, polymerase chain reaction (PCR) amplification, DNA melting, HRM and end-point fluorescence level analyses were performed on a Rotor-Gene 6000 real-time 5P HRM PCR Thermocycler (Corbett Research, Sydney, Australia) according to Ganopoulos et al. (Reference Ganopoulos, Argiriou and Tsaftaris2011). An initial set of five polymorphic SSR markers, conserved in different Prunus species, were selected (Table S1, available online). SSR-HRM experiments were repeated thrice. We tested three different samples (ramets) from the same rootstock denomination, in order to examine whether or not variations among individuals would interfere with the HRM analysis in distinguishing the sweet cherry rootstocks (data not shown).
Table 1 Average GCPs (±5.3) resulting from the HRM analysis of the BPPCT002 region of the six sweet cherry rootstocks at a ramp of 0.1°C

Results
We evaluated five different microsatellite markers in order to genotype and distinguish the most important sweet cherry rootstocks using the HRM analysis. Figure 1(a) depicts the normalized HRM melting curves of the six sweet cherry rootstocks using the SSR marker BPPCT002. Using the shape of the melting curves, we could reveal the differences between the rootstocks under investigation and show that all the rootstocks could be easily distinguished visually, for example ‘Maxma-14’ and ‘Piku-1’. The results obtained with the other markers used were similar, showing a clear discrimination of the varieties used (data not shown). The melting curve peaks range from 79.78 ± 0.3 (in Profile 1, Maxma-14) to 82.25 ± 0.2 (in Profile 6, Gisela-6), as shown in Table S2 (available online).
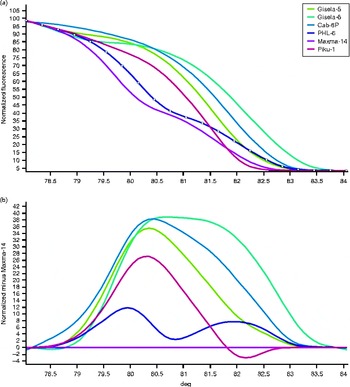
Fig. 1 (colour online) Microsatellite genotyping of the six sweet cherry rootstocks using the HRM analysis with the microsatellite marker BPPCT002. (a) Normalized melting curves and (b) difference graph of the six sweet cherry rootstocks using Maxma-14 as the genotype. The genotypes were assigned using a cut-off confidence value of 95%.
Figure 1(b) illustrates the difference graph of the representative set of the six Prunus rootstocks used when compared with the ‘Maxma-14’ rootstock forming the baseline. A closer examination of the HRM difference curve revealed part of the curve sitting outside the 95% CI curve. This suggests that all the examined rootstocks via the HRM curves are indeed different. As shown in Fig. 1(b), we estimated the confidence value of similarity between ‘Maxma-14’ and the other sweet cherry rootstocks used in the present study and showed that BPPCT002 was a sufficient microsatellite region to distinguish them. Table 1 presents the calculated genotype confidence percentages (GCPs) and a cut-off value of 95% was used to assign a genotype for each microsatellite region. In the case of BPPCT002, the highest GCP was found between the Cab-6P and Gisela-5 rootstocks, while the lowest was between Gisela-6 and a number of the other rootstocks, including Piku-1 and Maxma-14 (Table 1).
Discussion
The six genotypes used in this study presented different profiles among each other, but the same profile in the different ramets from the same rootstock. Therefore, there is no variation detected within the same rootstock denomination in the three different plants tested per rootstock. In contrast, discrimination of the different rootstock genotypes was fully achieved, despite their genetic relationship. The P. avium genome participates in three rootstocks (Maxma-14, Piku-1 and PHL-6), the P. cerasus genome also participates in three rootstocks (Cab-6P, Gisela-5 and Gisela-6) and the P. canescens genome in three rootstocks as well (Gisela-5, Gisela-6 and Piku-1). Nevertheless, their only partially common genetic background did not hinder their successful identification by the HRM approach.
Moreover, differences in ploidy levels among the rootstocks did not complicate the HRM approach, as it is the melting curve of the product that is taken into account by this analysis. In fact, in such cases, the interpretation of the HRM curves is probably simpler than the corresponding complex genetic marker banding patterns in electrophoresis or allele calling in electrophenograms.
Five of the rootstocks used in this study are the most commonly used in Greece, while one (PHL-6) has become very popular in the last few years (Chatzicharisis and Kazantzis, Reference Chatzicharisis and Kazantzis2011). Overall, the six rootstocks cover over 90% of sweet cherry cultivation in Greece, while there is a diminishing presence of old local wild-type rootstocks that were out-competed by the rootstocks reported and have traditionally fallen out of fashion (Chatzicharisis and Kazantzis, Reference Chatzicharisis and Kazantzis2011). While this research focused on the rootstocks that were almost exclusively used in Greece, the above results should be further verified by future analysis of a larger rootstock sample size in terms of species and interspecific hybrids used perhaps at the European scale. In addition, a larger study should include the analysis of more individuals (ramets) within each rootstock denomination in order to verify rootstock genotypic integrity.
To the best of our knowledge, this is the first report on the utilization of the HRM approach for rapid identification and discrimination of sweet cherry rootstocks by employing SSR markers. In this study, microsatellite characterization has provided a unique genetic fingerprint for each Prunus rootstock studied and allowed their discrimination based on their HRM profiles.
In conclusion, we have demonstrated here that a combined approach of using microsatellite markers and HRM is a useful method to identify commercially important sweet cherry rootstocks. Moreover, HRM is highly informative, as it was shown that it was possible to discriminate the main sweet cherry rootstocks by using only one microsatellite marker.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S1479262113000403
Acknowledgements
This research was co-financed by the European Union (European Social Fund) and Hellenic national funds through the Operational Program ‘Education and Lifelong Learning’ of the National Strategic Reference Framework – Research Funding Program: Heracletus-II. The Institute of Applied Biosciences/CERTH from the General Secretariat of Research and Technology of Greece is also gratefully acknowledged for continuous support.




