INTRODUCTION
Habitat quality, host community assemblage, host susceptibility/resistance and pathogen–pathogen interactions are interconnected in complex and dynamic ways (Woolhouse et al. Reference Woolhouse, Dye, Etard, Smith, Charlwood, Garnett, Hagan, Hii, Ndhlovu, Quinnell, Watts, Chandiwana and Anderson1997; Ostfeld et al. Reference Ostfeld, Keesing and Eviner2008; Beldomenico and Begon, Reference Beldomenico and Begon2010; Johnson et al. Reference Johnson, Preston, Hoverman and Richgels2013). Disease dynamics can rarely be explained by examining one component of a complex natural system although general patterns are often sought. Understanding the host community components and interactions associated with disease emergence and persistence may provide valuable information for unlocking mechanisms driving disease dynamics in the natural environment (Dobson, Reference Dobson2004; Lafferty, Reference Lafferty2010; Telfer et al. Reference Telfer, Lambin, Birtles, Beldomenico, Burthe, Paterson and Begon2010).
Anthropogenic disturbance and subsequent loss of both biodiversity and community structure have been associated with increases in disease emergence (Keesing et al. Reference Keesing, Belden, Daszak, Dobson, Harvell, Holt, Hudson, Jolles, Jones, Mitchell, Myers, Bogich and Ostfeld2010; Roche et al. Reference Roche, Dobson, Guegan and Rohani2012), as well as a reduction in disease occurrence (Lafferty, Reference Lafferty2012; Bush et al. Reference Bush, Reed and Maher2013; Young et al. Reference Young, Griffin, Wood and Nunn2013). There is evidence supporting both positive and negative general linear relationships among disease and factors such as diversity, density and relative abundance (Randolph and Dobson, Reference Randolph and Dobson2012). Additionally, empirical evidence is mounting that a host species’ relative abundance and density are also linearly associated with parasite prevalence and/or richness (Arneberg, Reference Arneberg2002; Froeschke et al. Reference Froeschke, van der Mescht, McGeoch and Matthee2013). Regardless of shape or direction and the indirect or direct influences, anthropogenic disturbance can affect disease dynamics in natural systems (Randolph and Dobson, Reference Randolph and Dobson2012; Salkeld et al. Reference Salkeld, Padgett and Jones2013).
To improve our understanding of the general patterns between host and parasite communities in relation to anthropogenic disturbance, we investigated parasite dynamics in terrestrial small mammal communities in and around Kibale National Park (KNP), Western Uganda. Specifically, we focused on 3 common forest dwelling rodent species – Lophuromys aquilus, Praomys jacksoni and Hylomyscus stella (Delany, Reference Delany1975; Struhsaker, Reference Struhsaker1997). We examined parasites of these host species and investigate correlations between parasite prevalence and habitat disturbance, host density and host species richness. We further evaluated the parasite communities harboured by each of these host species. These small mammals (or hosts) were collected from habitats experiencing variable intensities of habitat disturbance. The specific parasites investigated in our study included: gastrointestinal protozoans (Giardia spp. and Cryptosporidium spp.), blood-borne parasite (Trypanosoma spp.) and ectoparasites from the taxonomic orders Ixodida (ticks), Acarina (mites), Siphonaptera (fleas) and Phthiraptera (lice). This study investigates broad patterns pertaining to the relationships among parasite prevalence and community structure in habitats that vary by species richness, host density and disturbance intensity.
MATERIALS AND METHODS
Study area
KNP is a mid-elevation tropical moist forest located in the foothills of the Rwenzori Mountains in Western Uganda (0 13′–0 41′N, 30′19′–30′22′E) (Struhsaker, Reference Struhsaker1997; Chapman and Lambert, Reference Chapman and Lambert2000; Hartter, Reference Hartter2009). The park and surrounding areas represent a mosaic of habitats that have undergone various types and frequency of habitat disturbance (Hartter, Reference Hartter2009). Portions of KNP were logged at varied intensities in the 1960s resulting in a series of contiguous forest compartments of lightly logged, heavily logged and unlogged status within KNP (Struhsaker, Reference Struhsaker1997). Over the last four decades KNP and the surrounding forest fragments have supported research on the influence of habitat disturbance on a variety of forest dwelling species (Kasenene, Reference Kasenene1984; Isabirye-Basuta and Kasenene, Reference Isabirye-Basuta and Kasenene1987; Lwanga, Reference Lwanga1994; Dranzoa, Reference Dranzoa1998; Chapman et al. Reference Chapman, Balcomb, Gillespie, Skorupa and Struhsaker2000; Seavy and Apodaca, Reference Seavy and Apodaca2002; Gillespie and Chapman, Reference Gillespie and Chapman2008; Hartter et al. Reference Hartter, Ryan, Southworth and Chapman2011). Our sampling sites represent a broad gradient of anthropogenic disturbance in the region, which was historically a contiguous forested area. Sampling sites included, from least to most disturbed: (1) relatively pristine forest (known as CC), (2) low-intensity selectively logged forest (known as K14), (3) high-intensity selectively logged forest (known as K15), (4) forest–agricultural interface (known as forest edge), (5) forest fragments (referred to as Fragments 1 and 2) and (6) human settlements (Fig. 1). All sites were sampled twice except for K15 and the forest edge. Previous studies within KNP (Chapman et al. Reference Chapman, Balcomb, Gillespie, Skorupa and Struhsaker2000) and these forest fragments (Gillespie and Chapman, Reference Gillespie and Chapman2006) have extensively evaluated the gradient of habitat disturbance occurring in the areas studied in this investigation. These previous studies allow our specific study sites to be categorically placed along a gradient of disturbance (Gillespie et al. Reference Gillespie, Chapman and Greiner2005; Gillespie and Chapman, Reference Gillespie and Chapman2006, Reference Gillespie and Chapman2008).

Fig. 1. Map of KNP (contrast enhanced as darker areas of image) and surrounding areas outside the national park. The area in the map was all once associated with a contiguous mid-elevation tropical moist forest. Specific locations of trapping webs (200 m in diameter) are identified as: CC, pristine primary forest; K14, lightly logged forest; K15, heavily logged forest; Edge, forest edge that overlaps agriculture fields; Fragment 1, located near the village of Bugembe and surrounded by small scale agriculture and human dwellings; Fragment 2, located near the village of Kiko and surround by small-scale agriculture, trading centre, and monoculture in the form of tea plantations. Small mammals were also collected from human dwellings located in areas around KNP and the forest fragments.
Animal collection
Trapping webs were used in all habitats except within village homes to accurately estimate the abundance, density and structure of the small mammal community within each habitat (Anderson et al. Reference Anderson, Burnham, White and Otis1983). Each web was 200 m in diameter and covered 3·14 ha with 12 radii each containing 12 Sherman traps (3×3·5×9″, H.B. Sherman Traps, Inc, Tallahassee, FL), with the first 4 of these traps set at 5 m intervals and the 8 distal traps were set at 10 m intervals (Mills et al. Reference Mills, Yates, Ksiazek, Peters and Childs1999). The centre of the web contained 4 Sherman traps and 1 Tomahawk trap (19×6×6″, Tomahawk Live Trap Co., Tomahawk, WI). An additional 4 Tomahawk traps were each set 50 m from the centre in the cardinal directions. In total, 153 traps were used in each web. Trapping webs were operated for three consecutive nights on each trapping occasion for a total of 5049 trap nights at 7 sites. All sites were sampled on 2 occasions except the forest edge and heavily logged forest sites, which were sampled once. Traps were baited in evenings and animals were collected at sunrise the following morning to prevent trap-associated deaths. All traps were baited consistently with peanut butter and millet. All trapping was conducted in the dry season between May and July of 2009. Sites that had repeated sampling had at minimum a 6-week rest period between sampling efforts.
Terrestrial small mammal collection was approved by the Uganda Wildlife Authority, the Uganda National Council for Science and Technology, local authorities and homeowners at trap sites. Animal handling protocols were approved by Institutional Animal Care and Use Committees (IACUC) from Emory University (#062-2009) and the Centers for Disease Control and Prevention (CDC) (#1768).
Standard field methods for small mammal handling, necropsy and tissue collection techniques were followed (Mills et al. Reference Mills, Childs, Ksiazek and Peters1995). Necropsies were performed and tissue samples were collected for molecular identification of mammalian species and Trypanosoma spp. Feces were collected from the descending colon and preserved in 10% buffered formalin for detecting Giardia spp. and Cryptosporidium spp.. Skulls from a subset of small mammals sampled (n = 137) were prepared using standard procedures and identified to species using established mammalian guides (Delany, Reference Delany1975; Nagorsen and Peterson, Reference Nagorsen and Peterson1980; Thorn and Kerbis Peterhans, Reference Thorn and Kerbis Peterhans2009). Specimens were catalogued at the Field Museum in Chicago, IL, USA (Reference no. 210384–210540). Molecular identification using cytochrome B gene analysis was necessary for species indistinguishable by morphometrics (i.e. P. jacksoni and P. misonnei) (Peppers et al. Reference Peppers, Carroll and Bradley2002).
Immediately following humane euthanasia, each small mammal was placed in an individual plastic bag. The plastic bags containing euthanized small mammals were then opened and contents were placed in a clean white plastic tub (approximately 8×40×30 cm) (Bush, Reference Bush2009). Each individual and the contents of their plastic bag were processed within the tub for easy visualization and collection of ectoparasites. Ectoparasites were dislodged and collected by vigorous brushing of euthanized small mammals. Attached ectoparasites (i.e. ixodid ticks) were collected by parting the fur of each individual animal with forceps and visually inspecting the skin. Small mammals were processed until no additional ectoparasites were collected or for a maximum of 20 min in the case of heavily infested animals and all ectoparasites were placed into 70% ethanol. This process was conducted to maintain consistency in collection methods.
Parasite detection
All parasites were identified to the level of order. DNA was extracted from the spleen tissue (stored at −80 °C) of all the animals collected (n = 327) using the DNA EZ1 tissue kit (Qiagen, Hilden, Germany). To detect Trypanosoma spp. DNA we amplified the highly variable region of the 18S ribosomal RNA gene using previously described nested PCR methods (Noyes et al. Reference Noyes, Stevens, Teixeira, Phelan and Holz1999) and Platinum Taq polymerase (Invitrogen, Grand Island, New York, USA). We used external primers TRY927F and TRY927R, and internal primers SSU561F and SSU561R to confirm Trypanosome-positive individuals (Noyes et al. Reference Noyes, Ambrose, Barker, Begon, Bennet, Bown and Kemp2002).
Formalin-preserved feces were screened for Giardia spp. cysts and Cryptosporidium spp. oocysts using the MERIFLUOR immuno-fluorescent assay (Meridian BioScience Inc. Cincinnati, Ohio, USA). Fecal samples were concentrated into fecal pellets and resuspended in 1 g/mL concentrations and scored for presence/absence (Salzer et al. Reference Salzer, Rwego, Goldberg, Kuhlenschmidt and Gillespie2007).
All ectoparasites were shipped to Emory University in Atlanta, Georgia, USA where they were examined by light microscopy. Ectoparasites from each animal were quantified and categorized according to taxonomic order – Ixodida (ticks), Acarina (mites), Siphonaptera (fleas) or Phthiraptera (lice) (Lane and Crosskey, 1993). Only motile stages were identified for each ectoparasite, including larval, nymphal and adult stages for ticks and mites; nymphal and adult stages for lice; and adult stage for fleas.
Data analysis
Relative small mammal density for each habitat was calculated and measured by trapping success divided by trapping effort for each habitat where trapping webs were used for animal collection (omitting human settlements from this analysis). Additionally, relative species abundance was measured by the number of total L. aquilus, H. stella and P. jacksoni divided by the total animals collected at each site. Using individual-based rarefaction, species richness was estimated for each habitat. A rarefaction curve was generated to determine adequate sampling effort and species richness to correct for varied sample sized across habitats.
Parasite point prevalence (referred to as prevalence) is the proportion of infected/infested individuals at the time of sample collection and was calculated for each habitat and the most abundant rodent species (L. aquilus, H. stella and P. jacksoni). Correlations between parasite prevalence and species richness, small mammal density and relative species abundance were measured using linear regression models with the prevalence of each parasite as outcome variable. Additionally, correlations between habitat disturbance and parasite prevalence were evaluated using Spearman's rank-order correlation, since habitat disturbance is a qualitative and ordinal variable. Statistical analyses were performed using the stats package on the R version 3.0.1 (R Core Team, 2013).
To determine if parasite assemblages were more influenced by habitat or by taxonomic category of host (i.e. species), we considered each individual small mammal collected as a patch or community of parasites. Each patch was associated with parasite presence/absence in addition to a variable considered ‘parasite free’ to account for small mammals free of parasites. Using constrained correspondence analysis (a.k.a. canonical correspondence analysis) (CCA) we investigated the influence of the taxonomic classification of each individual and the habitat the individual was from to understand what serves as the best predictor for the parasites of each individual animal captured. CCA was performed for the habitat while conditioning for species and vice versa. The results of each CCA were analysed using an ANOVA-like permutation test for CCA to assess the significance of constraints. All analyses were performed using the Vegan package on the R statistical platform (version 3.0.1) (R Core Team, 2013) following established methods (Bellier, Reference Bellier2012).
RESULTS
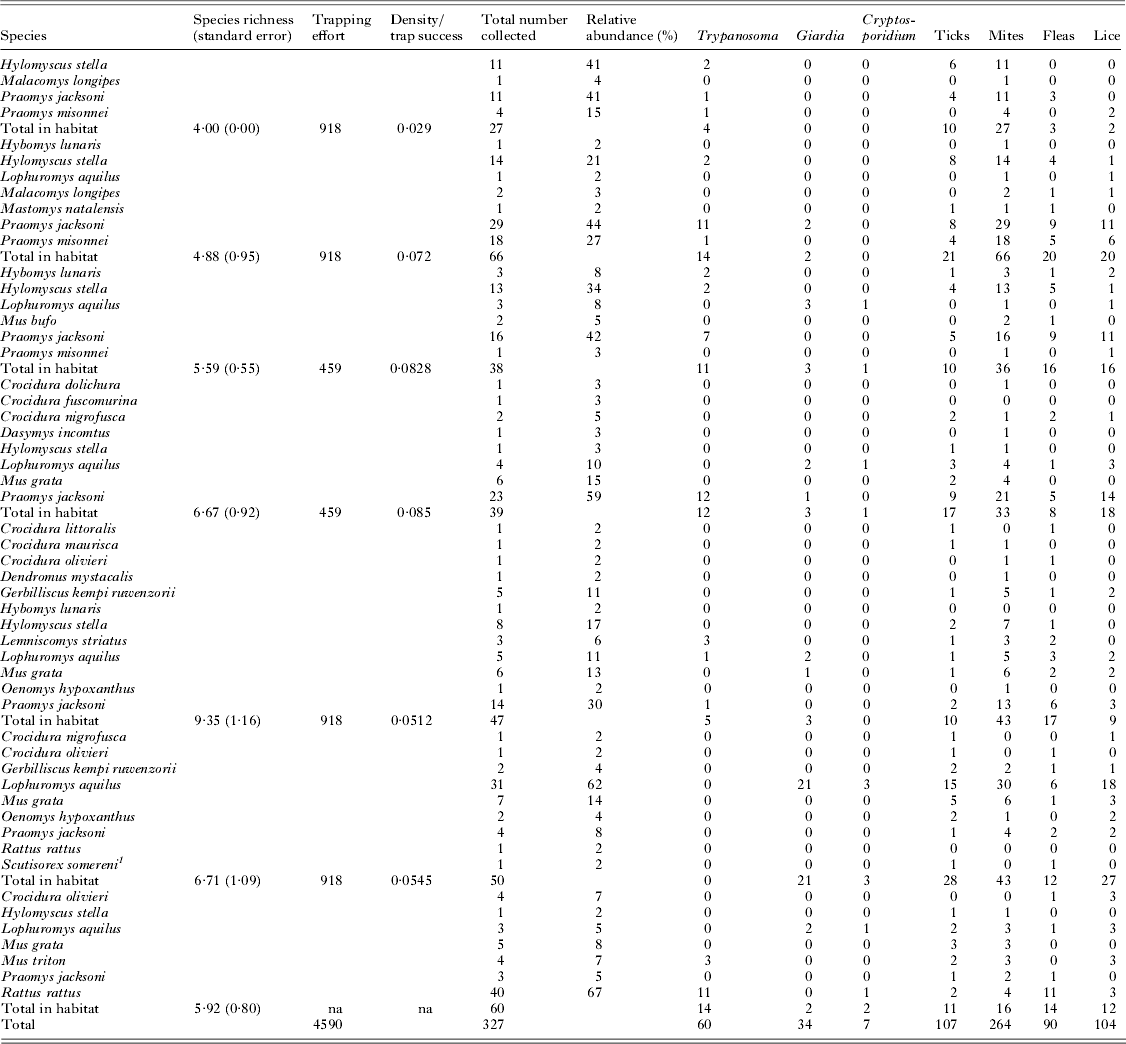
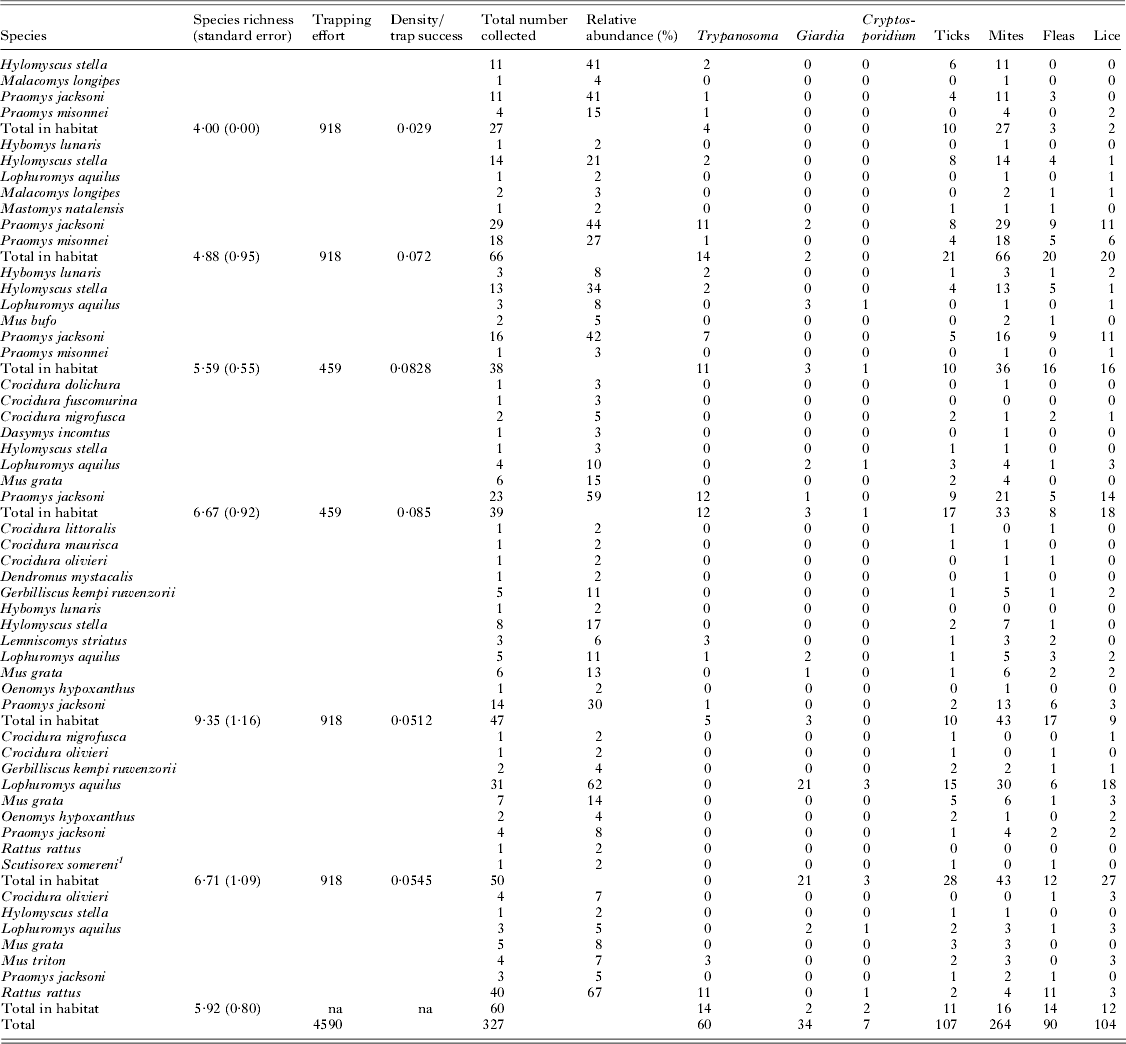
From May to July 2009, 327 small terrestrial mammals were collected in and around KNP (Fig. 1). Small mammal density, species richness (rarefied with standard error) and relative abundance of all 3 host species were calculated for each habitat, although density was not calculated for human settlements (Table 1). Mammals collected represented 23 species and parasites varied in prevalence across habitat and host species (Fig. 1). All parasites examined were found to be harboured by P. jacksoni and L. aquilus, while H. stella harboured all but cryptosporidium.
Table 1. In total, 327 small mammals representing 23 distinct species were collected from 7 habitats in and around Kibale National Park. Each habitat experienced varied levels of host density, species richness (rarefied) and disturbance level. Each individual small mammal was screened for a variety of parasites – Giardia spp., Cryptosporidium spp., Trypanosoma spp., mites, lice, fleas and ticks. The numbers in each parasite column (Giardia, Cryptosporidium, etc.) indicate the number of host from each individual species infected with each parasite group
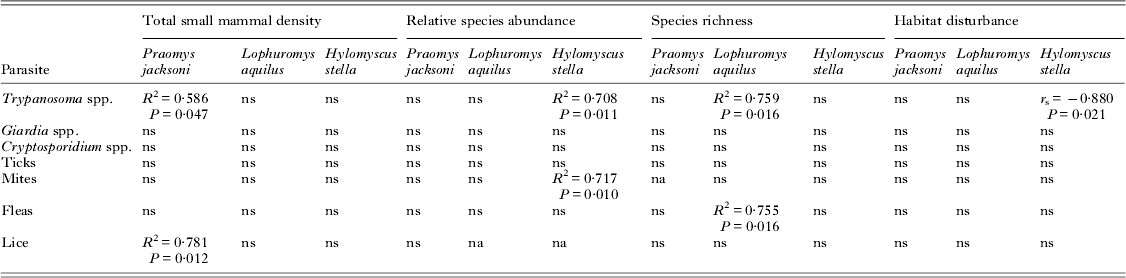
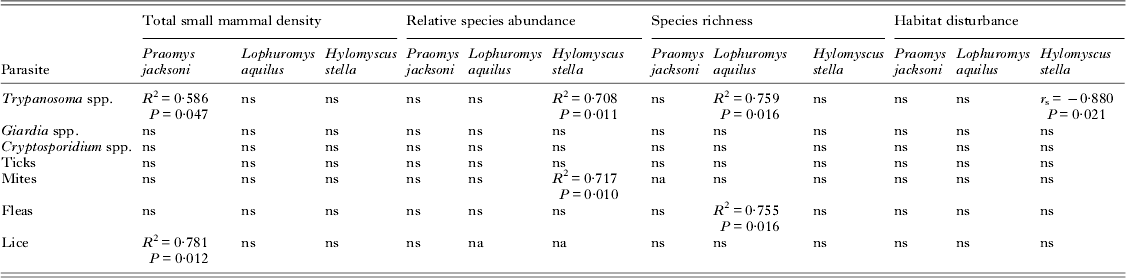
Linear regression was used to investigate parasite prevalence among the various species (Table 2). We identified a significant positive association between the overall small mammal density of a community and the prevalence of trypanosomes (R 2 = 0·586, P = 0·047) and lice (R 2 = 0·781, P = 0·012) among P. jacksoni. Additionally, there was a positive association among the relative abundance of H. stella and the prevalence of trypanosomes (R 2 = 0·708, P = 0·011) and mites (R 2 = 0·717, P = 0·010). Therefore, the more dominant H. stella become in the community of small mammals, the more individual H. stella are harbouring trypanosomes and mites. Parasite prevalence among L. aquilus was more closelyassociated with small mammal species richness with an association with flea infestation (R 2 = 0·755, P = 0·016) and trypanosome (R 2 = 0·759, P = 0·014). Relationships between habitat disturbance and parasite prevalence were examined using Spearman's rank-order correlation. There was a negative association between habitat disturbance and trypanosome prevalence among H. stella (r s = −0·880, P = 0·021). A Bonferroni correction was not applied to these analyses because of the increased criticism of its validity in ecological cases and the increased risk of a Type II error (Krasnov et al. Reference Krasnov, Korallo-Vinarskaya, Vinarski, Shenbrot, Mouillot and Poulin2008). But we cautiously interpreted our data in consideration of Type I error and the consideration that with Bonferroni correction these results would be considered insignificant despite the biological associations.
Table 2. Linear regression was used to investigate relationships between parasite prevalence and small mammal density, species richness and relative species abundance of the 3 most abundant rodent species (P. jacksoni, L. aquilus and H. stella). Additionally, correlation between habitat disturbance and parasite prevalence among these 3 host species was examined using Spearman's rank-order correlation test. Human settlements were omitted from the analysis of small host density.
Correspondence analysis was performed to consider the parasite communities (presence/absence) harboured by each individual. Using the parasite communities associated with each individual within the populations of P. jacksoni, L. aquilus and H. stella, we investigated the influence of habitat and/or taxonomic classification on the parasite community. We found that a host's parasite community was more closely associated with other individuals of that same host species than to other hosts within their same habitat but of various species. When the species was controlled and the habitat type was examined, no significant associations were identified (P = 0·784) (Fig. 2A). Alternatively, when habitat was controlled, the species was significantly clustered (P<0·0001) (Fig. 2B).

Fig. 2. (A) Correspondence analysis of habitats associated with KNP using parasite communities within each habitat harboured the most abundant rodent species – P. jacksoni, L. aquilus and H. stella. The 7 habitats are shown in the ordination plot represent a gradient of habitat disturbance (least to most disturbed is listed from top to bottom in the figure key) with each habitat represented by a specific colour indicated in the figure key and each point an individual. Some points are superimposed on each other. The correspondence analysis determined parasite communities on each individual small mammal were not associated with the habitat the host occupied. (B) Correspondence analysis of the parasite communities harboured by the 3 most abundant rodent hosts – P. jacksoni, L. aquilus and H. stella sampled from all habitats. Each species is represented by a specific colour indicated in the figure key and each point represents an individual. Ellipses represent 95% confidence interval of the species centroid, and non-overlapping ellipses are interpreted as significant differences between the species at α = 0·05. Some points are superimposed on each other. All 3 rodent species show distinct parasite community differences. The correspondence analysis determined parasite communities were significantly associated with taxonomic classification of their host species (P = 0·0001).
DISCUSSION
Generalized laws and theories, which are broadly applicable and repeatable under various conditions, were used to understand ecological interactions (Lawton, Reference Lawton1999; Lange, Reference Lange2005; Poulin, Reference Poulin2007). We investigated a broad range of habitat types that harbour diverse communities of host species as well as parasites. We focused our study on a single point in time and identified relatively a few patterns of parasitism among host species. In general, we did not identify any universal relationships or patterns. Despite the preliminary nature of our study, this work does provide additional empirical evidence for the importance of understanding host taxonomy and parasite specificity (Froeschke et al. Reference Froeschke, van der Mescht, McGeoch and Matthee2013).
Several patterns among the different species did emerge in this study, although at this point we can only speculate as to the dynamic drivers. Among the P. jacksoni population in forested Uganda, we found an association between trypanosomes and lice prevalence and increases in small mammal density. It is accepted that both lice and trypanosomes are moderately host-specific, usually infecting multiple hosts of restricted phylogenies or taxonomic classifications (Dobigny et al. Reference Dobigny, Poirier, Hima, Cabaret, Gauthier, Tatard, Costa and Bretagne2011; Froeschke et al. Reference Froeschke, van der Mescht, McGeoch and Matthee2013). Therefore, the association we found between parasite prevalence and total small mammal density (as opposed to relative abundance of P. jacksoni) may be associated with spill-over of trypanosomes and lice from other dominant host species. This pattern may also be indicative of a decline in host resistance in competitive environments (Johnson et al. Reference Johnson, Preston, Hoverman and Richgels2013). Interestingly, trypanosome prevalence among H. stella was associated with increases in relative abundance and habitat disturbance which may indicate H. stella as a potential primary native host for trypanosomes in addition to possible spill-over from other invasive host species in the disturbed habitats. One of the most common species of trypanosome-infecting rodents is Trypanosoma lewisi which is a globally invasive parasite of rodents and transmitted from invasive Rattus rattus into wild populations (Dobigny et al. Reference Dobigny, Poirier, Hima, Cabaret, Gauthier, Tatard, Costa and Bretagne2011). Lophuromys aquilus experienced a positive association of both trypanosomes and fleas and overall small mammal richness, which may compete with hypotheses that predict an increase in host species richness leads to declines in host-specific parasite prevalence. This relationship between trypanosomes and fleas among L. aquilus and overall small mammal richness may also be explained by spill-over if these parasites are less host-specific and have the ability to infect a wide range of hosts. Additionally, we found an association between mite prevalence on H. stella and relative abundance of H. stella, which is not a surprising association since mites are recognized as being predominately host-specific and we would suspect their infestation rate to increase as the available hosts increase (Fain, Reference Fain1994). We did not find any linear associations among ticks, Giardia or Cryptosporidium.
Our study highlights the importance of taxonomic classification of host species to understand their parasite communities, with environmental factors being a secondary influence on a host's parasite community. Our results support recent findings that relationships between biodiversity and pathogen transmission are idiosyncratic and highly dependent on the host species and parasites studied (Salkeld et al. Reference Salkeld, Padgett and Jones2013). It is possible that general patterns between parasite prevalence and host dynamics/environment would emerge in more simplistic community assemblages of small mammals experiencing less extensive habitat disturbance. Additionally, there are possible factors and interactions unaccounted for in our work, which may be more influential on parasite prevalence than those examined in our present study. Such factors in more disturbed habitats could likely include pathogen spill-over from human and domestic animal sources, the influence of invasive species (and their parasites), and alterations in individual host susceptibility to parasites (Daszak et al. Reference Daszak, Cunningham and Hyatt2000; Torchin et al. Reference Torchin, Lafferty, Dobson, McKenzie and Kuris2003; Dobson et al. Reference Dobson, Lafferty, Kuris, Hechinger and Jetz2008). General linear patterns associated with parasitism among different host communities are likely more easily identified when these communities examined maintain a certain level of shared species and experience a more gradual gradient of habitat disturbance. To more accurately investigate relationships between host diversity and parasite prevalence/richness in KNP, we would need to have a larger sampling effort with substantial replications over multiple sites and seasons. The sampling in this study was from 7 distinct habitats experiencing distinct small mammal communities with an absence of paired samplings. Replicate sites would need to first be identified and then sampled at various time points to more accurately answer questions related to the dilution effect and amplification of parasites in forested Uganda.
The coarseness of the parasite data limits our ability to identify the host species responsible for the maintenance of specific parasites within these communities and parasite–host specificity. Future studies could provide further taxonomic characterizations of these parasites to the species level. This finer examination of parasites, particularly the ectoparasites, would certainly identify even greater parasite richness and identify parasite–host dynamics specific to KNP (Alvarado-Otegui et al. Reference Alvarado-Otegui, Ceballos, Orozco, Enriquez, Cardinal, Cura, Schijman, Kitron and Gurtler2012; Salyer et al. Reference Salyer, Gillespie, Rwego, Chapman and Goldberg2012). Despite the coarseness of the data and the innate limitations, host taxonomy is still significant in determining the parasites an individual harbours. Further identification of parasite species will only strengthen this finding. Additionally, given recent identification of viral anti-bodies circulating in small mammals around KNP, future work using serologic assays would broaden our knowledge of disease life history for each individual small mammal (Salzer et al. Reference Salzer, Carroll, Rwego, Li, Falendysz, Shisler, Karem, Damon and Gillespie2013).
Ecosystems are complex with an intricate network of hosts and parasites. General patterns would be expected to be less common as complexity increases in these natural systems. In the absence of a larger sampling effort, this impact of habitat disturbance, host community density and species richness in Western Ugandan small mammal communities appear less influential in determining parasite prevalence and parasite community. Our data provide empirical evidence that within the small mammal communities in Western Uganda, very few patterns emerged to explain these ecological drivers of parasite prevalence. However, we did find strong associations between individual small mammal taxonomy and the community of parasites they harbour. This study provides further evidence that the parasite communities found in the mosaic landscape in Western Uganda are strongly influenced by the taxonomy of the dominant host species and the natural history of the parasites they harbour.
ACKNOWLEDGEMENTS
We thank the Uganda Wildlife Authority, Uganda National Council for Science and Technology, Makerere University Biological Field Station and local authorities for permission to conduct this study. IACUC committees at Emory University (#062-2009) and the CDC (#1768) approved all animal collection and handling protocols. Additionally, animals from human dwellings were collected with permission from local authorities (Local Chairman) and homeowners. Thanks to R. R. Lash for cartographic contributions and C. Akora and I. Mwesige who provided assistance in the field and K. Cross for assistance in the laboratory. We also thank R. Stevens, W. Stanley, J. Mills, A. Peralta, I. Damon and C. Hughes for helpful comments, logistical and/or analytical assistance. The views expressed in this paper are solely those of the authors and do not represent those of the CDC, US Government or any other entity which the authors may be affiliates. Lastly, we are very grateful to two anonymous reviewers for their helpful comments and expertise.
FINANCIAL SUPPORT
This research was supported in part by the Emory Global Health Institute, Emory University Environmental Sciences Department and the appointment of J.S.S. to the Research Participation Programme administered by Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement with CDC.