INTRODUCTION
To look around, the diversity of life would appear to be populated by animals, plants and fungi. Appearances can be deceiving. Bacteria aside, the vast diversity of cells on the planet today is composed of unicellular eukaryotic microbes, or protists. These predominantly single-celled creatures can be at the same time beautiful, bizarre and deadly. Plasmodium falciparum, the causative agent of cerebral malaria which kills close to a million people a year (Snow et al. Reference Snow, Guerra, Noor, Myint and Hay2005), may be the most notorious protistan pathogen. It is hardly the only one. From our top (the brain-eating amoeba Naegleria fowleri) to our tail (gut parasites Giardia and Entamoeba), a myriad array of microbial eukaryotes – may inhabit, parasitise, and ravage the human body. Understanding the biology of these organisms and how they kill is a critical task impacting not only human health but global development, as many diseases caused by protistan pathogens have their biggest impacts in Africa, South America and South-East Asia (see the World Health Organization webpage for the most recent and motivating statistics, http://www.who.int/vaccine_research/diseases/en/).
While the traditional approach to parasitology involves focused experimental work in the individual organisms, a complementary way forward is to take a broader sweep, knitting together genetic, medical and cell biological information across a framework of eukaryotic relationships. This allows for comparisons between distantly related parasites, yielding insight into convergently evolved pathogenic mechanisms (e.g. the regulations of antigenic variation in Plasmodium and Trypanosoma (Duraisingh et al. Reference Duraisingh, Voss, Marty, Duffy, Good, Thompson, Freitas-Junior, Scherf, Crabb and Cowman2005; Horn and Barry, Reference Horn and Barry2005)) or similarities that can be exploited in a single treatment (e.g. the use of metronidazole against anaerobic microbes). It also allows for comparisons between the parasites and their free-living relatives. This can uncover the evolutionary path taken to parasitism and highlight lab-safe model organisms for study (e.g. the free-living soil amoeba Naegleria gruberi instead of its deadly cousin Naegleria fowleri (Fritz-Laylin et al. Reference Fritz-Laylin, Prochnik, Ginger, Dacks, Carpenter, Field, Kuo, Paredez, Chapman, Pham, Shu, Neupane, Cipriano, Mancuso, Tu, Salamov, Lindquist, Shapiro, Lucas, Grigoriev, Cande, Fulton, Rokhsar and Dawson2010)). Finally, comparisons amongst parasitic sister-taxa allow for knowledge translation between experimental models and less studied related organisms. A result in Plasmodium might be applicable to Eimeria: a finding in Trypanosoma brucei may be of relevance in Leishmania aetheopica.
This comparative approach means delving into fields initially appearing tangential to parasitology. Our current understanding of eukaryotic systematics is based both on molecular and microscopic evidence. Consequently molecular phylogenetics and genomics are playing an increasingly important role. Evolutionary cell biology and ancient eukaryotic evolution are two of the high-profile fields dependent on eukaryotic systemics. They, in turn, are crucial for the interpretation of parasitological and cell biological data in this comparative framework.
In this review, we will discuss the latest literature on higher-level protist systematics and provide an updated scheme of the major divisions of eukaryotic diversity. Each of these divisions will be described, highlighting some of the parasitic organisms and the current and upcoming genome projects. Finally, we will explore two of the most pressing controversies in eukaryotic systematics and discuss the anticipated role that next generation sequencing might play in the evolutionary study of parasites and eukaryotes in general.
SHIFTING VIEWS ON EUKARYOTIC SYSTEMATICS: FROM ‘PRIMITIVE’ PROTOZOA TO DIVERSE SUPERGROUPS
The tree of eukaryotes perhaps most familiar to many parasitologists is that of a collection of crown eukaryotes (animals, plants, fungi and many algae) with a ladder-like sequential divergence of prominent parasites such as Giardia, Trichomonas, Encephalitozoon and Trypanosoma from the base. This view of eukaryotic relationships, and the place of parasites in it, was based on analyses of SSU rDNA genes and single protein coding genes performed during the early to mid-1990s (Sogin, Reference Sogin1991; Sogin and Silberman, Reference Sogin and Silberman1998). Such a phylogeny very much supported the Archezoa hypothesis (Cavalier-Smith, 1983, Reference Cavalier-Smith, Schenk and Schwemmler1987), a prevailing paradigm at the time (and still one sometimes held today) that these “amitochondriate” protists represent basal eukaryotic lineages that diverged before the acquisition of the mitochondrial endosymbiont and other key eukaryotic innovations (e.g. introns, Golgi bodies, peroxisomes). If true, such parasites would be considered primitive and could be used as possible routes to investigate ancestral states of cellular systems.
Despite its elegance and logic, the Archezoa hypothesis and the crown/base view of eukaryotic relationships have been rejected based on several lines of evidence. First of all, mitochondrially-derived organelles (i.e. hydrogenosomes and mitosomes) have been found in nearly all of the proposed “amitochondriate” organisms (van der Giezen, Reference Van Der Giezen2009). Initially, this was demonstrated by the identifying of gene sequences of mitochondrial origin (HSP60, HSP70, IscU) and later, the localizing of these gene products (by immuno-microscopy) to double membrane-bound organelles found in these taxa (Clark and Roger, Reference Clark and Roger1995; Bui et al. Reference Bui, Bradley and Johnson1996; Roger et al. Reference Roger, Clark and Doolittle1996, Reference Roger, Svard, Tovar, Clark, Smith, Gillin and Sogin1998; Tovar et al. Reference Tovar, Leon-Avila, Sanchez, Sutak, Tachezy, Van Der Giezen, Hernandez, Muller and Lucocq2003). Secondly, it has been shown that systematic phylogenetic error, such as ‘long branch attraction’, has had a major effect on the positioning of these organisms in the tree (Philippe and Germot, Reference Philippe and Germot2000; Dacks et al. Reference Dacks, Marinets, Doolittle, Cavalier-Smith and Logsdon2002). The less conserved gene sequences were clustered together regardless of whether the divergence was due to rapid evolution (as in many parasitic taxa) or a protracted period of time in which to accumulate independent mutations (as in the prokaryotic sequences used as outgroups for the eukaryotic analyses). This artifact caused otherwise highly divergent protists to be mistaken for basal eukaryotes. When accounted for, by algorithms and models that take different modes of sequence evolution into account (Holder and Lewis, Reference Holder and Lewis2003), the ‘primitive’ parasites and other proposed basal eukaryotes were either clearly linked with relatives elsewhere in the tree (as in the case of Microsporidia and fungi (Keeling and Fast, Reference Keeling and Fast2002)) or were unresolved.
The period following the demise of the Archezoa hypothesis was one of taxonomic agnosticism and caution with respect to interpretation of molecular sequence data and the evolution of eukaryotes. Such data were still of immense value but a growing number of researchers abandoned the search for a single gene that would resolve relationships, great and small, in favour of a strategy whereby different genes were used to test hypotheses about particular taxonomic relationships (e.g. EF2 demonstrating the monophyly of red and green algae (Moreira et al. Reference Moreira, Le Guyader and Phillippe2000) or actin phylogenies uniting the cercozoans and foraminiferans (Keeling, Reference Keeling2001)). These resolved relationships would then contribute to a consensus view of the eukaryotic tree as a whole. It was also a time for a renewed appreciation of ultrastructural data and the realization of the need for this to be interpreted in the light of molecular results (Taylor, Reference Taylor1999).
These ideas crystallized in a seminal paper by a consortium of taxonomic experts in the diverse protist organisms. An interim taxonomy of eukaryotes was thus provided by Adl et al. (Reference Adl, Simpson, Farmer, Andersen, Anderson, Barta, Bowser, Brugerolle, Fensome, Fredericq, James, Karpov, Kugrens, Krug, Lane, Lewis, Lodge, Lynn, Mann, Mccourt, Mendoza, Moestrup, Mozley-Standridge, Nerad, Shearer, Smirnov, Spiegel and Taylor2005) based on morphological, ultrastructural and molecular data and split the tree into six ‘supergroups’: Opisthokonta, Amoebozoa, Excavata, Archaeplastida, Rhizaria and Chromalveolata. By and large, these divisions have held up and form the basis for the supergroups that we will describe below.
Nonetheless, work carried out in the 5 years since the Adl et al. paper necessitates changes in the systematic viewpoint and eventually in the taxonomy. In some cases, the first gene sequences for key taxa have been obtained, shifting their affiliations such as the placement of the amoeboid protist Fonticula with opisthokonts (Brown et al. Reference Brown, Spiegel and Silberman2009), or the placement of various incertae sedis taxa in the tree (e.g. Stephanopogon with heteroloboseans (Yubuki and Leander, Reference Yubuki and Leander2008)).
Moreover, the burgeoning availability of gene sequences from diverse eukaryotes, either through genome sequences or from sequence surveys of expressed genes, have allowed for a new approach to eukaryotic molecular systematics: increased use of multi-gene concatenated phylogenies. Analyses of gene sequences can not only be affected by systematic artifact, but stochastic artifact as well, where there is simply insufficient sequence information in a given matrix to distinguish between the different possible tree topologies. By stringing tens or hundreds of proteins end to end, and treating them as a single data matrix, stochastic error can be reduced. This approach has been very powerful in resolving various issues in eukaryotic systematics, providing tremendous support for some of the eukaryotic supergroups (Bapteste et al. Reference Bapteste, Brinkmann, Lee, Moore, Sensen, Gordon, Durufle, Gaasterland, Lopez, Muller and Philippe2002; Rodriguez-Ezpeleta et al. Reference Rodriguez-Ezpeleta, Brinkmann, Burey, Roure, Burger, Loffelhardt, Bohnert, Philippe and Lang2005; Burki et al. Reference Burki, Inagaki, Brate, Archibald, Keeling, Cavalier-Smith, Sakaguchi, Hashimoto, Horak, Kumar, Klaveness, Jakobsen, Pawlowski and Shalchian-Tabrizi2009; Hampl et al. Reference Hampl, Hug, Leigh, Dacks, Lang, Simpson and Roger2009). It has also allowed a few orphan lineages to find homes, such as centrohelids and telonemids (Burki et al. Reference Burki, Inagaki, Brate, Archibald, Keeling, Cavalier-Smith, Sakaguchi, Hashimoto, Horak, Kumar, Klaveness, Jakobsen, Pawlowski and Shalchian-Tabrizi2009). A complementary approach to focusing on extensive concatenation is increased taxonomic representation with fewer genes in the matrix. This technique has confirmed the validity of many of the supergroups, but also raised doubts as to others including the monophyly of the Chromalveolata (Parfrey et al. Reference Parfrey, Grant, Tekle, Lasek-Nesselquist, Morrison, Sogin, Patterson and Katz2010); the authors are not alone in raising these concerns.
Originally proposed as an assembly of cryptomonads, alveolates, stramenopiles and haptophytes, the Chromalveolata account for the overwhelming majority of recorded algal species (Cavalier-Smith, Reference Cavalier-Smith1999; Simon et al. Reference Simon, Cras, Foulon and Lemee2009). However, new molecular sequence analyses do not support the monophyly of these groups to the exclusion of others. The issue is whether two new, but well-supported groups containing ‘chromalveolate’ taxa – the CCTH clade (Burki et al. Reference Burki, Inagaki, Brate, Archibald, Keeling, Cavalier-Smith, Sakaguchi, Hashimoto, Horak, Kumar, Klaveness, Jakobsen, Pawlowski and Shalchian-Tabrizi2009; Okamoto et al. Reference Okamoto, Chantangsi, Horak, Leander and Keeling2009) and SAR clade (Cavalier-Smith, Reference Cavalier-Smith2010) – should be treated as supergroups. Based on their diversity, we treat them as such, noting that this has no implications for taxonomic rank.
The final advance that has arisen since the Adl et al. (Adl et al. Reference Adl, Simpson, Farmer, Andersen, Anderson, Barta, Bowser, Brugerolle, Fensome, Fredericq, James, Karpov, Kugrens, Krug, Lane, Lewis, Lodge, Lynn, Mann, Mccourt, Mendoza, Moestrup, Mozley-Standridge, Nerad, Shearer, Smirnov, Spiegel and Taylor2005) taxonomy is in the resolution between the supergroups. In two separate concatenated gene phylogenies, resolution was obtained separating the excavates, amoebozoans and opisthokonts from an assemblage of archaeplastids, Rhizaria, stramenopiles, alveolates, and CCTH groupings (Burki et al. Reference Burki, Shalchian-Tabrizi and Pawlowski2008; Hampl et al. Reference Hampl, Hug, Leigh, Dacks, Lang, Simpson and Roger2009). Although this resolution is unrooted, and thus it is unclear if either of these assemblages are true clades, it still provides an important framework upon which to polarize various traits and deduce cellular states of the ancestral eukaryote. It particularly emphasizes that diverse microbial eukaryotes, embedded in the various eukaryotic supergroups (Table 1), have independently adopted the parasitic life-style (Fig. 1).

Fig. 1. Unrooted tree of eukaryotes. This cartoon schematic of eukaryotic diversity shows the classification scheme for the 6 supergroups and their relative relationships described in the body of this paper. The tree is based on the results of numerous large-scale genomic and phylogenetic analyses as well as comparative ultrastructural data described within each text section. For complete references see the supplementary materials for each division. ‘Jolly Roger’ flags beside taxonomic groups denote the presence of parasites of agricultural or human importance within that group.
Table 1. Where are they now? The current eukaryotic affinities of some of the best-known parasites

EUKARYOTIC SUPERGROUPS
With this historical overview in mind, we now present a primer on the diverse and well-supported major eukaryotic divisions (Fig. 1). The text here is abbreviated: full descriptions of every group, and extensive references, are provided in the supplementary material (Supplementary material 1 – see http://journals.cambridge.org/PAR). The defining features of groups are given briefly here; these are synapomorphies only in the cases where they are listed as such, and are less clearly-defined in other cases – a lot of systematic research remains to be done. Defining features are derived from the published studies cited at the end of each group in the supplementary material. We use informal names where possible, and present an indented hierarchy, explicitly so as not to imply a formal taxonomic scheme with ranks. The supplementary section (Supplementary material 1 – see http://journals.cambridge.org/PAR) explains this rationale in detail.
We have placed emphasis at the beginning of each supergroup description on the synapomorphies that define the group, both molecular and ultrastructural where possible. We have inset the sub-divisions in each group, again listing the major synapomorphies and parasitological relevance of example species. Information on currently public genome projects is given: nuclear where possible, gene survey or organelle when this is the best sampling available. However, due to the fast moving state of these data and emerging new projects, no websites for data access are provided. Instead readers are urged to check the Genomes Online Database (http://www.genomesonline.org/) or the NCBI listing of genome projects for the most up to date information.
OPISTHOKONTS (James et al. Reference James, Kauff, Schoch, Matheny, Hofstetter, Cox, Celio, Gueidan, Fraker, Miadlikowska, Lumbsch, Rauhut, Reeb, Arnold, Amtoft, Stajich, Hosaka, Sung, Johnson, O'rourke, Crockett, Binder, Curtis, Slot, Wang, Wilson, Schussler, Longcore, O'donnell, Mozley-Standridge, Porter, Letcher, Powell, Taylor, White, Griffith, Davies, Humber, Morton, Sugiyama, Rossman, Rogers, Pfister, Hewitt, Hansen, Hambleton, Shoemaker, Kohlmeyer, Volkmann-Kohlmeyer, Spotts, Serdani, Crous, Hughes, Matsuura, Langer, Langer, Untereiner, Lucking, Budel, Geiser, Aptroot, Diederich, Schmitt, Schultz, Yahr, Hibbett, Lutzoni, Mclaughlin, Spatafora and Vilgalys2006; Philippe and Telford, Reference Philippe and Telford2006; Hibbett et al. Reference Hibbett, Binder, Bischoff, Blackwell, Cannon, Eriksson, Huhndorf, James, Kirk, Lucking, Thorsten Lumbsch, Lutzoni, Matheny, Mclaughlin, Powell, Redhead, Schoch, Spatafora, Stalpers, Vilgalys, Aime, Aptroot, Bauer, Begerow, Benny, Castlebury, Crous, Dai, Gams, Geiser, Griffith, Gueidan, Hawksworth, Hestmark, Hosaka, Humber, Hyde, Ironside, Koljalg, Kurtzman, Larsson, Lichtwardt, Longcore, Miadlikowska, Miller, Moncalvo, Mozley-Standridge, Oberwinkler, Parmasto, Reeb, Rogers, Roux, Ryvarden, Sampaio, Schussler, Sugiyama, Thorn, Tibell, Untereiner, Walker, Wang, Weir, Weiss, White, Winka, Yao and Zhang2007; Shalchian-Tabrizi et al. Reference Shalchian-Tabrizi, Minge, Espelund, Orr, Ruden, Jakobsen and Cavalier-Smith2008)
This supergroup encompasses animals, fungi and their protistan relatives (Fig. 2). Most flagellated taxa have one posteriorly-inserting, posteriorly-directed flagellum, with a barren second basal body; mitochondrial cristae are flattened. There is a synapomorphic insertion in the EF1-alpha gene. Opisthokonts are divided into two principal lineages: holozoa and holomycetes. Prominent parasitic taxa exist in both divisions with the parasitic nematodes and Microsporidia being only two of the many examples. Genome sequencing efforts in this group are numerous and extensive.

Fig. 2. Opisthokonts. Panels a–b show holozoans, whereas panels c–g show Holomycetes. (a) Metazoan: Malurus cyaneus; (b) Choanoflagellate: Choanoeca sp. ; (c) Nucleariid: Nuclearia sp.; (d) Ascomycete fungus: Saccharomyces cerevisiae; (e) Fungal spore; (f) Fungal hyphae; (g) Basidiomycete fungus; unidentified mushroom species. Scale bar in d: For a, 10 cm; e, 20 μm; f, 10 cm; g, 20 μm. Panel a is used with permission from John Walker. All other images in this and subsequent figures are taken from the Micro*scope website and used under the Creative Commons Licence. http://starcentral.mbl.edu/microscope/portal.php
Holozoa: Animals, choanoflagellates and several related protists. Many lineages have unbranched, non-tapering tentacles; in choanoflagellates and some animal cells these form a collar surrounding the emergent flagellum. There is a conserved gene fusion of ubiquitin and rps30. A close relationship between Metazoa and choanoflagellates is well supported.
Metazoa (Animals): An extremely diverse group of multicellular, usually motile organisms, with extensive cell differentiation; diploid except for eggs and sperm, with meiosis preceding sexual reproduction. Several gene families (e.g. PRD and ANTP homeobox transcription factors) are uniquely associated with animals. Many species are parasitic, and a few are even known to acquire chloroplasts via kleptoplastidy from algae. Genome sequences include basally divergent lineages (e.g. the sponge Amphimedon queenslandica; the placozoan Trichoplax adhaerens; the cnidarian Nematostella viridis) and numerous higher animals (Homo sapiens, Drosophila melanogaster ibid.).
Choanoflagellates: Unicellular and colonial collar-flagellates, with a single anterior flagellum inside a funnel collar supported by actin filaments. Genome sequences: Monosiga brevicollis.
Capsaspora owczarzaki: Amoebae with very long filose pseudopodia. Capsaspora is a symbiont of the parasitic snail Biomphalaria glabrata.
Ichthyosporids: (also Mesomycetozoea). Unicellular parasites of vertebrates (dermocystids) and marine invertebrates (icthyophonids), with a large central vacuole and thick cell walls.
Holomycetes: A diverse clade, consisting of fungi, and two basally divergent protist lineages (nucleariids and fonticulids). Pseudopodia, where present, are tapering and may be branched.
Nucleariids: Aflagellated amoebae with radiating fine pseudopodia, which consume whole prey cells. Genome sequences: Nuclearia simplex (mitochondrial only).
Fonticulids: Coprophilic cellular slime moulds. Mitochondrial cristae are discoidal.
Fungi: Mycelial and unicellular opisthokonts, with chitinous cell walls. All studied lineages (except for Microsporidia) synthesize lysine via the α-aminoadipate pathway, and uniquely amongst eukaryotes contain genes encoding non-ribosomal peptide synthetases. The phagotrophic microsporidia and rozellids diverge basally from osmotrophic taxa.
Ascomycetes: Fungi with a mycelial habit and no flagellated stages. Karyogamy, meiosis and membrane division occurs in the ascus, a sac-like cell; ascospores develop in the cell. There is a dikaryotic (functionally diploid) mycelium stage in the life cycle. Ascomycetes include symbionts of lichens, insects and plants, and pathogens or parasites of plants (e.g. Magnaporthe grisea) and animals (e.g. Pneumocystis carinii, Candida albicans) Genome sequences include Saccharomyces cerevisiae, Schizosaccharomyces pombe, Neurospora crassa, Magnaporthe grisea, Pneumocystis carinii and Candida albicans.
Basidiomycetes: Club fungi, mostly with a dikaryotic, mycelial habit and no flagellated stages. Karyogamy and meiosis occur in the basidium, a cell produced from the mycelium; basidiospores are released and develop exogenously into hyphae. Basidiomycetes include several symbiotic and parasitic taxa, (e.g. the crop pathogen Ustilago maydis, and the human pathogen Cryptococcus neoformans). Genome sequences include saprotrophic (Phanerochaete chrysosporium), ectomycorrhizal (Laccaria bicolor) and parasitic species (Ustilago maydis, Cryptococcus neoformans).
Zygomycetes: A paraphyletic array of fungi with long, haploid multinucleate mycelia, and no flagellated stages. Sexual reproduction occurs via the fusion of gametangia on the hyphae. Zygomycetes include parasites of arthropods and of other fungi, and several are potentially lethal human pathogens (e.g. Rhizopus oryzae, Mortierella verticillata). Genome sequences: Rhizopus oryzae; Mortierella verticillata and Smittium culisetae (both mitochondrial only).
Nephridiophagids: Parasites of the malpighian tubules of insects, with sporous, uninucleate amoeboid and multinucleate plasmodial forms.
Glomeromycetes: Mycorrhizal symbionts of plants, with an asexual life cycle. Spores germinate outside the host; on contact with the host, mycelia differentiate into complex tree-like arbuscules with reduced cell walls. Genome sequences: Glomus intraradices (mitochondrial only).
Chytridiomycetes: Coenocytic fungi that form unwalled flagellated zoospores. Hydrogenosomes are present in some species. Chytridiomycetes include parasites of diatoms and amphibians (e.g. Batrachochytrium dendrobatidis). Genome sequences: Batrachochytrium dendrobatis; Monoblepharella, Harpochytrium, Hyaloraphidium, Spizellomyces and Rhizophydium (all mitochondrial only).
Blastocladiomycetes: Filamentous fungi that form uniflagellate zoospores. The nucleus is covered with a distinctive cap of ribosomes. Species may be saprotrophs or parasites of plants, green algae, invertebrates and other fungi. Genome sequences: Blastocladiella emersonii and Allomyces macrogynus (both mitochondrial only).
Microsporidia: Amitochondriate, aflagellated, intracellular parasites of ciliates and animals (e.g. the bee parasite Nosema apis; the human pathogen Encephalitozoon cuniculi). Non-canonical Golgi bodies present. Nuclear genomes are the most compact of all studied eukaryotes. Genome sequences: Encephalitozoon cuniculi and Encephalitozoon intestinalis.
Rozellids: Parasites of chytrids, blastocladiomycetes, oomycetes and coleochaete algae, with uniflagellate and wall-less amoeboid stages. Encysted and flagellated cells contain conspicuous polyphosphate granules.
Opisthokonts incertae sedis
Corallochytrium: Aflagellated marine saprotrophs, which reproduce via multiple rounds of binary fission, and release daughter cells through a pore in the cell wall.
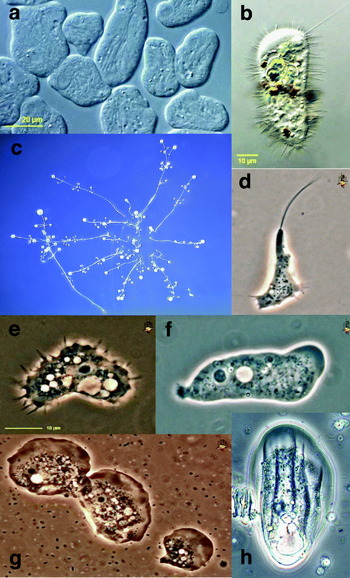
AMOEBOZOA (Page, Reference Page1987; Shadwick et al. Reference Shadwick, Spiegel, Shadwick, Brown and Silberman2009)
This supergroup is composed predominantly of amoebae and amoeboid flagellates (Fig. 3). Some species have flagella and/ or subpseudopodia, and many have branching, irregular mitochondrial cristae. The amoebozoa include a number of parasitic taxa (e.g. Entamoeba histolytica, Acanthamoeba castellanii). The supergroup was originally most clearly identified from multigene phylogenies, with details of membership emerging from more taxon-rich SSU rRNA and actin phylogenies; unrooted phylogenies frequently support a close relationship between amoebozoans and opisthokonts. Some traditional hypotheses of relationships within amoebozoans (e.g. Lobosea, Mycetozoa, Conosa, Centramoebae) currently have no support as monophyletic clades, and internal relationships are incompletely defined. For ease of retrieval, taxa are organized below into amoebae, slime moulds and flagellated amoebae: this is an artificial distinction.
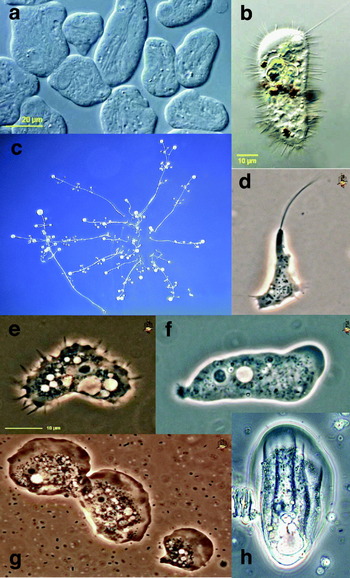
Fig. 3. Amoebozoa. (a) Archamoeba: Entamoeba histolytica; (b) Archamoeba: Mastigamoeba setosa; (c) Myxogastrid slime mould: Polysphondylium spp.; (d) Myxogastrid slime mould: Didymium dachnaya; (e) Amoeba Acanthamoeba castellanii; (f) Tubulinid amoeba: Saccamoeba spp.; (g) Flabellinid amoeba: Vanella spp.; (h) Thecamoebid amoeba: Thecamoeba spp. Scale bar in a: For c, 200 μm; d–h, 10 μm.
‘Amoebae’
Tubulinea: A group of amoebae with diverse morphologies, recovered by molecular phylogeny, including Tubulinids, Arcellinids and Leptomyxids. Pseudopodia are frequently tubular and mono-directional cytoplasmic flow has been observed in a wide range of species. The hydra parasite Hydramoeba has been proposed to be a tubulinid.
Flabellinida/Discosea: A group of flattened amoebae, including Vannellids and Dactylopodids, that have cytoplasm with an anterior hyaline (glassy) zone, polydirectional cytoplasmic flow and radiating pseudopodia. Lineages within the dactylopodid genus Neoparamoeba are believed to contain perkinsid endoparasites and are themselves associated with amoebic gill disease in farmed salmon.
Acanthamoebae: Amoebae with clear, eruptive pseudopodia at front end of the cell, and numerous slender, tapering subpseudopodia (acanthopodia) giving the cell a spiny appearance. Widely distributed and ecologically dominant in freshwater and soil habitats, acanthamebae contain the human parasite Acanthamoeba castellanii, which can cause amoebic keratitis, uveitis and encephalitis. Genome sequences: Acanthamoeba castellanii.
Cochliopodids: Amoebae with a flexible dorsal layer of trumpet-shaped carbohydrate scales, and a distinctive electron-dense body near the Golgi apparatus.
Thecamoebae: A debated clade of amoebae with a thin pellicle and a thick cell coat. Many forms are predators of other amoebae, and Sappinia may be a causative agent of encephalitis.
‘Slime moulds’
Dictyostelids: Cellular, haploid, aflagellated filopodial slime moulds. When starved, these amoebae can aggregate in reponse to molecular signals generated by other individuals, forming a differentiated slug, or that fuse to form a zygote, which ingests aggregating haploid amoebae. Genome sequences: Dictyostelium discoideum.
Myxogastrids: Acellular slime moulds with haploid flagellate and filose amoeboid stages, which fuse to form a diploid plasmodium composed of veins. The mitochondrion contains a filamentous nucleoid and branching tubular cristae. Genome sequences: Physarum polycephalum.
‘Protostelids’: A collection of groups including protosteliids sensu stricto, soliformoviids, protosporangiids, cavosteliids, ceratiomyxids and schizoplasmodiids; principally acellular, with a fruiting body containing one to four spores, with a cellulose-containing stalk and a basal disc.
‘Flagellated amoebae’
Archamoebae: Microareophilic or anaerobic protists with an unusually clear cytoplasm, helical arrays of ribosomes, and small, non-respiratory mitochondria-like organelles.
Entamoebae: Amoebae with clear eruptive anterior pseudopodia and mitosome-like organelles (degenerate mitochondria). Stacked dictyosomes are absent. Most are intestinal parasites of vertebrates, including Entamoeba histolytica, the causative agent of amoebic dysentery in humans; one is a gingival parasite. Genome sequences: Entamoeba histolytica.
Pelobionts: Amoeboid flagellates containing a single basal body connected to a cone and ribbon of microtubules; with a distinctive languid or slow flagellar beat. One species is an endosymbiont in amphibians. Stacked dictyosomes are absent. Mitochondria-like organelles with metabolism intermediate between hydrogenosomes and mitosomes are present.
EXCAVATES (Simpson, Reference Simpson2003; Hampl et al. Reference Hampl, Hug, Leigh, Dacks, Lang, Simpson and Roger2009; Parfrey et al. Reference Parfrey, Grant, Tekle, Lasek-Nesselquist, Morrison, Sogin, Patterson and Katz2010)
The excavates are an assemblage of predominantly heterotrophic flagellates, many of which live in oxygen-poor environments, and may contain non-aerobic alternatives to mitochondria (Fig. 4). Most excavate lineages contain a distinctive longitudinal feeding groove where suspended food particles are collected from a current generated by the beating of posteriorly directed flagella. Two major divisions are currently recognized: the principally amitochondriate metamonads and the predominantly mitochondriate discoba (less formally discobans or occasionally ‘discoballs’). Excavates include a number of major human parasites (e.g. Trypanosoma brucei, Trichomonas vaginalis, Giardia intestinalis). Genome sequences are available for the above-mentioned species, as well as for several Leishmania species and for the free-living Naegleria gruberi.

Fig. 4. Excavates. Panels a, e–i show discobans, whereas Panels b–d show metamonads. (a) Jakobid: Reclinomonas americana; (b) Diplomonad: Giardia intestinalis; (c) Carpediemonas marsupialis; (d) Oxymonad: Pyrsonympha sp.; (e) Heterolobosean: Percolomonas cosmopolitus (flagellated form); (f) Percolomonas cosmopolitus (amoeboid form); (g) Kinetoplastid: Bodo designis; (h) Euglenid: Euglena mutabilis; (i) Kinetoplastid trypanosome: Trypanoplasma sp. Scale bar in a: For a–c, e–g, 10 μm; i, 5 μm.
Metamonads
Parabasalids: Amitochondriate flagellates containing a striated root with attached Golgi dictyosomes that extends posteriorly from the flagellar apparatus. The feeding groove is secondarily absent. Parabasalids contain hydrogenosomes, degenerate mitochondrially-derived organelles. Many species are gut commensals of insects (e.g. Mixotricha paradoxa, hypermastigids) or parasites of vertebrates (trichomonads e.g. Trichomonas vaginalis). Genome sequences: Trichomonas vaginalis.
Carpediemonads: Four flagellate genera – Carpediemonas, Kipferlia, Dysnectes and Hicanonectes – identified from low-oxygen sediments, which resolve paraphyletically at the base of the fornicates. Nomenclature, rank and divisions are not agreed. Cells frequently contain acristate organelles, resembling the hydrogenosomes of parabasalids.
Diplomonads+Enteromonads: Small amitochondriate excavate flagellates, many of which have a doubled cell structure, containing two nuclei, each attached to a flagellar apparatus supporting a feeding groove. There is a mitosome organelle homologous to mitochondria and parabasalid hydrogenosomes. Stacked dictyosomes are absent, although Golgi homologues have been characterized. Diplomonads contain several parasites of humans (e.g. Giardia lamblia, a causative agent of water-borne enteric disease), and fish (Spironucleus salmoncidus). Genome sequences: Giardia lamblia.
Retortamonads: Amitochondriate excavates with four flagella arising from four basal bodies at the anterior end of the feeding groove. Stacked dictyosomes are absent. The overwhelming majority of studied species are parasitic; Chilomastix is a gut commensal of humans. The retortamonads, diplomonads + enteromonads and carpediemonads together form the taxonomic grouping Fornicata.
Oxymonads: Amitochondriate flagellates that lack identifiable Golgi bodies, peroxisomes or feeding grooves, and contain a distinctive axostyle made of multiple parallel sheets of microtubules. Oxymonads are found primarily as symbionts of termites or wood-eating cockroaches and may utilise non-canonical genetic codes.
Trimastix: Free-living amitochondriate excavates; four flagella insert orthogonally at the anterior end of the feeding groove. The oxymonads and Trimastix together form the taxonomic grouping Preaxostyla.
Discoba
Heterolobosea: Free-living, heterotrophic excavates, which contain eruptive pseudopodia and lack Golgi stacks. Some genera (e.g. Psalteriomonas) contain hydrogenosomes, and some mitochondriate species (e.g. Naegleria gruberi) may be facultatively anaerobic. Naegleria fowleri is an opportunistic pathogen that can infect the human central nervous system. Genome sequences: Naegleria gruberi.
Jakobids: Mitochondriate, heterotrophic excavates, with a sole vane on the dorsal surface of the posterior flagellum. The mitochondria of some lineages (e.g. Reclinomonas americana) retain highly unreduced genomes and shared genetic features with bacteria. Genome sequences: Reclinomonas americana (mitochondrial only).
Euglenozoa: A diverse group of flagellates, including euglenids, kinetoplastids (trypanosomes and bodonids), diplonemids and symbiontids, unified by the presence of a feeding apparatus (cytostome) that may be highly complex and by two heterodynamic flagella that contain paraxial rods or lattices. The mitochondrial genomes may be arranged in minicircles (kinetoplasts in kinetoplastids) or small circular chromosomes (diplonemids and euglenids). Predominantly heterotrophic, one lineage of euglenids (including Euglena gracilis) contains secondary, green algal-derived chloroplasts. Diplonemids include the facultative crustacean parasite Rhynchopus; kinetoplastids include parasites and endosymbionts of amoebozoa, fish and mammals, most notably the medically important trypanosomes Trypanosoma brucei (causative agent of African Sleeping Sickness), Trypanosoma cruzi (Chagas’ disease) and Leishmania mexicana (kala azar). Genome sequences: multiple Trypanosoma species including Trypanosoma brucei, Trypanosoma cruzi, multiple Leishmania species including Leishmania major, Leishmania infantum and Leishmania brazilensis (trypanosomes); Bodo saltans (bodonid, in preparation); Euglena gracilis and Euglena longa (euglenids, chloroplasts only).
Excavates incertae sedis
Malawimonads: A single genus, Malawimonas, of free-living mitochondriate excavates, containing an anterior flagellum that inserts apically and a posterior flagellum that inserts at the head of the feeding groove. The mitochondrial genome contains bacteria-like features. Phylogenetic analyses recover affinities to both metamonads and discoba. Genome sequences: Malawimonas jakobiformis (mitochondrial only).
ARCHAEPLASTIDS (Saunders and Hommersand, Reference Saunders and Hommersand2004; Rodriguez-Ezpeleta et al. Reference Rodriguez-Ezpeleta, Brinkmann, Burey, Roure, Burger, Loffelhardt, Bohnert, Philippe and Lang2005; Becker and Marin, Reference Becker and Marin2009)
Three phyla, Viridiplantae, Rhodoplastida and Glaucophyta (Fig. 5), that are unified by the presence of primary plastids believed to have arisen from a single endosymbiotic event with a cyanobacterium. This supergroup is also referred to as Plantae. Archaeplastid monophyly is supported by nuclear multigene phylogenies and discrete features recovered in chloroplastids and rhodoplastids (e.g. a cytosolic FBA duplication and the type I transcription factor pBRp); the branching relationships between the three constituent phyla are not fully resolved. The archaeplastids include numerous parasitic taxa, both multicellular parasitic plants and unicellular algae. Genome sequences have been produced for several agriculturally important plants (e.g. rice, grape, soya) as well as green algae. The recently published genome of Volvox allowed investigation of another independent example of multicellularity. Only a single red algal genome is available but several are in preparation.

Fig. 5. Archaeplastids. Panel a shows a glaucophyte; Panels b–c show red algae; Panels d–o show green plants and algae. (a) Glaucophyte: Cyanophora paradoxa; (b) Rhodophyte: Porphyra yezoensis; (c) Rhodophyte: Porphyridium sp.; (d) Chlorophyte model organism: Chlamydomonas reinhardtii; (e) Chlorophyte: Eudorina sp.; (f) Chlorophyte: Volvox carteri; (g–i) Ulvophyte: Wittrockiella sp.; (j) ‘Prasinophyte’: Pyramimonas sp. (k) ‘Prasinophyte: Nephroselmis olivacea; (l) Zygnemophyte desmid: Micrasterias sp.; (m) Streptophyte: Mesostigma viride; (n) Zygnemophyte desmid: Closterium sp.; (o) Embryophyte tree: Acacia melanoxylon. Scale bar in a: for a, c, d, i–n, 5 μm; b, 5 cm; e, 20 μm; f–h, 100 μm; o, 1 m.
Viridiplantae: Green algae and plants, also referred to as chloroplastids, with primary chloroplasts that contain thylakoids arranged in stacks, and DNA arranged in numerous small nucleoids. Uniquely, starch is deposited principally in the chloroplast stroma; this has been linked to the conserved duplications of genes involved in starch biosynthesis. Viridiplantae are divided into chlorophytes and streptophytes; the terms prasinophytes and charophytes refer to paraphyletic assemblies within each clade. Four secondary endosymbioses of chlorophytes by other eukaryotes are known: in the euglenids, the chlorarachniophytes, the dinoflagellate genus Lepidodinium and the katablepharid Hatena arenicola.
Chlorophytes
Chlorophyceae: Haplobiontic chlorophytes, with a transition region in the flagellum consisting of a short proximal, and a long distal, stellate structure, the latter containing a thick transverse plate structure. Genome sequences: Chlamydomonas reinhardtii and Volvox carteri; Botryococcus braunii and Dunaliella salina (in preparation).
Ulvophytes: Predominantly diplobiontic filamentous chlorophytes, containing an extremely long transitional region. Species have been identified as epibionts on trees, red algae and sloth fur; trentepohliales are photosymbionts of orange lichen. Genome sequences: Pseudendoclonium akinetum (mitochondria and chloroplasts) and Oltmannsiellopsis viridis (mitochondrial only).
Trebouxiophytes: Coccoid and filamentous chlorophytes lacking a defined synapomorphy. Species have been identified as epibionts and endobionts of plants, dinoflagellates, marine invertebrates, and lichens. Two non-photosynthetic genera, Helicosporidium and Coccomyxa, are parasites of marine invertebrates; Prototheca is the causative agent of protothecosis in humans. Genome sequences: Chlorella; Helicosporidium and Prototheca (mitochondria and chloroplast).
‘Prasinophytes’
Chlorodendrales: Flagellated prasinophytes, covered by an outer layer of stellate and inner layer of diamond-shaped, scales. The group includes Tetraselmis convolutae, an acoel endosymbiont.
Pycnococcales: Flagellated and coccoid prasinophyes, covered by an outer layer of rod-shaped or stellate and inner layer of square or pentagonal, scales. A member of the pycnococcales is a katablepharid endosymbiont. Genome sequences: Nephroselmis olivacea (chloroplast only).
Mamiellophytes: Very small flagellated or coccoid prasinophytes that may be covered with a spider web of flattened scales. Genome sequences: Ostreococcus tauri, Micromonas RCC299.
Pyramimonadales: Swimming scaly prasinophytes with four, eight or sixteen flagella arising from an inversely pyramidal apical pit. Genome sequences: Pyramimonas (chloroplast only).
Prasinococcids: Naked coccoid prasinophytes containing prasinoxanthin; mitochondrial membranes intrude into the pyrenoid. A sister-group to all other chlorophytes.
Streptophytes
Mesostigma viride: Asymmetrical unicellular biflagellated and filamentous streptophytes, covered with distinctive maple-leaf shaped scales.
Chlorokybus atmophyticus: Two- to four-celled sarcinoid packets surrounded by a thick layer of mucilage, lacking plasmodesmata, that divide by the formation of a thin septum. With Mesostigma viride, forms the sister-group to all other streptophytes. Genome sequences: chloroplast only.
Klebsormidiophytes: Charophytes forming unbranched filaments without holdfasts or plasmodesmata; zoospores are released through a pore in the cell wall.
Zygnemophytes: Unicellular, colonial and unbranched filamentous charophytes, lacking plasmodesmata, with a cell wall composed of crystalline cellulose microfibrils. Genome sequences: Spirogyra pratensis (EST only).
Coleochaetales: Branched filamentous charophytes, which bear sheathed hairs; zoospores have unique pyramidal, diamond-shaped scales on the flagellum and body. Some species may be epiphytes of charalean algae. Genome sequences: Coleochaete orbicularis (EST only); Chaetosphaeridium globosum (mitochondria and chloroplasts).
Charales: Charophytes with extremely complex body structures; thalli contain a central axis of multinucleate, internodal cells and whorls of branches radiating from uninucleate, node cells. Genome sequences: Chara vulgaris (chloroplast only).
Embryophytes: Land plants: vascular plants, mosses, hornworts and liverworts. Cell walls contain the complex hemicellulose rhamnogalacturonan II. Over 200 angiosperm genera (e.g. Viscum – mistletoe; Cuscuta – dodder; Rafflesia), are known to be parasites of other plants. Genome sequences: many, ranging from model angiosperms (e.g. Arabidopsis thaliana) to important crop species (rice, soya), representative gymnosperms and bryophytes (Pinus taeda, Physcomitrella patens), as well as non-photosynthetic parasites (Epifagus virginiana; chloroplast only).
Rhodoplastids: Red algae sensu lato, also referred to as Rhodoplantae. Unicellular and multicellular archaeplastids which lack flagella and centrioles at all life history stages; chloroplasts contain DNA molecules arranged in multiple small blebs; thylakoids are non-aggregated and embedded with phycobilisomes. Rhodoplastids utilise a form ID rubisco obtained from a proteobacterial donor. Two principle monophyletic divisions are known: cyanidiophytes and rhodophytes.
Cyanidiophytes: Unicellular red algae tolerant of extreme environments, with thick proteinaceous cell walls; carbohydrates are principally stored as glycogen. Genomes are highly reduced and are depleted of transpososons, introns and several otherwise broadly conserved eukaryotic gene families. Genome sequences: Cyanidioschizon merolae; Galdieria sulphuraria (EST only).
Rhodophytes
Rhodellophytes: Unicellular rhodophytes with a single, highly lobed plastid, surrounded by lipid droplets; storage carbohydrates are predominantly semi-amylopectins with some amyloses.
Porphyridiophytes: Unicellular rhodophytes with a single branched or stellate chloroplast lacking an encircling thylakoid in the plastid; storage carbohydrates are principally composed of semi-amylopectins, with some amyloses.
Stylonematophytes: Unicellular, pseudofilamentous or filamentous rhodophytes; cytoplasmic storage carbohydrates are absent.
Compsopogonophytes: Rhodophytes with a biphasic life history (gametophytes and sporophytes), and a central, thylakoid-free region in each chloroplast. One order, the Erythropeltidales, are principally found as epibionts of marine macroalgae.
Bangiophytes: Rhodophytes with a biphasic life history, which uniquely produce carposporangia and spermatangia in distinct packets by successive divisions. Genome sequences: Porphyra umbilicalis (in preparation); Porphyra purpurea (EST only).
Florideophytes: Branched filamentous rhodophytes with a triphasic life history (gametophytes, carposporophytes, tetrasporophytes), and a distinctive reproductive apparatus, consisting of terminal or lateral carpogonia bearing a long extension for the attachment of spermatangia. Several genera (e.g. Asterocolax, Harveyella, Holmsella) are parasites of other closely related Florideophytes.
Glaucophytes: Small eukaryotes with a plasma membrane subtended by sacs or shields. The blue-green chloroplasts are putatively more primitive than other primary plastid lineages, retaining carboxysomes (protein-encased bacterial pyrenoids) and a peptidoglycan cell wall. No parasitic taxa are known. Three groups are recognised: Cyanophorales, Glaucocystales and Gloeochaetales. Genome sequences: Cyanophora paradoxa (cyanophorale, chloroplast only).
THE SAR CLADE (Leander and Keeling, Reference Leander and Keeling2003; Andersen, Reference Andersen2004; Burki et al. Reference Burki, Shalchian-Tabrizi, Minge, Skjaeveland, Nikolaev, Jakobsen and Pawlowski2007; Bass et al. Reference Bass, Chao, Nikolaev, Yabuki, Ishida, Berney, Pakzad, Wylezich and Cavalier-Smith2009)
The SAR clade (also referred to as Harosa) is an assembly of the stramenopiles, alveolates and Rhizaria, each of which contain photosynthetic, mixotrophic and heterotrophic members (Fig. 6). Nuclear multigene phylogenies robustly support SAR clade monophyly, and suggest that the rhizarians basally diverge from stramenopiles and alveolates. Synapomorphies are limited, although a novel duplication of the GTPase Rab1 has recently been identified in all three phyla. The SAR clade contains a number of pathogenic and parasitic genera of major anthropic interest, including Plasmodium (causative agents of malaria) and Phytophthora (crop pathogens). Genome sequencing in this group has focused on parasitic taxa such as the apicomplexans and oomycetes and ecologically prominent ones such as the diatoms. The first sequence of a rhizarian (Bigellowiella natans) is in progress.

Fig. 6. SAR. Panels a–h shows stramenopiles, i–o show alveolates, and p–x show Rhizaria. (a) Stramenochrome, chrysophyte: Dinobryon sp.; (b) Stramenochrome, pedinellid: Ciliophrys sp.; (c) Stramenochrome, synurid: Mallomonas sp.; (d) Bicosoecid: Cafeteria roenbergensis; (e) Slabyrinthulid: Labyrinthula sp.; (f) Sloomycete – unidentified oomycete; (g) Stramenochrome, diatom: Pleurasigma sp.; (h) Stramenochrome, phaeophyte: Laminaria digitata; (i) Ciliate: Aspidisca sp.; (j) Ciliate: Chilodonella sp.; (k) Ciliate: Dileptus sp.; (l) Apicomplexan: Plasmodium vivax; (m) Apicomplexan: Colpodella vorax; (n) Dinoflagellate: Peridinium sp.; (o) Dinoflagellate: Oxyrrhis marina; (p) Filosan – unidentified cercomonad; (q) Endomyxan: Vampyrella sp.; (r) Filosan, thecofilosan: Protaspis tegere; (s) Filosan: Chlorarachnion reptans; (t) Filosan, desmothoracid: Clathrulina elegans; (u) Filosan: Euglypha sp.; (v) Filosan – Ebria sp.; (w) Endomyxan – unidentified foraminiferan; (x) Radiozoan – unidentified acantharean. Scale bar in b: for a–c, e, f, g, i, j, q–v, 10 μm; for d, l, m, p, 5 μm; h, 10 cm; w, x, 100 μm.
Stramenopiles: A diverse clade of photosynthetic and non-photosynthetic unicellular and multicellular organisms. Flagella, where present, are of uneven length, and the long flagellum carries tripartite tubular hairs. Several stramenopile taxa are parasites and pathogens of Metazoa (e.g. Aureococcus, Blastocystis) and plants (Phytophthora). Six major lineages are known, currently divided into three moderately-supported groups: labyrinthulomycetes; bicosoecids, placidids and slopalinids; and sloomycetes and stramenochromes.
Bicosoecids: Heterotrophic biflagellates with an ingestion area supported by an L-shaped microtubular loop. Predominantly free-living, although some taxa have been identified as chrysophyte epibionts.
Labyrinthulids: Saprotrophic and heterotrophic stramenopiles with a characteristic secretory organelle (sagenogenetosome) that produces an ectoplasmic network involved in adhesion and feeding. Marine, freshwater and terrestrial free-living and epibiotic species are known; a parasitic relationship has been observed between the species Thraustochytrium caudivorum and its flatworm host, and the soil-borne Labyrinthula terrestris has been implicated in late blight of turf grass.
Placidids: Heterotrophic gliding flagellates, with two unequal flagella containing a double helix in the transitional region, and a distinctive u-shaped microtubular root.
Sloomycetes: Oomycetes sensu lato (hypochytrids, oomycetes, Developayella): rhizoidal stramenopiles, with a flagellated zoospore stage, cell walls generally made of cellulose, and glycogen and mycolaminarin as storage products. A number of genera (e.g. Phytophthora, Aphanomyces, Sclerophthora) are major biotrophic pathogens of higher plants. Genome sequences: Phytophthora infestans (the causative agent of potato blight), Phytophthora ramorum (sudden oak death) and Phytophthora sojae (soybean pathogen).
Slopalinids: Opalinids sensu lato (opalinids, proteromonads, Blastocystis): flagellated stramenopiles, with a ridged cell surface supported by microtubular ribbons and a crestal amorphous fibre, and with characteristic struts extending from the flagellar basal body to the cell surface. Some species are intestinal commensals of cold-blooded vertebrates, and Blastocystis has been suggested to be an opportunistic parasite associated with HIV infection. Genome: Blastocystis.
Stramenochromes: Brown algae sensu lato: a diverse array of flagellated (e.g. chrysophytes), coccoid (diatoms), amoeboid (dictyochophytes) and multicellular (phaeophytes) phototrophs, and one entirely non-photosynthetic lineage (actinophryids). Chloroplasts, where present, contain a distinctive girdle lamella and are surrounded by three or four membranes, the outermost of which is contiguous with the ER. Stramenochromes utilise aureochromes, a unique class of blue light receptor. Blooms of some marine pelagophytes form harmful brown tides, some chrysophytes and xanthophytes are epibiotic or soil-borne symbionts of plants, and at least two diatom lineages have been uptaken as tertiary chloroplastic endosymbionts by dinoflagellates. Genome sequences: Aureococcus anophageferrens (pelagophyte); Thalassiosira pseudonana, Phaeodactylum tricornutum and Fragilariopsis cylindrus (all diatoms); Ectocarpus siliculosus (phaeophyte); Kryptoperidinium foliaceum and Durinskia baltica (diatom-derived dinoflagellate endosymbionts; chloroplast); Ochromonas danica (chrysophyte, EST only).
Alveolates: Predatory, phototrophic or parasitic organisms, containing a contiguous layer of cortical alveoli under the cell membrane and an unique family of associated proteins, alveolins. Includes seven lineages that are currently divided into three well-supported groups: the ciliates, which are basal to all other alveolates; apicomplexans, colpodellids and chromerids; and dinoflagellates, perkinsids and ellobiopsids. Many parasitic taxa are known, most notably within the Apicomplexa.
Ciliates: Heterotrophic aerobic and anaerobic alveolates, with cilia arranged in lines over the surface, and a complex cell cortex. Each cell contains several small germline nuclei, of which one differentiates to form a large somatic macronucleus. Free-living in marine, freshwater, soil and epiphytic environments; symbiotic species are known, e.g. entodiniomorphids (intestinal commensalists/ parasites of mammals) and Ichthyophthirius multifiliis (fish parasite). Some taxa have algal photosymbionts; anaerobic species may have methanogenic bacteriosymbionts. Genome sequences: Tetrahymena thermophila, Paramecium tertauralia, Ichthyophthirius multifiliis (EST only).
Apicomplexa: Intracellular, intestinal or coelomic parasites of metazoa, including Plasmodium (causative agent of malaria). Defined by the presence of an apical complex consisting of a closed conoid, a polar ring, rhoptries and micronemes, involved in host cell attachment and invasion. Apicoplasts, non-photosynthetic relict plastids, bound by four membranes, and containing a genome with extremely reduced content, may be present. Genome sequences: species of Plasmodium, Toxoplasma (toxoplasmiosis), Theileria (cattle parasite; East Coast disease), Babesia (cattle parasite; tick fever) and Cryptosporidium (AIDS-associated intestinal parasite).
Colpodellids: Free-living predatory flagellates, with hairs or bulbs on the anterior flagellum, and an apical feeding complex containing an open conoid, which attaches to prey and allows the myzocytotic uptake of cytoplasm.
Chromerids: Free-living, immotile photosynthetic alveolates, containing distinctive, cone-shaped golden-brown chloroplasts contacted at the apex by an intracellular cilium. The chloroplast contains a circular or long linear genome, utilises a form II rubisco, and uniquely amongst photosynthetic alveolates does not contain chlorophyll c. One species, Chromera velia, has been identified; a photosynthetic flagellate CCMP3155 has been isolated that groups with chromerids and apicomplexa, but the relationships between these lineages is uncertain. Genome sequences: Chromera velia (chloroplast only).
Dinoflagellates: Phototrophic, mixotrophic and heterotrophic alveolates, with a coiled transverse flagellum held in a central girdle, and a longitudinal flagellum in a longitudinal furrow. Many species contain a secondary, red-algal derived chloroplast containing peridinin and a form II rubisco; in addition, a diverse array of serial chloroplast acquisitions are known. The distinctive, haploid nucleus contains permanently condensed chromosomes and lacks standard histones; the genomes of peridinin-containing chloroplasts consist of multiple small subgenomic minicircles. Some free-living species form harmful red tide blooms; others are endobionts of marine invertebrates (e.g. zooxanthellae, the primary producers of coral ecosystems). Genome sequences: Symbiodinium spp., Alexandrium tamarense (both EST only).
Perkinsids: Intracellular parasites of molluscs and dinoflagellates, with a row of bipartite hooks or thick hairs on one side of the anterior flagellum. An apical complex, containing an open conoid, an anterior and a posterior ring, is used to penetrate host cells. Perkinsids are non-photosynthetic; there is moderate genetic and ultrastructural evidence for the retention of plastids. Genome sequences: Perkinsus marinus (EST only).
Ellobiopsids: Multinucleate parasites, principally of pelagic crustaceans, that superficially resemble fungi; each individual consists of one or more external tube-like structures, and a nutrient-absorbing root and trophic/ generative structures inside the host.
Rhizaria: A major group of eukaryotes with fine root-like, reticulate or filose pseudopodia; there are no defining synapomorphies, but monophyly is strongly supported by molecular phylogenies. Groups below are divided into Cercozoa, Foraminifera, Radiozoa and incertae sedis taxa; the relationships between and within these groups are incompletely resolved. Rhizaria include parasites of algae, plants, fungi and invertebrates. Multigene studies robustly support a position for Rhizaria at the base of the SAR clade; whether the Rhizaria historically contained secondary, red algal-derived chloroplast lineages is currently under debate.
Cercozoa: A diverse assemblage of flagellates and amoebae that may form filose or reticulate pseudopodia, and may harbour endosymbionts; currently identified on the basis of molecular phylogenies. Cercozoa share with Foraminifera an insertion of one or two amino acids at the monomer–monomer junctions of polyubiquitin. Cercozoa are currently divided into Filosa and Endomyxa.
Filosa
Cercomonads: Flagellates that produce filose, finger-shaped and branching pseudopodia; the anterior flagellum beats stiffly in a cone shape and the posterior flagellum trails behind the cell.
Chlorarachniophytes: Reticulate amoebae, flagellates, and/ or individual filose amoebae, with a secondary, green algal-derived chloroplast that retains a highly reduced nucleomorph (relict algal nucleus). Genome sequences: Bigelowiella natans (chloroplast and nucleomorph, nuclear in preparation).
Clautriavia and Auranticordis: Large, multi-lobed tetraflagellates (Auranticordis) or small gliding uniflagellates (Clautriavia), with a cell surface bearing pores and muciferous bodies. A. quadriverberis has been reported to contain photosynthetic endosymbionts, which may be of cyanobacterial origin.
Desmothoracids (=Clathrulinids): Heliozoan protists where the cell body is surrounded by axopodia that protrude through a perforated capsule made of silica and organic matter.
Euglyphids: Filose amoebae with a test of regularly-shaped siliceous plates held together by organic cement. One member, Paulinella chromatophora, has endosymbiotic, cyanobacterial-derived cyanelles unrelated to archaeplastid chloroplasts. Genome sequences: Paulinella chromatophora (two strains; cyanelles only).
Glissomonads: Small flagellates that glide on a trailing posterior flagellum and have a short, waving anterior flagellum.
Limnofila: Predominantly amoeboid with very fine, filose pseudopodia bearing granules (extrusomes); most lineages contain two flagellar stubs that stop at the transitional region.
Massisteria: Small irregular amoebae from which radiate thin pseudopodia bearing extrusomes; the pseudopodia may branch and anastomose. There are two flagella, which are normally inactive.
Metopion: Small disc-shaped biflagellates with a very shallow ventral groove at the anterior end, and long posterior and, in some taxa, stumpy anterior trailing flagella.
Metromonas: Small lozenge-shaped gliding flagellates with a long posterior and a stumpy anterior trailing flagellum. The posterior flagellum can form a hook shape and attach to the substrate.
Pansomonads: Heterotrophs with alternating sedentary amoeboid and motile biflagellate stages. Flagella are hairy, heterodynamic and free from the cell body.
Sainouroids: Small, lozenge-shaped, gliding flagellates with a long posterior and a stumpy anterior trailing flagellum.
Thaumatomonads: Rounded heterotrophic flagellates with a short anterior scaly flagellum, a long naked posterior flagellum and the ability to produce filose pseudopodia. Many species are covered in siliceous scales or spines. One species contains bacterial endosymbionts.
Thecofilosea: Filose amoebae; cells are surrounded by an organic flexible tectum or rigid test (coverings) with one or two apertures for filopodia.
Cryothecomonas: Oval-shaped biflagellates, covered with a delicate theca, with a pronounced ventral groove from which pseudopodia emerge. Cryothecomonas is an intracellular parasite of diatoms.
Ebriids: Large marine flagellates with a prominent basket-shaped internal siliceous skeleton.
Phaeodarea: Radiolarian or heliozoan deep-sea protists with an elaborately decorated skeleton and needles of amorphous silica mixed with organic and inorganic components. The life cycle is complex and includes flagellated stages.
Protaspis: Marine flagellates shaped like elongated ovals with parallel lateral sides, with two heterodynamic flagella emerging through funnels.
Pseudodifflugia: Filose amoebae with a rigid agglutinated test. There may be up to three nuclei per cell.
Endomyxa
Ascetosporea: Arthropod parasites that produce simple spores.
Haplosporidia: Invertebrate parasites or hyper-parasites, which include pathogens of significant commercial importance (e.g. Haplosporidium nelsoni, agent of MSX disease in oysters). Most taxa have a distinctive open spore case.
Paramyxids: Economically important parasites of bivalve molluscs (e.g. Marteilia, parasite of Sydney rock oysters), crustaceans and annelids; which make multicellular spores by endogenous budding.
Paradinids: Marine parasites of crustaceans, (e.g. Paradinium, a parasite of spot prawns), which are usually seen as large bag-like spores with a ridged surface.
Filoreta: Amoebae that form extensive multinucleate reticulate plasmodial networks. Cells are connected by cytoplasmic strands that vary from very fine projections to sheet-like expanses and lack prominent granules.
Gromia: Large marine rhizopods with filose, non-granular pseudopodia and a large ovoid proteinaceous test. There is a motile stage with two flagella.
Phytomyxids: Plasmodiophorids and phagomyxids; parasites of vascular plant roots and stramenopiles, which form multinucleate plasmodia, biflagellate zoospores, and an invasive attacking stage that has unique modifications of the ER in the intracellular protrusion, the stachel and rohr. The group includes important agricultural pests e.g. Spongospora subterranea (potato powdery scab) and Plasmodiophora brassicae (cabbage club root disease).
Vampyrellids: Large globular, multinucleate, reticulate amoebae; with long, thick cytoplasmic arms ending in fan-like flat pseudopodia giving rise to filopodia. Principally parasites of algae and fungi.
Foraminifera: Highly diverse marine, freshwater and terrestrial rhizopods, with large reticulate networks of granular pseudopodia that exhibit unique bidirectional rapid cytoplasmic streaming. There is alternation of generations and very complex and variable morphology in the life cycle. The group is important in palaeontology: many taxa have chamber-bearing tests used as stratigraphic markers. The pseudopodia frequently contain algal endosymbionts. Genome sequences: Reticulomyxa filosa (EST only).
Radiozoa: Organisms with radiating arms, microtubule-supported axopodia, which extend outwards from a central cell body though a porous organic capsule to connect with a frothy external layer that contains digestive vacuoles and symbionts. There may be a siliceous or strontium skeleton surrounding the central cell body. Radiozoa lack a polyubiquitin insertion, distinguishing them from other rhizarians.
Acantharea: Radiolarian marine protists with radiating axopodia and spicules of strontium sulphate. The cell is surrounded by a capsule of fibrillar material which interconnects myonemes that control the direction of the spicules. Endosymbiotic algae may be present.
Polycystinea: Radiolarian marine protists with axopodia and a siliceous skeleton ranging from spicules to lattices with radiating spines. There is a complex life cycle including biflagellated swarmers and vegetative colonies. The peripheral ectoplasm may bear symbionts.
Sticholonche: Kidney-shaped, bilaterally symmetrical marine protists with parallel rows of locomotory axopodia and rosettes of flattened siliceous spicules.
THE CCTH CLADE (Smith and Patterson, Reference Smith and Patterson1986; Edvardsen et al. Reference Edvardsen, Eikrem, Green, Andersen, Moon-Van-Der-Staay and Medlin2000; Burki et al. Reference Burki, Inagaki, Brate, Archibald, Keeling, Cavalier-Smith, Sakaguchi, Hashimoto, Horak, Kumar, Klaveness, Jakobsen, Pawlowski and Shalchian-Tabrizi2009; Okamoto et al. Reference Okamoto, Chantangsi, Horak, Leander and Keeling2009)
The final major eukaryotic division is the CCTH clade (also named Hacrobia), a group of free-living, heterotrophic, mixotrophic and autotrophic organisms (Fig. 7). Evidence for the group comes from both single-gene and multigene phylogenies. No synapomorphies have been reported, although similar flagella, sub-lamellar vesicles and ejectisomes are observed in most phyla, and cryptomonads and haptophytes share a derived, bacterial isoform of the chloroplast-targeted protein rpl36. Cryptomonads, picobiliphytes and kathablepharids are believed to be closely related; there are conflicting data regarding the phylogenetic positions of haptophytes, centrohelids and telonemids. No parasitic taxa are known. A genome sequence is available for the haptophyte Emiliania huxleyi; a sequence for the cryptomonad Guillardia theta is in preparation.

Fig. 7. CCTH. (a) Telonemid: Telonema subtile; (b) Haptophyte: Pavlova pinguis; (c) Haptophyte, coccolithophorid: Emiliania huxleyi; (d) Cryptophyte; Cryptomonas sp.; (e) Kathablepharid: Kathablepharis sp.; (f) Centrohelid: Heterophrys sp. Scale bar in f: for a, 5 μm; b, 20 μm; c–f, 10 μm.
Cryptomonads: Mostly autotrophic cells distinguished by characteristic arrays of bipartite flagellar hairs, a geometric cell coat and flattened mitochondrial cristae. The secondary, red algal-derived chloroplasts retain a nucleomorph (relict algal nucleus). Cryptomonad-derived plastids have been observed in dinoflagellate species; these may be tertiary endosymbionts, or may be obtained by kleptoplastidy. Genome sequences: Guillardia theta (in progress).
Kathablepharids: Free-living heterotrophic flagellates, with a distinctive, spiralled organic sheath, complex conical feeding apparatus and a peripheral ER. Feeding may occur via engulfment or myzocytotic consumption of prey cytoplasm and individuals may swarm to engulf prey. Five genera are currently known; none have true chloroplasts, although one species, Hatena, exists with a green algal photosymbiont.
Picobiliphytes: Small (6 μm long) planktonic organisms of unknown general appearance, distributed throughout sub-arctic, temperate and tropical waters, described only from environmental studies. Picobiliphytes have an organelle similar in fluorescence profile to the plastids of red algae and cryptomonads, and have been suggested to retain a nucleomorph.
Haptophytes: Free-living mixotrophic or autotrophic flagellates, distinguished by a haptonema (a locomotory, attachment and feeding organelle supported by microtubules), and distinctive lamellae in the red algal-derived chloroplasts; species may be naked or covered in calcareous, organic or in one case siliceous scales. Several species (e.g. Phaeocystis globosa) are agents of fish-killing planktonic blooms. Genome sequences: Emiliania huxleyi, Phaeocystis globosa (in progress).
Centrohelids: Heterotrophic, amoeboid protists with multiple radiating arms that contain distinctive ball-and-cone extrusomes, a radiating system of endocytic vesicles, and many Golgi bodies dispersed through the cell. Some species are observed to form swarms that fuse to form a single multinuclear cell during feeding.
Telonemids: Predatory flagellates with a complex, multilayered lamina and vesicles containing paracrystalline objects, beneath the cell surface. One genus is known, but is diverse with a cosmopolitan distribution.
EUKARYOTES INCERTAE SEDIS (Foissner et al. Reference Foissner, Blatterer and Foissner1988; Brugerolle et al. Reference Brugerolle, Bricheux, Philippe and Coffea2002; Kim et al. Reference Kim, Simpson and Graham2006)
Some eukaryotic taxa remain unplaced: through being inadequately described, genuinely difficult to place or genuinely not closely related to any other group. The taxa listed here are a small proportion of incertae sedis eukaryotes, selected on the basis of recent publication and current phylogenetic interest.
Apusozoa: Small, gliding biflagellates, with a trailing posterior flagellum, two basal bodies, and a dorsal organic sheath. Apusozoa currently consist of Ancyromonas/Planomonas (with a single-layered sheath and flattened mitochondrial cristae) and apusomonads (with a double-layered sheath and tubular cristae); support for this grouping is incomplete. Apusozoa share some ultrastructural and discrete genomic features with bikonts, but predominantly resolve in molecular analyses as close relatives of the opisthokonts (Fig. 1).
Breviates: A group containing only one cultured species, Breviata anathema, an amoeboid flagellate with filose branching pseudopodia at anterior and posterior ends, and a large, branching mitochondrion-like structure without cristae (Fig. 1). Breviates have been proposed in some recent multi-gene analyses to be a sister-taxon to the amoebozoans.
Collodictyonids (=Diphylleids): Flagellates with a deep ventral feeding groove and a distinctive flagellar transition zone containing an electron-dense sleeve around the central microtubules, and unusual horseshoe-shaped dictyosomes.
Colponema: Small kidney-shaped flagellates with subsurface alveoli and a ventral feeding groove, that can emit fine filopodia and is the region of phagocytosis. Ultrastructure suggests possible relationships with the alveolates.
Hemimastigophora: Multiflagellated protists with diagonally symmetrical dorsal and ventral subsurface plates, and distinctive concentric extrusomes are present. Ultrastructure suggests a possible relationship with euglenids.
Palpitomonas: Heterotrophs with two long flagella and a single mitochondrion that surrounds the Golgi apparatus. Molecular phylogenies and ultrastructural similarities suggest a close relationship to the archaeplastids or the CCTH clade.
For a more detailed treatment of eukaryotic systematics, see the supplementary section (see http://journals.cambridge.org/PAR).
CURRENT CONTROVERSIES: WHERE TO ROOT THE TREE AND HOW TO CLASSIFY THE ALGAE
Despite the advances in eukaryotic systematics seen in recent years, there remain two immediate and exciting areas of controversy. Firstly, as detailed above, the conceptualization of the eukaryotic tree still held by many parasitologists and cell biologists is of a separation of crown eukaryotes (animals, plants and fungi) and simple/primitive archaezoan organisms such as Giardia, Trypanosoma or Trichomonas (Cavalier-Smith, Reference Cavalier-Smith1987; Sogin and Silberman, Reference Sogin and Silberman1998). As is obvious from the new and well-supported grouping of eukaryotes into component supergroups, the various parasites do not group together at the base of the tree (Fig. 1), and yet many authors still operate under modified versions where their organism of interest still represents a primitive eukaryote and thus a living fossil for study (Morrison et al. Reference Morrison, Mcarthur, Gillin, Aley, Adam, Olsen, Best, Cande, Chen, Cipriano, Davids, Dawson, Elmendorf, Hehl, Holder, Huse, Kim, Lasek-Nesselquist, Manning, Nigam, Nixon, Palm, Passamaneck, Prabhu, Reich, Reiner, Samuelson, Svard and Sogin2007; Koopmann et al. Reference Koopmann, Muhammad, Perbandt, Betzel and Duszenko2009; Yadav et al. Reference Yadav, Mandal, Rao and Bhattacharya2009). The Archezoa idea remains simple and intuitive, and thus alluring; but perhaps one of the major reasons why it is still in currency is the lack of strong alternate hypotheses for the root of eukaryotes. Although it is possible to interpret individual pieces of evidence for the following rooting hypotheses, there is no preponderance of evidence for any of the proposed scenarios, making this perhaps the greatest open question in eukaryotic cellular evolution today.
Proposed in 2002, the bikont-unikont rooting was built on the idea that a bifurcation exists between an amoebozoan-opisthokont group (unikonts), and the rest of eukaryotes (bikonts) (Stechmann and Cavalier-Smith, Reference Stechmann and Cavalier-Smith2002). It was presumed that the ancestral bikonts possessed two basal bodies and two flagella, whereas ancestral unikonts were presumed to only possess one of each. These two groups were also thought to undergo different types of flagellar transformation upon cell division. The posterior flagellum became the anterior in the next generation in unikonts, while the opposite was proposed as the case for bikonts. Molecular data were also put forward to support this split in the form of a gene fusion between dihydrofolate-reductase (DHFR) and thymidylate synthase (TS) (Philippe et al. Reference Philippe, Lopez, Brinkmann, Budin, Germot, Laurent, Moreira, Muller and Le Guyader2000), where the fusion was thought to be exclusive to bikonts (Stechmann and Cavalier-Smith, Reference Stechmann and Cavalier-Smith2002), as well as the presence of type II myosin only present in unikonts (Richards and Cavalier-Smith, Reference Richards and Cavalier-Smith2005).
This rooting was dependent on the clean distribution of the bikont versus unikont basal body and flagellar arrangement. However, this was not universally supported by the data available in the literature even at the time the unikont/ bikont split was proposed (see Supplementary Table 1 – http://journals.cambridge.org/PAR), suggesting that more information would be needed before these flagellar data could be regarded as a good heuristic for this particular rooting proposal. Also, the recently proposed classifications of the incertae sedis taxa apusomonads as sister to opisthokonts (Kim et al. Reference Kim, Simpson and Graham2006), and Breviata as sister to amoebozoans (Minge et al. Reference Minge, Silberman, Orr, Cavalier-Smith, Shalchian-Tabrizi, Burki, Skjaeveland and Jakobsen2009), call into question the rooting, as they each possess the ‘wrong’ type of flagellar apparatus (Karpov and Zhukov, Reference Karpov and Zhukov1986; Walker et al. Reference Walker, Dacks and Embley2006). This rooting hypothesis was also supported by the distribution of the DHFR-TS fusion (Stechmann and Cavalier-Smith, Reference Stechmann and Cavalier-Smith2002); however, the presence in apusomonads of this fusion gene challenges the validity of this conclusion.
Several more recent rooting hypotheses have been proposed. Based on the distribution pattern of rare conserved amino acids in orthologous genes, Rogozin et al. (Rogozin et al. Reference Rogozin, Basu, Csuros and Koonin2009) deduced that the root of eukaryotes lies between the archaeplastids and all other taxa. This rooting would imply that the acquisition of the cyanobacterial primary endosymbiont sparked the first divergence in the eukaryotic line. This rooting hypothesis certainly needs further investigation and continued inclusion of taxa in such analyses.
Additionally, a root lying between euglenozoans and the rest of eukaryotes (neokaryotes) has been proposed (Cavalier-Smith, Reference Cavalier-Smith2010). This rooting places kinetoplastids at the base of the eukaryotic tree due to the nature of their cytochrome biosynthesis pathway, which uses the cytochrome c/c1 biogenesis mechanism proteins rather than haem lyase, a trait shared with bacteria. Phylogenetic trees of the mitochondrial pore TOM, and the trypanosomatid porin VDAC, support this root (Pusnik et al. Reference Pusnik, Charriere, Maser, Waller, Dagley, Lithgow and Schneider2009). This is further suggested by the presence of the archaeal Cdc6 replication initiator instead of the canonical eukaryote DNA replication ORC in trypanosomes (Godoy et al. Reference Godoy, Nogueira-Junior, Paes, Cornejo, Martins, Silber, Schenkman and Elias2009); however, more evidence is needed to demonstrate the stability of such a root.
Even if the root of eukaryotes was clear, controversy would remain surrounding the precise phylogenetic position and history of selected parasitic eukaryotes, a cause for identity crisis in high-profile pathogens such as Plasmodium, Phytophthora, and Blastocystis. The nature of the conceptual overlap between the contentious Chromalveolata and the SAR and CCTH clades has important implications for the evolution of complex plastids in eukaryotes.
Numerous authors have historically suggested from ultrastructure, that different combinations of cryptomonads, stramenopiles, haptophytes and alveolates, the only phyla containing red algal-derived chloroplast lineages, might be closely related (Whittaker, Reference Whittaker1969; Lucas, Reference Lucas1970; Cavalier-Smith, Reference Cavalier-Smith1981). However, the four phyla were first unified under the ‘chromalveolate hypothesis’ by Cavalier-Smith (Cavalier-Smith, Reference Cavalier-Smith1999), which suggested that the red algal-derived chloroplasts in each lineage arose from a common, ancestral secondary endosymbiosis, and were secondarily lost in non-photosynthetic members of each phylum. This hypothesis had been supported by chloroplast gene phylogenies, which strongly recover support for the monophyly of chromalveolate chloroplasts, suggesting that they originated from a single secondary endosymbiotic event (Yoon et al. Reference Yoon, Hackett, Ciniglia, Pinto and Bhattacharya2004; Li et al. Reference Li, Nosenko, Hackett and Bhattacharya2006; Iida et al. Reference Iida, Takishita, Ohshima and Inagaki2007). In addition, there is clear evidence (based on the retention of diminished, non-photosynthetic chloroplasts, endosymbiont-derived genes and from basally divergent photosynthetic lineages) of secondary endosymbiont loss in non-photosynthetic alveolates (Moore et al. Reference Moore, Obornik, Janouskovec, Chrudimsky, Vancova, Green, Wright, Davies, Bolch, Heimann, Slapeta, Hoegh-Guldberg, Logsdon and Carter2008; Reyes-Prieto et al. Reference Reyes-Prieto, Moustafa and Bhattacharya2008; Slamovits and Keeling, Reference Slamovits and Keeling2008; Joseph et al. Reference Joseph, Fernandez-Robledo, Gardner, El-Sayed, Kuo, Schott, Wang, Kissinger and Vasta2010).
The ‘chromalveolate’ hypothesis has been challenged on two grounds. Firstly, recent analyses have suggested that a much greater number of non-photosynthetic lineages resolve with the ‘chromalveolates’ than was previously imagined: molecular phylogenies strongly support the unification of cryptomonads and haptophytes with the non-photosynthetic centrohelids, katablepharids, and telonemids, in the ‘CCTH clade’ (Burki et al. Reference Burki, Inagaki, Brate, Archibald, Keeling, Cavalier-Smith, Sakaguchi, Hashimoto, Horak, Kumar, Klaveness, Jakobsen, Pawlowski and Shalchian-Tabrizi2009; Okamoto et al. Reference Okamoto, Chantangsi, Horak, Leander and Keeling2009); more surprisingly, multigene phylogenies and the conserved presence of a novel Rab GTPase have suggested that the primarily non-photosynthetic Rhizaria cluster with the plastid-containing stramenopiles and alveolates in the ‘SAR clade’ (Burki et al. Reference Burki, Shalchian-Tabrizi, Minge, Skjaeveland, Nikolaev, Jakobsen and Pawlowski2007; Elias et al. Reference Elias, Patron and Keeling2009; Cavalier-Smith, Reference Cavalier-Smith2010). As there is no evidence currently supporting secondary loss of chloroplasts in these lineages, it must be asked whether an ancient endosymbiosis event, followed by multiple secondary losses, is necessarily more parsimonious than the independent or serial acquisition of chloroplasts by photosynthetic chromalveolates.
Secondly, and perhaps more critically, it is not known whether the CCTH and SAR clades are in fact sister-groups, or whether the chromalveolates sensu lato are paraphyletic: while some nuclear multigene phylogenies do support chromalveolate monophyly (Hackett et al. Reference Hackett, Yoon, Li, Reyes-Prieto, Rummele and Bhattacharya2007; Burki et al. Reference Burki, Inagaki, Brate, Archibald, Keeling, Cavalier-Smith, Sakaguchi, Hashimoto, Horak, Kumar, Klaveness, Jakobsen, Pawlowski and Shalchian-Tabrizi2009; Nozaki et al. Reference Nozaki, Maruyama, Matsuzaki, Nakada, Kato and Misawa2009), many recover a closer relationship between the CCTH clade and the archaeplastids (e.g. (Burki et al. Reference Burki, Shalchian-Tabrizi, Minge, Skjaeveland, Nikolaev, Jakobsen and Pawlowski2007, 2008; Hampl et al. Reference Hampl, Hug, Leigh, Dacks, Lang, Simpson and Roger2009; Reeb et al. Reference Reeb, Peglar, Yoon, Bai, Wu, Shiu, Grafenberg, Reyes-Prieto, Rummele, Gross and Bhattacharya2009)). As such, a number of alternative hypotheses for the origin of CCTH and SAR clade plastids have been suggested, principally based on the tertiary transfer of a recent secondary endosymbiont within and between the two lineages (Sanchez Puerta and Delwiche, Reference Sanchez Puerta and Delwiche2008; Bodyl et al. Reference Bodyl, Mackiewicz and Stiller2009; Baurain et al. Reference Baurain, Brinkmann, Petersen, Rodriguez-Ezpeleta, Stechmann, Demoulin, Roger, Burger, Lang and Philippe2010).
Two questions must be resolved for the taxonomic validity and status of the ‘chromalveolates’ to be decided. Firstly, and most critically, is whether the nuclear lineages of the CCTH and SAR clades are sister-taxa. This question has already proven to be exceptionally difficult, due to both the presumed ancient and deep divergence of the archaeplastids, CCTH and SAR clades (Yoon et al. Reference Yoon, Hackett, Ciniglia, Pinto and Bhattacharya2004; Okamoto and McFadden, Reference Okamoto and Mcfadden2008) which may obscure clear phylogenetic signals. Recently, Baurain et al. (Reference Baurain, Brinkmann, Petersen, Rodriguez-Ezpeleta, Stechmann, Demoulin, Roger, Burger, Lang and Philippe2010) have demonstrated via an ingenious series of statistical analyses that, even accounting for branch length and divergence date, support for the monophyly of the mitochondrial and nuclear lineages of cryptomonads, haptophytes and stramenopiles is substantially weaker than might be expected were they monophyletic; however, a test of comparable rigour has not yet been applied to a CCTH+archaeplastid clade, and it is possible that this result may merely be evidence of an extremely weak phylogenetic signal in chromalveolate taxa. Secondly, if the chromalveolates are indeed monophyletic, it must be determined whether non-photosynthetic ‘chromalveolates’ contain sufficient clearly algal-derived genes to suggest an ancestral chloroplast history, and whether this chloroplast was likely acquired prior to the radiation of the chromalveolates, as opposed to independently acquired by multiple lineages. This has been rendered especially complicated by the recent suggestion that some or all chromalveolate taxa may have possessed a cryptic, green algal-derived secondary endosymbiont (Frommolt et al. Reference Frommolt, Werner, Paulsen, Goss, Wilhelm, Zauner, Maier, Grossman, Bhattacharya and Lohr2008; Moustafa et al. Reference Moustafa, Beszteri, Maier, Bowler, Valentin and Bhattacharya2009). It is entirely possible that an ancestral, secondary endosymbiosis could have occurred, thus cementing the status of the chromalveolates as a kingdom, that is not retained in any extant chromalveolates.
The recent developments and challenges to established theories concerning the global evolution of the eukaryotes, such as the Archaezoa and chromalveolate hypotheses, and the unikonts-bikont root, reflect a general trend away from defining phylogeny solely on discrete synapomorphies such as rare genetic events (gene fusions and indels), single ultrastructural features and endosymbioses. Traditional hypotheses have been founded on the explicit assumption that these traits would not undergo horizontal transfer, convergent evolution, or reversion (e.g. Stechmann and Cavalier-Smith, Reference Stechmann and Cavalier-Smith2002), but broader taxonomic sampling and more detailed phylogenetic studies have frequently drawn evidence to the contrary such as the validity of a cytosolic duplication of GAPDH as a chromalveolate-defining synapomorphy (Fast et al. Reference Fast, Kissinger, Roos and Keeling2001; Obornik et al. Reference Obornik, Janouskovec, Chrudimsky and Lukes2009; Takishita et al. Reference Takishita, Yamaguchi, Maruyama and Inagaki2009). Although there is certainly a place for the use of rare and discrete events in defining phylogeny, it seems ever more necessary that these be supported and used in conjunction with large-scale, multigene phylogenetic data.
CONCLUSIONS
Recent systematics of protistan parasites, and microbial eukaryotes in general, has revealed the considerable complexity in the relationship between circumscriptions of taxa based on light microscopy and those incorporating molecular data. The future of this field will be in the continued application of molecular phylogenetics, based on extensive sequence datasets, with the careful incorporation of ultrastructural and rare genomic event data to highlight possible analytical artifacts and help us understand the connection to observations at the light microscopical level and above. The technique of concatenating hundreds of protein sequences into mega-alignments for analysis has proven exceedingly powerful and has both clarified contentious issues as well as served up some real surprises. Nonetheless, it has still failed to overcome the bugbear in reconstructing higher-level eukaryotic relationships. Concatenated analyses are still only partly able to mitigate ‘long-branch attraction’ effects. Indeed, unchecked, these can be exacerbated as random noise and may accumulate faster than phylogenetic signals (Brinkmann et al. Reference Brinkmann, Van Der Giezen, Zhou, Poncelin De Raucourt and Philippe2005; Philippe and Telford, Reference Philippe and Telford2006), and there is no agreed or entirely effective way to mitigate these artifacts.
One potential way to address this issue is to include sequences from organisms whose genes are more slowly evolving. These are often free-living relatives of parasitic taxa (e.g. chromerid relatives of apicomplexans; Saprolegniales, free-living relatives of parasitic oomycetes). While once this was a hugely laborious prospect, with the advent of next-generation sequencing, it will increasingly become feasible. Already, EST-projects can contribute large amounts of data, but in the next few years it should be increasingly possible for eukaryotic genome projects to be produced by a few interested laboratories rather than being expensive endeavours; limited to genome sequencing centres only (e.g. Diguistini et al. Reference Diguistini, Liao, Platt, Robertson, Seidel, Chan, Docking, Birol, Holt, Hirst, Mardis, Marra, Hamelin, Bohlmann, Breuil and Jones2009; Steuernagel et al. Reference Steuernagel, Taudien, Gundlach, Seidel, Ariyadasa, Schulte, Petzold, Felder, Graner, Scholz, Mayer, Platzer and Stein2009). The bottleneck then will not be sequence acquisition but rather the careful annotation of cellular systems that will need to be done manually by expert researchers, if the projects are to be more than simple gene catalogues. Nonetheless, the addition of these slow-evolving gene sequences should significantly help to resolve some of the issues pertaining to the lack of resolution between the supergroups and provide insight into some of the more group-specific taxonomic issues. Of particular interest to parasitologists, it will also mean that genomes of the neglected parasitic diseases will be sequenced, organisms that may not have the large-scale global impact as Plasmodium or Entamoeba, but are nonetheless deserving of our attention.
Next-generation sequencing may not in itself overcome problems of lateral gene transfer and thus the potentially artifactual grouping of phylogenetically distant lineages. In fact, analyses of the genomes and proteomes of model eukaryotes have in some cases suggested that lateral gene transfer, especially between endosymbionts and host lineages may be extensive (e.g. Nosenko and Bhattacharya, Reference Nosenko and Bhattacharya2007; Moustafa et al. Reference Moustafa, Beszteri, Maier, Bowler, Valentin and Bhattacharya2009; Worden et al. Reference Worden, Lee, Mock, Rouze, Simmons, Aerts, Allen, Cuvelier, Derelle, Everett, Foulon, Grimwood, Gundlach, Henrissat, Napoli, Mcdonald, Parker, Rombauts, Salamov, Von Dassow, Badger, Coutinho, Demir, Dubchak, Gentemann, Eikrem, Gready, John, Lanier, Lindquist, Lucas, Mayer, Moreau, Not, Otillar, Panaud, Pangilinan, Paulsen, Piegu, Poliakov, Robbens, Schmutz, Toulza, Wyss, Zelensky, Zhou, Armbrust, Bhattacharya, Goodenough, Van De Peer and Grigoriev2009). This is particularly problematic when considering parasites and nascent endosymbiont lineages, where gene transfer events may blur the boundaries between host and symbiont genomes, as in the apicomplexans and the well studied arthropod parasite Wolbachia (Huang et al. Reference Huang, Mullapudi, Lancto, Scott, Abrahamsen and Kissinger2004; Dunning Hotopp et al. Reference Dunning Hotopp, Clark, Oliveira, Foster, Fischer, Torres, Giebel, Kumar, Ishmael, Wang, Ingram, Nene, Shepard, Tomkins, Richards, Spiro, Ghedin, Slatko, Tettelin and Werren2007). Already, some authors have suggested portmanteau nomenclature for taxa with distinctive and relatively unreduced symbionts (e.g. ‘dinotoms’ for the diatom-containing dinoflagellates Kryptoperidinium foliaceum and Durinskia baltica; Imanian et al. Reference Imanian, Pombert and Keeling2010). Although next generation sequencing is unlikely to precipitate a widespread renaming of eukaryotes to accommodate endosymbiotic gene transfer, it does raise the broader conceptual question of the extent to which the eukaryotes may be considered as a reductive, neatly branching tree.
Regardless of the underlying technical and conceptual issues, next generation sequencing will also open doors for understanding the parasites themselves as analyses are done comparing gene expression by deep-sequencing of cDNA from organisms in various infective conditions (e.g. Cantacessi et al. (Reference Cantacessi, Mitreva, Campbell, Hall, Young, Jex, Ranganathan and Gasser2010a, Reference Cantacessi, Mitreva, Jex, Young, Campbell, Hall, Doyle, Ralph, Rabelo, Ranganathan, Sternberg, Loukas and Gasserb)). Such studies will be particularly insightful if paired with gene expression analyses from close free-living relatives of the parasites, giving a final example of the utility of eukaryotic systematics to parasitologists. As we stand on the cusp of a new era of genome studies in protozoan parasites, we can anticipate exciting new results and new insights into the evolution of these creatures that demand our respect, our curiosity and the best of our efforts to defeat.
ACKNOWLEDGMENTS
We applaud the initiative that was Micro*Scope and is now EOL. We wish to thank David Patterson, Alastair Simpson, Andrew Roger, Mark Field and Chris Howe for general discussion; and Bob Andersen, David Bass, Michael Melkonian, Charley O'Kelly, Fred Spiegel and Gary Saunders for specific help on aspects of this manuscript, as well as general insight. Finally, we want to thank Emily Herman for help with production of Fig. 1.
FINANCIAL SUPPORT
This work was supported by fellowships from Darwin College Cambridge and the John Stanley Gardiner Fund of the University of Cambridge; and grants from The Royal Society and the Templeton Foundation, to GW; and an NSERC Discovery Grant to JBD.