INTRODUCTION
Monogeneans of the family Diplozoidae are blood-feeding freshwater ectoparasites inhabiting the gills of cyprinid fishes. They can be significantly virulent to their hosts, causing mechanical damage to gill filaments accompanied by a risk of secondary bacterial and mycotic infections or hypochromic microcytic anaemia (Kawatsu, Reference Kawatsu1978; Buchmann and Brescini, Reference Buchmann and Brescini2006). The alimentary tract of these worms is morphologically adapted to blood uptake, although the mechanisms of blood processing in monogeneans are largely unknown. The foregut is composed of an oral opening with prominent buccal suckers, with an eversible pharynx that leads via an oesophagus into several dead-end side caeca (Smyth and Halton, Reference Smyth and Halton1983; Valigurová et al. Reference Valigurová, Hodová, Sonnek, Koubková and Gelnar2011; Konstanzová et al. Reference Konstanzová, Koubková, Kašný, Ilgová, Dzika and Gelnar2015). Both sections of the gut are morphologically and functionally well differentiated. Numerous gland cells with proposed extracellular digestive function open into the foregut lumen. The chemical nature of their secretion is unknown, but it has been expected that they may produce haemolysins or peptidases, thus could be involved in preliminary degradation of complex foodstuff (e.g., blood cells) to smaller components suitable for endocytosis (Smyth and Halton, Reference Smyth and Halton1983; Hodová et al. Reference Hodová, Matějusová and Gelnar2010; Valigurová et al. Reference Valigurová, Hodová, Sonnek, Koubková and Gelnar2011). The basic principles of the digestive process have been described for a few monogenean species and they were based mainly on ultrastructural and histochemical studies (Jennings, Reference Jennings1959; Tinsley, Reference Tinsley1973; Halton and Stranock, Reference Halton and Stranock1976; Hodová et al. Reference Hodová, Matějusová and Gelnar2010; Valigurová et al. Reference Valigurová, Hodová, Sonnek, Koubková and Gelnar2011; Konstanzová et al. Reference Konstanzová, Koubková, Kašný, Ilgová, Dzika and Gelnar2015). According to these records, the terminal phase of digestion occurs inside specialized types of gut cells within the lysosomal cycle (Smyth and Halton, Reference Smyth and Halton1983). From this view, the digestion in diplozoids is more similar to haematophagous mites such as ticks (Sonenshine, Reference Sonenshine1991), than to other blood-feeding platyhelmints with their extracellular digestion (Dalton et al. Reference Dalton, Skelly and Halton2004). The protein part of haemoglobin is broken down into peptides and amino acids (aa); the toxic haem is oxidized to haematin, exocytosed into the gut lumen and regurgitated by the worm through its oral opening (Llewellyn, Reference Llewellyn1954; Jennings, Reference Jennings1959; Smyth and Halton, Reference Smyth and Halton1983; Konstanzová et al. Reference Konstanzová, Koubková, Kašný, Ilgová, Dzika and Gelnar2015). Current knowledge of monogenean peptidases involved in digestion and other processes is poor. So far, only two reports have been published concerning peptidases of marine monogeneans Neobenedenia spp. feeding on epidermal tissues and mucus of their hosts. A gene encoding cysteine peptidase cathepsin L in Neobenedenia melleni was characterized and cloned (Rao and Yang, Reference Rao and Yang2007) and some activity of serine peptidases was found in homogenates of adult Neobenedenia girellae by zymography (Hirazawa et al. Reference Hirazawa, Umeda, Hatanaka and Kuroda2006). Their functions are unknown.
In general, processing of blood in sanguinivorous helminths and mites relies on an evolutionary conserved network of cysteine and aspartic peptidases (e.g., cathepsins B, L, F, C, legumain and cathepsin D) identified in trematodes – Schistosoma mansoni, Schistosoma japonicum, Fasciola hepatica (Caffrey and Ruppel, Reference Caffrey and Ruppel1997; Dalton et al. Reference Dalton, Neill, Stack, Collins, Walshe, Sekiya, Doyle, Mulcahy, Hoyle, Khaznadji, Moire, Brennan, Mousley, Kreshchenko, Maule and Donnelly2003; Sajid et al. Reference Sajid, McKerrow, Hansell, Mathieu, Lucas, Hsieh, Greenbaum, Bogyo, Salter, Lim, Franklin, Kim and Caffrey2003; Caffrey et al. Reference Caffrey, McKerrow, Salter and Sajid2004; Delcroix et al. Reference Delcroix, Sajid, Caffrey, Lim, Dvořák, Hsieh, Bahgat, Dissous and McKerrow2006; Robinson et al. Reference Robinson, Dalton and Donnelly2008), nematodes – Haemonchus contortus, Ancylostoma caninum, Necator americanus (Williamson et al. Reference Williamson, Brindley, Knox, Hotez and Loukas2003a , Reference Williamson, Lecchi, Turk, Choe, Hotez, McKerrow, Cantley, Sajid, Craik and Loukas2004), and in ticks – Ixodes ricinus, Boophilus microplus (Renard et al. Reference Renard, Garcia, Cardoso, Richter, Sakanari, Ozaki, Termignoni and Masuda2000; Sojka et al. Reference Sojka, Franta, Horn, Hajdušek, Caffrey, Mareš and Kopáček2008). We suppose that similar molecular tools for protein hydrolysis are employed inside the digestive tract of monogeneans.
Our study represents the first direct evidence of the presence of acid endopeptidases in a haematophagous monogenean – Eudiplozoon nipponicum; these were identified and partially characterized by biochemical, proteomic and molecular methods.
METHODS
Parasites
Living adult worms of E. nipponicum were carefully removed from gills of common carps (Cyprinus carpio) immediately after slaughter in a commercial facility of Rybářství Třeboň, Plc. The fish originated from localities (ponds) in South Bohemia, Czech Republic.
Sample preparation
Freshly collected worms were repeatedly gently washed in sterilized tap water, placed in Eppendorf tubes, frozen immediately on dry ice and stored at −80 °C. Soluble protein extracts (solPE) were prepared by homogenization of worms in 10 mm acetate buffer pH 5 using motor-driven teflon pellet pestle followed by 3 cycles of sonication on ice (10 W, 30 s) and centrifugation at 16 000 g for 20 min. Supernatants were collected. Excretory/secretory products (ESP) were obtained by incubation of washed adult live worms in 10 mm phosphate buffer pH 7·4 or 10 mm acetate buffer pH 6 for 3–4 h at room temperature (RT) in Eppendorf tubes. The ability of the ectoparasitic worms to survive for several hours in selected low osmolarity buffers was verified by previous overnight incubation. Full mobility and no obvious damage of the worms were observed. ESP were concentrated on Amicon Ultra filters (MWCO 10 kDa; Millipore). Protein concentration in all samples was measured by Quant-iT Protein Assay Kit (Invitrogen). The samples were stored at −80 °C.
Living adult worms were washed several times in sterile tap water and then placed in TRIzol® Reagent (Invitrogen) for stabilization of RNA and processed or stored at −80 °C. After homogenization, total RNA was extracted following the TRIzol protocol and concentration measured in NanoDrop 1000 (Thermo Scientific). First-strand cDNA synthesis was carried out with 0·3 µg of total RNA using oligo-dT18 primer from SuperScript™ III First-Strand Synthesis kit (Invitrogen).
Peptidolytic activities, pH optima and inhibition assays
Enzyme activities in solPE and ESP were measured with a set of synthetic fluorogenic peptide substrates (Bachem): Z-Phe-Arg-AMC (FR) was used for detection of papain-like cysteine peptidases (acidic pH) and trypsin-like serine peptidases (neutral to basic pH), Z-Arg-Arg-AMC (RR) selectively for cathepsin B and Z-Ala-Ala-Asn-AMC (AAN) for legumain (asparaginyl endopeptidase). Abz–Lys–Pro–Ala–Glu–Phe–Nph–Arg–Leu (KPAEFnFRL), the specific substrate for cathepsin D, was kindly provided by Dr. Martin Horn, Institute of Organic Chemistry and Biochemistry, AS CR Prague (Abz = 2-aminobenzoyl; Nph and nF = nitrophenylalanine). All assays were performed in black 96-well flat bottom plates (Nunc, Thermo Scientific). Samples (1 and 5 µg of protein per well of solPE and ESP, respectively) were pre-incubated (5 min, RT) in various buffers of pH in the range 2–10 for substrates FR, RR, AAN and pH 2–6 for KPAEFnFRL (0·1 m phosphate buffer pH 2; 0·1 m citrate pH 3; 0·1 m citrate/0·2 m phosphate pH 4–7; 0·2 m Tris–HCl pH 8; 0·2 m glycine/NaOH pH 9–10). Final volume was 100 µL. All buffers contained 2 mm dithiothreitol (DTT, Sigma-Aldrich), except for KPAEFnFRL substrate. The reactions were started by addition of particular peptidyl substrate (final concentration 50 µ m) in 100 µL of the same buffer. Kinetics of the release of free fluorophors was detected by Infinite M200 flurometer (TECAN) at 28 °C for 60 min in 1 min intervals. Excitation/emission wavelengths for AMC substrates and nF substrate were 355/460 nm and 330/410 nm, respectively. Controls contained equal volume of appropriate buffer instead of the sample. Measurements were performed in triplicates. The quantification of cysteine and aspartic peptidase activities was performed in particular pH optima with FR and KPAEFnFRL substrates, respectively. Results were expressed as nm of substrate cleaved per minute in the presence of 1 µg of solPE total protein.
Peptidase inhibitors (50 µ m final concentration, Sigma-Aldrich) were used for inhibition of particular peptidase activities: E-64 (L-trans-epoxysuccinyl–leucylamido [4-guanidino] butane), an irreversible inhibitor of papain-like cysteine peptidases; CA-074 (N-(L-3-trans-propylcarbamoyloxirane-2-carbonyl)-L-isoleucyl-L-proline]), an irreversible inhibitor of cathepsin B; iCL (Arg–Lys–Leu–Leu–Trp–NH2), a reversible inhibitor of cathepsin L; pepstatin A (isovaleryl–Val–Val–Sta–Ala–(3S,4S)-4-amino-3-hydroxy-6-methylheptanoic acid), inhibitor of aspartic peptidases. Inhibitors were mixed with samples prior to the addition of substrates and incubated for 15 min. Inhibition tests were performed in pH optimum of peptidase activity for each particular substrate. All measurements were performed in triplicates, repeated at least three times. Values in graphs are expressed as means with standard deviations.
Degradation of haemoglobin
Bovine haemoglobin (10 µg, Sigma Aldrich) diluted in 20 µL of particular buffers in the range of pH 3-6 (see above) containing 2 mm DTT was incubated with 3 µg of ESP or 1·5 µg of solPE proteins for 16 h at 30 °C. Control reactions contained only haemoglobin. The contribution of individual aspartic or cysteine peptidase activities to degradation of haemoglobin was evaluated by inhibition assays at pH 3 and pH 5. Pepstatin, E-64, CA-074 and iCL were used at concentrations of 10 µ m. Resulting hydrolysates were mixed with reducing electrophoretic sample buffer and separated by SDS–PAGE in 4–15% gradient precast gels (Bio-Rad) which were stained with Coomassie Brilliant Blue R-250 (CBB) and scanned on GS-800 Calibrated Densitometer (Bio-Rad).
Active site-labelling of cysteine peptidases
Fluorescent Green BODIPY-DCG-04 (Greenbaum et al. Reference Greenbaum, Baruch, Hayrapetian, Darula, Burlingame, Medzihradszky and Bogyo2002), an analogue of E-64 inhibitor which binds irreversibly to the active site of papain-like peptidases, was incubated with ESP (15 µg of total protein) for 1 h (20 µ m DCG-04, 5 mm DTT, 5 mm MgCl2) at RT in the dark. Controls were incubated for 30 min with cysteine peptidase inhibitors (10 µ m) E-64, iCL or CA-074 prior to addition of the probe. SDS–PAGE of labelled samples was performed as described above. Fluorescent signal was recorded on a fluorescence scanner (Molecular Imager FX, Bio-Rad) using excitation/emission wavelengths 488/530 nm. Finally, the gels were stained by Silver Stain Plus™ Kit (Bio-Rad).
Amplification of partial DNA sequences of cysteine peptidases
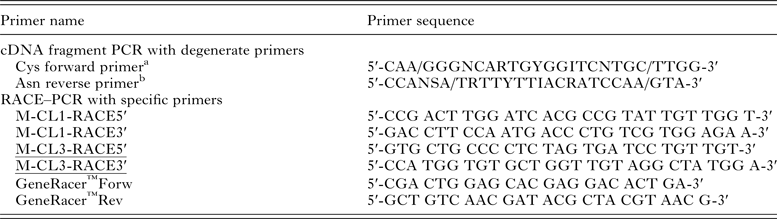
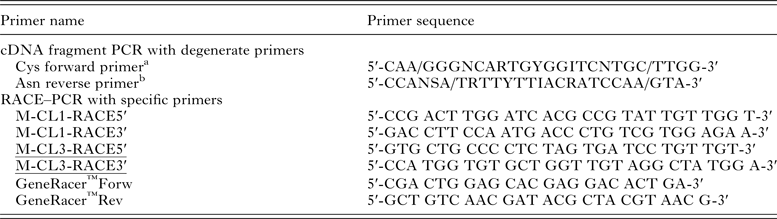
cDNA obtained by reverse transcription from adult E. nipponicum was amplified by PCR using PPP Master Mix (Top-Bio) and degenerate primers (Table 1) designed according to the consensus sequences corresponding to the coding sequences of cysteine peptidases of several parasitic species (especially according to the active site with a cysteine residue – Cys forward primer and with an asparagine residue – Asn reverse primer) (Eakin et al. Reference Eakin, Bouvier, Sakanari, Craik and McKerrow1990; Heussler and Dobbelaere, Reference Heussler and Dobbelaere1996; Li et al. Reference Li, Moon, Park, Na, Hwang, Oh, Cho, Kong, Kim and Chung2006). The PCR protocol was as follows: one cycle 5 min initial denaturation at 94 °C, then 35 cycles of denaturation at 94 °C for 1 min, primer annealing 45 °C for 1 min, extension at 72 °C for 1 min and finally one cycle 10 min final extension at 72 °C. Control reactions were performed using the same primers and cDNA obtained from gills of a non-infected carp. The amplified gene fragments of the expected length (ca. 500 bp) were electrophoresed and isolated from agarose gel (1·5%) with Gel Extraction Kit (Qiagen), sub-cloned into pCR2·1-TOPO cloning vector (Invitrogen) and transformed into TOP10 E. coli (Invitrogen). pCR2·1-TOPO constructs were isolated using Qiaprep Purification Kit (Qiagen) and sequenced with M13 forward and reverse primers (DNA Sequencing Laboratory, Faculty of Science, Charles University in Prague). Obtained partial DNA sequences were compared with NCBI database using BLASTX (http://blast.ncbi.nlm.nih.gov/).
Table 1. Sequences of primers used for amplification of E. nipponicum cathepsin L genes.
a TGC nucleotide triplet encoding cysteine.
b RTT nucleotide triplet encoding asparagine.
Rapid amplification of cDNA ends (RACE–PCR)
In order to amplify the 5′/3′ ends and to obtain the entire gene sequences, two pairs of gene-specific primers (M-CL1-RACE5′ + M-CL1-RACE3′, M-CL3-RACE5′ + M-CL3-RACE3′; see Table 1) were designed according to the obtained ca 500 bp sequence fragments (see above). First-strand cDNA for RACE prepared according to manufacturer′s instructions of GeneRacer™Kit (Invitrogen) was used as a template. The PCR reaction (25 µL) contained 12·5 µL of the EmeraldAmp PCR master mix (Clontech), 1 µL of the RACE cDNA (100 ng µL−1), 1 µL of the M-CL1/3-RACE5′/3′ (10 µ m), 3 µL of the GeneRacer primers Fwd/Rev (10 µ m), 7·5 µL of ddH20. The profile of amplification of 5′/3′ ends was: 1× 94 °C = 120 s, 5 × (94 °C = 30 s + 72 °C = 60 s), 5 × (94 °C = 30 s + 70 °C = 60 s), 35 × (94 °C = 30 s + 60 °C = 90 s + 70 °C = 90 s) and finally 1 × 72 °C = 10 min. PCR products from 5′ and 3′ RACE were cloned into pCR2·1-TOPO cloning vector as described above and submitted for sequencing with M13 forward and reverse primers (DNA Sequencing Laboratory, Faculty of Science, Charles University in Prague).
Sequence analysis
The obtained full-length cDNA sequences were blasted against sequences available in the GenBank database (NCBI) by BLASTX (http://blast.ncbi.nlm.nih.gov/). The presence of a signal sequence was predicted by the SignalP 4·1 Server (http://www.cbs.dtu.dk/services/SignalP/). The theoretical position of a pro-region was determined by multiple sequence alignment of cathepsins L from other organisms: human cathepsin L [GenBank: NP_001903·1], cathepsin L of Clonorchis sinensis [GenBank: ABK91809·1], cathepsin L of C. carpio [GenBank: BAD08618·1] and cathepsin L of Neobenedenia melleni [GenBank: ABK62794·1]. Multiple sequence alignments of aa sequences were carried out using the program Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Molecular mass and theoretical pI of the deduced proteins were determined by the Compute pI/Mw software available at the Expasy site (http://www.expasy.ch/tools/pitool.html). Detection of potential N-glycosylation sites was performed using an online tool at NetNGlyc 1·0 Server (http://www.cbs.dtu.dk/services/NetNGlyc).
Protein identification from 1D gel bands by LC–MS/MS
ESP were separated by SDS–PAGE as described above and stained by CBB. Bands (range 25–40 kDa) were manually excised, destained and incubated each with trypsin (Promega). LC–MS/MS analyses were done using RSLCnano system (Thermo Fisher Scientific) on-line connected to Impact II Q-TOF mass spectrometer with CaptiveSpray nanoBooster ion source (Bruker). Prior to LC separation, tryptic digests were online concentrated and desalted using trapping column (100 µm × 30 mm) filled with 3·5 µm X-Bridge BEH 130 C18 sorbent (Waters). After washing of trapping column (0·1% FA), the peptides were separated (300 nl/min) using Acclaim Pepmap100 C18 column (2 µm particles, 75 µm × 500 mm; Thermo Fisher Scientific) by the 0·1% FA in water : 0·1% FA in 80% ACN gradient program.
MS data were acquired in a data-dependent strategy with 3 s long cycle time. Mass range was set to 150–2200 m/z and precursors were selected from 300 to 2000 m/z. Acquisition speed of MS and MS/MS scans was 2 and 4–16 Hz, respectively. Speed of MS/MS spectra acquisition was based on precursor intensity (low and high absolute thresholds were 10 000 and 100 000 cts, respectively). Mascot (version 2.4·1) MS/MS ion searches were done against in-house database with sequences of expected recombinant proteins. Search results obtained against in-house database were checked for false positive identifications using database search against the whole SwissProt database (version 2014_07). Mass tolerances for peptides and MS/MS fragments were 15 ppm and 0·05 Da, respectively. Oxidation of methionine, deamidation (N, Q) and propionamide (C) as variable modifications and three enzyme miss cleavages were set for all searches.
RESULTS
Activity profiling
The results of fluorometric analyses of peptidolytic activities in solPE and in ESP, pH optima and inhibition tests are shown in Fig. 1. The highest activity in both sample types was attributed to papain-like cysteine peptidases in acid buffers of pH 3–5 (optimum pH 5) with FR substrate (Fig. 1A and B). It dropped down to only ca. 20% in pH 6 and vanished above pH 7. Inhibitors E-64 and iCL suppressed the activity to zero and CA-074 by ca. 25 and 80% in the case of solPE and ESP, respectively (Fig. 1a and b).

Fig. 1. The pH profiles and inhibition assays of acid endopeptidase activities. The pH optima of peptidolytic activities in soluble protein extracts (solPE) and E/S products (ESP) of E. nipponicum and their inhibition at corresponding pH optimum were measured with a spectrum of fluorogenic substrates: (A, a, B, b) Z-FR-AMC; (C, c, D, d) Z-RR-AMC; (E, e) KPAEF-nFRL and with selective peptidase inhibitors (E-64, CA-074, iCL, pepstatin A). The values are expressed as percentage of maximum activity in the sample (100% at pH optimum).
With cathepsin B-selective substrate RR, the activity in solPE peaked in pH 5 and it was negligible in pH 6 and above (Fig. 1C). However, in ESP the highest activity appeared in pH 6, dropped to ca. 50% in pH 7 (Fig. 1D). In the case of both sample types, E-64 and iCL inhibited the peptidolytic activities nearly to zero. CA-074 reduced the activities in ESP by ca. 75% and by ca. 95% in solPE (Fig. 1c and d).
The activity of aspartic cathepsin D with was found in the range of pH 2–4 (optimum in pH 3) in solPE of worms in the presence of KPAEFnFRL (Fig. 1E); this was inhibited by ca. 90% by pepstatin A (Fig. 1e). In ESP, the activity of cathepsin D was recorded occasionally in some samples (not shown). The ratio of cathepsin L + B to cathepsin D activity in solPE was approx. 4:3 (0·45 vs 0·33 nm min−1 µg−1 of protein). No activity of cysteine asparaginyl endopeptidase (legumain) was detected with AAN substrate in any sample (not shown). There was no difference in activity profiles between the samples of ESP collected at pH 7·4 and pH 6 (not shown). Therefore, a pooled mixture of samples was used for the experiments.
Degradation of haemoglobin
ESP as well as solPE efficiently degraded bovine haemoglobin during overnight (16 h) incubation within the pH range of 3–5 and less at pH 6 (Fig. 2A). No substantial degradation occurred above pH 7 (not shown). At pH 5 and 3, haemoglobinolytic activity was markedly (but not completely) inhibited by the general inhibitor of papain-like cysteine peptidases E-64. Lower level of inhibition was observed with CA-074, iCL and pepstatin alone; the latter two were less effective at pH 3 in the case of ESP. Complete inhibition at both pH values was reached only when using a mixture of E-64 and pepstatin (Fig. 2B and C). No marked differences in inhibition profiles were observed between the samples containing ESP and solPE.

Fig. 2. The pH profile and inhibition of haemoglobin degradation by ESP and solPE. Electrophoretograms of bovine haemoglobin (Hb) after overnight incubation with E/S products and soluble protein extracts of E. nipponicum. (A) Degradation in the presence of ESP (lanes E3-E6) and solPE (H3-H6) at pH values 3-6. Controls of Hb in particular pH (C3-C6) contained neither ESP nor solPE. (B) Inhibition of haemoglobinolysis in the presence of ESP in pH 5 and pH 3. (C) Inhibition of heamoglobinolysis in the presence of solPE in pH 5 and pH 3. Headings above each pair of lanes (left lane = pH 5, right lane = pH 3) indicate the type of sample: haemoglobin = controls of Hb without ESP or solPE; no inhibitor = controls of Hb with ESP or solPE; other samples contained inhibitors as labelled. M = markers of molecular size. Bands of Hb monomers are boxed in grey. Arrowhead points to alpha/beta heterodimers of Hb.
Active site-labelling of cysteine peptidases
Using the irreversible affinity probe DCG-04, the presence of papain-like cysteine peptidases was demonstrated in the samples of ESP. After SDS–PAGE, three protein bands of molecular sizes between 25 and 35 kDa were demonstrated in the gels. The interaction of DCG-04 with the active sites of cysteine peptidases was efficiently inhibited by the general inhibitor of cysteine peptidases (E-64) and by the inhibitor of cathepsin L (iCL), and only partially by the inhibitor of cathepsin B (CA-074) (Fig. 3).

Fig. 3. Active site-labelling of papain-like peptidases in ESP by DCG-04 affinity probe. Electrophoretogram of ESP incubated with fluorescent DCG-04 and inhibitors of particular cysteine peptidases; signal (lanes 1–4) was recorded on a fluorescence scanner, excitation/emission wavelengths 488/530 nm. Lane 1 - control without any inhibitor; lane 2 - E-64; lane 3 - iCL; lane 4 - CA-074; lane 5 - silver staining of the gel. Arrowheads point to the three protein bands labelled by the probe. M = markers of molecular size.
Amplification of E. nipponicum cathepsin L genes
Two amplified DNA fragments (ca. 500 bp) obtained by PCR using E. nipponicum cDNA as a template and primers shown in Table 1 were identified as partial sequences of cathepsin L-like cysteine peptidases. In control PCR reactions with DNA template from a non-infected carp and with the same set of primers, a sequence encoding cathepsin L was also amplified. This showed 94·8% sequence identity with cathepsin L gene from C. carpio [GenBank: BAD08618·1] and only 55·19 and 49·35% with the fragments amplified from cDNA of E. nipponicum.
Applying RACE–PCR on the basis of fragments mentioned above, whole sequences of two E. nipponicum cathepsin L genes were obtained, termed EnCL1 and EnCL3. Their nucleotide sequences have been deposited in the NCBI GenBank database under accession numbers [GenBank: KP793605] and [KP793606]. The EnCL1 gene has an open reading frame (ORF) of 954 bp encoding a proenzyme consisting of 317 aa (Fig. 4). Deduced propeptide region contains 93 aa and the catalytic domain (mature enzyme) 224 aa residues. No signal leader sequence has been identified. The calculated theoretical molecular weights of the zymogen/mature protein are 35/24·4 kDa and pI 5·86/6·08. EnCL1 exhibits the highest similarity (53%) to cathepsin L from the Chinese liver fluke Clonorchis sinensis [GenBank: ABK91809·1]. The second obtained cathepsin L sequence from E. nipponicum was termed EnCL3, as it has the greatest similarity to cathepsin L3 precursor of Schistosoma mansoni [GenBank: ABV71063·1]. The ORF of EnCL3 is made up of 1107 bp and encodes a pre-proenzyme of 368 aa (Fig. 4). The sequence is composed of a signal leader sequence (24 aa), an unusually long propeptide (120 aa) and a mature (catalytic) domain (224 aa). Expected Mr/pI values of the zymogen and mature enzyme are 38·0/4·8 and 24·1/4·15, respectively.

Fig. 4. Multiple sequence alignment of Eudiplozoon nipponicum cathepsins L with orthologs from other organisms. Neobenedenia melleni [GenBank:ABK62794·1]; Cyprinus carpio [GenBank:BAD08618·1]; Schistosoma mansoni CL3 [GenBank:ABV71063·1]; Clonorchis sinensis [GenBank:ABK91809·1]; Eudiplozoon nipponicum CL1 [GenBank:KP793605]; E. nipponicum CL3 [GenBank:KP793606]. The position of pro-region cleavage site is marked by arrowheads. ERFNIN- and GNFD-like motifs are in bold and indicated by underlined headings. Catalytic triad of the active site (C, H, N) is marked by arrows. Conserved motifs around active site residues are shaded in light grey. Tripeptides of potential N-glycosylation sites are boxed. Amino residue (A) at the base of the S2 pocket is boxed in black.
The conserved ‘ERFNIN’ motif typically present in prosequences of various papain-like cysteine peptidases, e.g. cathepsins L, K, S, but not in cathepsin B (Karrer et al. Reference Karrer, Peiffer and DiTomas1993), was found in EnCL3; this was slightly modified in the case of EnCL1 (ERFNVN). Another sequence motif, ‘GNFD’, that may be involved in intra-cellular trafficking and intramolecular processing of some papain-like cysteine peptidases (Vernet et al. Reference Vernet, Berti, de Montigny, Musil, Tessier, Ménard, Magny, Storer and Thomas1995; Dvořák et al. Reference Dvořák, Mashiyama, Sajid, Braschi, Delcroix, Schneider, McKerrow, Bahgat, Hansell, Babbitt, Craik, McKerrow and Caffrey2009), is modified in the prosequences of EnCL1 and EnCL3 to ‘ANLD’ and ‘TNFD’, respectively. The catalytic domains of both enzymes include the typical catalytic triad (EnCL1/3 numbering: Cys118, His264, N284/Cys169, His314, N335) of papain-like cysteine peptidases and the relatively conserved flanking sequences around the catalytic residues (QGQCGSCWAFS, LD HA/GVL, YWIVKNS/TW) (Turk et al. Reference Turk, Turk and Turk2000) (Fig. 4). No potential N-linked glycosylation sites have been found throughout the sequences of both proenzymes.
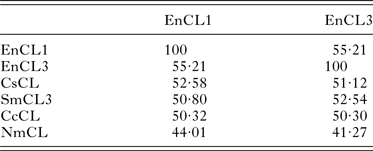
The sequence identity between EnCL1 and EnCL3 is 55·21% on the aa level. Identities among E. nipponicum cathepsins L and selected aa sequences of cathepsins L from other organisms are summarized in Table 2. EnCL1 shares the highest number of identical aa positions (52·58%) with cathepsin L of C. sinensis [GenBank: ABK91809·1] and EnCL3 with cathepsin L3 of S. mansoni [GenBank: ABV71063·1] (52·54%). Identities of EnCL1 and EnCL3 with the only published whole sequence of cathepsin L from a monogenean, N. melleni, [GenBank: ABK62794·1] are only 44·01 and 41·27%, respectively.
Table 2. Amino acid identities between cathepsins L of E. nipponicum and orthologues from other organisms.
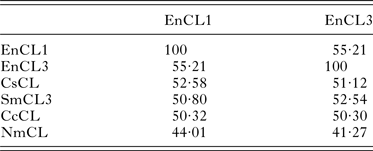
Identities expressed in [%]. EnCL1, Eudiplozoon nipponicum [GenBank:KP793605]; EnCL3, E. nipponicum [GenBank:KP793606]; CsCL, Clonorchis sinensis [GenBank:ABK91809·1]; SmCL3, Schistosoma mansoni [GenBank:ABV71063·1]; CcCL, Cyprinus carpio [GenBank:BAD08618·1]; NmCL, Neobenedenia melleni [GenBank:ABK62794·1].
Mass spectrometry analysis of ESP of E. nipponicum
Both cathepsins L1 and L3, the genes of which have been amplified, were confirmed in ESP of adult E. nipponicum by mass spectrometry. Fifteen peptide sequences from trypsin-digested protein bands matched to the sequence of EnCL1 [GenBank: KP793605] and three peptides matched to EnCL3 [GenBank: KP793606] (Fig. 5). EnCL1 peptides were identified in nine of ten bands between ca. 24–35 kDa; EnCL3-derived peptides were recorded only above 35 kDa.

Fig. 5. Identification of cathepsins L1 and L3 in ESP of E. nipponicum by mass spectrometry. Each grey line corresponds to a single identified peptide; the sequences of some peptides partially overlap with others. (A) Fifteen tryptic peptides identified in ESP matched to the sequence of cathepsin L1 [GenBank: KP793605]. (B) Three peptides matched to the sequence of cathepsin L3 [GenBank: KP793606].
DISCUSSION
In contrast to the comprehensive knowledge concerning peptidases from various groups of blood-feeding parasites, only very limited information is available about proteolytic enzymes of haematophagous monogeneans. These neglected helminths in terms of biochemical studies have been thought to digest haemoglobin and other proteins intracellularly in the acidic lysosomes/phagolysosomes within the specialized types of gut cells, unlike other blood-feeding helmints such as schistosomes (Dalton et al. Reference Dalton, Skelly and Halton2004), but similarly to ticks (Smyth and Halton, Reference Smyth and Halton1983; Sonenshine, Reference Sonenshine1991). However, it is still not clear whether the first phase of digestion in monogeneans may take place in the extracellular space of the lumen of the gut, with the participation of secreted proteases.
We aimed to identify and partially characterize dominant cysteine and aspartic endopeptidases which can be potentially involved in blood digestion or other processes in the adults of E. nipponicum. solPE of worms and ESP were compared in terms of peptidolytic activities. We expected that ESP may be a potential source of peptidases released into the gut lumen (either from gut epithelium or accessory glands that are opened into the front part of the digestive system), and from there excreted outside the worm by continuous regurgitation of the rests of digestion (Smyth and Halton, Reference Smyth and Halton1983; Hodová et al. Reference Hodová, Matějusová and Gelnar2010; Valigurová et al. Reference Valigurová, Hodová, Sonnek, Koubková and Gelnar2011). However, the presence of peptidases originated in other tissues of the worm cannot be excluded, especially in the case of solPE from whole worms.
Activity profiling revealed that the major peptidase activity can be attributed to the cysteine class, clan CA, family C1 (MEROPS classification). This activity tested with Z-FR-AMC as a substrate at pH 5 was fully inhibited by E-64 (general inhibitor of papain-like cysteine peptidases) and the iCL, but only partially by an inhibitor specific for cathepsin B (CA-074). It is known, that in a reducing environment (presence of DTT), cathepsin L can also be inhibited by CA-074 (Steverding, Reference Steverding2011; Steverding et al. Reference Steverding, Sexton, Wang, Gehrke, Wagner and Caffrey2012). Besides, although iCL (the pentapeptide RKLLW) has been stated as a highly potent iCL (Brinker et al. Reference Brinker, Weber, Stoll, Voigt, Müller, Sewald, Jung, Wiesmüller and Bohley2000), it was able to diminish significantly the activity of recombinant cathepsin B2 of Trichobilharzia regenti in an unrelated previous experiment (Jedličková, unpublished). With the substrate Z-RR-AMC, which is often considered as specific for cathepsin B, the relatively high peptidase activity in ESP was also fully inhibited by E-64 and iCL, but only partially by CA-074 (at pH 6). This brought some confusion on the interpretation of the relative proportion of cathepsin L/B activities.
Cathepsin B- and cathepsin L-like peptidases have been usually distinguished by their ability to degrade the substrate with an arginine in P2 position. Human cathepsin B is able to hydrolyse the substrate, whereas cathepsin L is not – a situation which is not always applicable to orthologous enzymes in other organisms (Sajid and McKerrow, Reference Sajid and McKerrow2002; Choe et al. Reference Choe, Leonetti, Greenbaum, Lecaille, Bogyo, Brömme, Ellman and Craik2006). The substrate specificity of cysteine cathepsins is thought to be determined by interactions in the S2 pocket, particularly the glutamic acid residue in cathepsin B (Glu205 in human CB) and the equivalent alanine residue in cathepsin L (Ala205 in human CL) localized at the bottom of the S2 pocket. The Glu residue can accommodate and stabilize the polar guanidino group of Arg in the substrate, but Ala in this position cannot bind to Arg (Sajid and McKerrow, Reference Sajid and McKerrow2002). This was confirmed for mammalian cathepsins L/B and for peptidases of numerous organisms (including parasites). However, many other parasite peptidases do not possess ‘mammalian’ residues in this position. For example, the ability to cleave Z-RR-AMC was described for cathepsin L-like peptidase ‘cruzain’ from Trypanosoma cruzi (Gillmor et al. Reference Gillmor, Craik and Fletterick1997) and cathepsin L of Entamoeba histolytica (Brinen et al. Reference Brinen, Que, McKerrow and Reed2000). On the other hand, cathepsin B from Leishmania major (Chan et al. Reference Chan, Selzer, McKerrow and Sakanari1999) and cathepsin B1·4 from T. regenti (Dvořák et al. Reference Dvořák, Delcroix, Rossi, Vopálenský, Pospíšek, Šedinová, Mikeš, Sajid, Sali, McKerrow, Horák and Caffrey2005) cannot hydrolyse this ‘typical’ cathepsin B substrate. According to the sequence data of EnCL1 and EnCL3, cathepsins L identified in the ESP of E. nipponicum have Ala (Ala234 in EnCL1, Ala285 in EnCL3) at the bottom of the S2 pocket, and therefore should likely not be able to cleave substrates with Arg in P2 position; however, in the light of the facts mentioned above, this must be verified experimentally with recombinant enzymes.
Cysteine peptidases in both solPE and ESP of E. nipponicum exhibited the greatest activity at pH 5 with the substrate Z-FR-AMC; in the presence of Z-RR-AMC the optima were pH 5 and pH 6 for solPE and ESP, respectively. The latter discrepancy could be caused by the presence of different other peptidases in solPE which is a more complex sample in terms of protein composition in comparison to ESP. The results corresponded, e.g. to pH optima measured for cysteine peptidases in secretions of adult S. mansoni (Dalton et al. Reference Dalton, Clough, Jones and Brindley1996); it has been suggested that the activity against Z-FR-AMC at lower pH is predominantly due to cathepsin L, while cathepsin B activity is less significant under these conditions. The presence of at least two different cathepsins L in our samples might explain the high activity in a broader range of acidic pH 3–5. Similarly to our results, the complex sample of protein extract from the gut of I. ricinus ticks showed the activity in the presence of the Z-FR-AMC at acidic pH 4-6 (Horn et al. Reference Horn, Nussbaumerová, Šanda, Kovářová, Srba, Franta, Sojka, Bogyo, Caffrey, Kopáček and Mareš2009). The fact that this substrate was not hydrolysed at pH > 7 suggests that trypsin-like serine peptidases were not present in the samples.
Summarizing the results, we dare to assume that the majority of cysteine peptidase activity in the samples was of cathepsin L-like nature. In addition, a smaller proportion of the activity was cathepsin B-like and sensitive to inhibition by iCL.
To verify the presence of other peptidases in solPE and ESP, substrates suitable for detection of cysteine asparaginyl endopeptidase (legumain) and aspartic cathepsin D were used. Surprisingly, no activity of legumain was detected. Legumain has been found in other blood-feeding helminths as well as ticks (Dalton et al. Reference Dalton, Hola-Jamriska and Brindley1995; Caffrey et al. Reference Caffrey, Mathieu, Gaffney, Salter, Sajid, Lucas, Franklin, Bogyo and McKerrow2000; Oliver et al. Reference Oliver, Skuce, McNair and Knox2006; Sojka et al. Reference Sojka, Hajdušek, Dvořák, Sajid, Franta, Schneider, Craik, Vancová, Burešová, Bogyo, Sexton, McKerrow, Caffrey and Kopáček2007; Abdul et al. Reference Abdul, Tsuji, Miyoshi, Khyrul Islam, Huang, Motobu and Fujisaki2007). Its function is connected not only with hydrolysis of ingested host proteins, but it also plays an important role in activation of other peptidases by cleaving their pro-sequences (Sajid et al. Reference Sajid, McKerrow, Hansell, Mathieu, Lucas, Hsieh, Greenbaum, Bogyo, Salter, Lim, Franklin, Kim and Caffrey2003; Caffrey et al. Reference Caffrey, McKerrow, Salter and Sajid2004; Sojka et al. Reference Sojka, Franta, Horn, Hajdušek, Caffrey, Mareš and Kopáček2008). There is a certain possibility that this enzyme occurred in our samples as an inactive zymogen or an inactive variant. Significant activity of cathepsin D (ca. ¾ of that of cathepsins L + B) was observed in solPE, whereas only a minute activity was usually recorded in some samples of ESP (not shown). Also cathepsin D acts in the digestive process in other blood-feeding parasites (Brindley et al. Reference Brindley, Kalinna, Wong, Bogitsh, King, Smyth, Verity, Abbenante, Brinkworth, Fairlie, Smythe, Milburn, Bielefeldt-Ohmann, Zheng and McManus2001; Verity et al. Reference Verity, Loukas, McManus and Brindley2001; Banerjee et al. Reference Banerjee, Liu, Beatty, Pelosof, Klemba and Goldberg2002; Williamson et al. Reference Williamson, Brindley, Abbenante, Datu, Prociv, Berry, Girdwood, Pritchard, Fairlie, Hotez, Zhan and Loukas2003b ; Boldbaatar et al. Reference Boldbaatar, Sikalizyo Sikasunge, Battsetseg, Xuan and Fujisaki2006; Sojka et al. Reference Sojka, Franta, Frantová, Bartošová, Horn, Váchová, O'Donoghue, Eroy-Reveles, Craik, Knudsen, Caffrey, McKerrow, Mareš and Kopáček2012) and we expect its involvement in blood digestion by monogenean parasites, too. The non-presence or low abundance of activities of these two crucial enzymes in some samples could be also possibly explained by alleged instability of their molecules and low production into the gut lumen.
In vitro degradation of haemoglobin by E. nipponicum peptidases is optimal at acidic pH, which corresponds with the pH optima of cysteine and aspartic peptidases. The reaction was completely blocked by a mixture of E-64 and pepstatin, less by E-64 and only partially by individual inhibitors. These results clearly confirmed that cysteine cathepsins L + B and aspartic cathepsin D are involved in haemoglobinolysis in E. nipponicum.
Affinity labelling with the active-site specific probe DCG-04 confirmed active cysteine peptidases in ESP, proved in three bands (between ca. 25–37 kDa), indicating the presence of various cysteine peptidases or intermediates of their activation by limited proteolysis (the sizes in gel approximately agree with theoretical MWs of proenzymes and mature parts of EnCL1 and EnCL3). The binding of the probe to all three bands was fully inhibited by both E-64 and iCL, whereas CA-074 inhibited only the band of lowest MW. This can be considered as another proof of the presence of both cathepsins L and B in ESP of E. nipponicum.
PCR with degenerate primers and RACE-PCR led to the amplification of E. nipponicum gene specific DNAs which were sequenced and identified as cathepsin L genes (named EnCL1 and EnCL3) [GenBank: KP793605, KP793606]. The pro-peptide of EnCL1 and EnCL3 contains some highly conserved regions typical for cathepsins L. One such a region is a variation of the EX3RX2(V/I)FX2NX3IX3N (‘ERFNIN’) motif (present as ‘ERFNVN’ in EnCL1) (Karrer et al. Reference Karrer, Peiffer and DiTomas1993). The residues in this motif are probably important in stabilizing the globular structure of the pro-peptide (Coulombe et al. Reference Coulombe, Grochulski, Sivaraman, Ménard, Mort and Cygler1996; Groves et al. Reference Groves, Coulombe, Jenkins and Cygler1998). Another conserved motif is the GXNXFXD (‘GNFD’) present as ‘ANLD’ in EnCL1 and as ‘TNFD’ in EnCL3, with residues supposed to be involved in the control of intramolecular processing of cysteine peptidase precursors and/or participate in folding (Vernet et al. Reference Vernet, Berti, de Montigny, Musil, Tessier, Ménard, Magny, Storer and Thomas1995; Dvořák et al. Reference Dvořák, Mashiyama, Sajid, Braschi, Delcroix, Schneider, McKerrow, Bahgat, Hansell, Babbitt, Craik, McKerrow and Caffrey2009). Processing is believed to be facilitated by a low pH. When pH is lowered, protonation of Asp36 (papain numbering) may cause a conformational change of the cysteine peptidase precursor in which the propeptide is bound less tightly into the active site, making it more susceptible for clip-off (Coulombe et al. Reference Coulombe, Grochulski, Sivaraman, Ménard, Mort and Cygler1996; Jerala et al. Reference Jerala, Zerovnik, Kidric and Turk1998). Amino acid differences in these conserved motifs in EnCL1 and EnCL3 should not affect the function.
An asparagine residue, found between the pro-peptide and mature domain of gut cysteine peptidases – cathepsins from other blood-feeding parasites, e.g. schistosomes and ticks, is responsible for recognition and cleavage by legumain for trans-activation (Sajid et al. Reference Sajid, McKerrow, Hansell, Mathieu, Lucas, Hsieh, Greenbaum, Bogyo, Salter, Lim, Franklin, Kim and Caffrey2003; Sojka et al. Reference Sojka, Hajdušek, Dvořák, Sajid, Franta, Schneider, Craik, Vancová, Burešová, Bogyo, Sexton, McKerrow, Caffrey and Kopáček2007). However, this Asn is absent in both EnCL1 and EnCL3, suggesting that pro-EnCL1 and pro-EnCL3 are rather activated by autocatalysis.
The analysis of EnCL1 sequence using SignalP 4·1 software did not indicate the presence of an N-terminal signal sequence necessary for vesicle or extracellular targeting. The absence of such signal sequence has been described for a few cathepsins L from other organisms, e.g. the assassin bug Rhodnius prolixus cathepsin L-like protein (Lopez-Ordoñez et al. Reference Lopez-Ordoñez, Rodriguez and Hernández-Hernández2001). Additionally, it has been reported that human cathepsin L can be directed to a secretion pathway due to an aa sequence (-SXPXV) located at the carboxy (C) terminus of the enzyme (Chauhan et al. Reference Chauhan, Ray, Kane, Willingham and Gottesman1998). Since the secretion of EnCL1 was supported by our result from mass spectrometry analysis, it seems that it may be realized in an alternative way; a non-signal peptide triggered protein secretion was also predicted by SecretomeP 2·0 software tool (http://www.cbs.dtu.dk/services/SecretomeP/), but just at threshold values of the analysis. So far, it is not clear whether some cryptic (unknown) intramolecular signals are involved in secretion of this enzyme and only speculations can be made on another possibility – extracellular transport in exosomes. On the other hand, EnCL3 possesses a typical N-terminal signal sequence (24 aa). EnCL3 zymogen has an atypical long pro-peptide (120 aa) similarly as the human blood fluke S. mansoni SmCL3 (130 aa) [GenBank: ABV71063·1]. It has been suggested that these extensions may have other functions to the proenzyme, for example in protein trafficking or as binding sites for other proteins (Dvořák et al. Reference Dvořák, Mashiyama, Sajid, Braschi, Delcroix, Schneider, McKerrow, Bahgat, Hansell, Babbitt, Craik, McKerrow and Caffrey2009).
Both EnCL1 and EnCL3 have no potential N-linked glycosylation sites in their sequences, similarly to cathepsin L of the monogenean parasite N. melleni [GenBank: ABK62794·1]. In general, glycosylation with mannose 6-phosphate has been shown to be an important sorting signal for routing mammalian proteins into lysosomes. However, there are several experimental evidences for mannose 6-phosphate independent trafficking of proteins into lysosomes (Ni et al. Reference Ni, Canuel and Morales2006; Braulke and Bonifacino, Reference Braulke and Bonifacino2009). According to this, it would be possible that EnCL occurrence in ESP is not connected with the discharge of phagolysosome residual content into the gut lumen or that the enzyme may be targeted into lysosomes in an alternative manner.
Both cathepsins L were confirmed in E. nipponicum ESP in areas of various molecular sizes by means of mass spectrometry. Individual bands may represent partial processing or autolysis of the enzymes. According to the results, it seems that EnCL1 represents an abundant peptidase in ESP of E. nipponicum. In addition, the dominant protein spots found in E. nipponicum solPE separated by 2D electrophoresis were also identified by mass spectrometry as EnCL1 (data not shown).
Digestion of proteins in blood-feeding parasites represents a major proteolytic process. We showed that the most abundant haemoglobinolytic endopeptidase activities in E. nipponicum belong to the cysteine class, with cathepsin L-like activity predominating over cathepsin B-like activity. Proteomic investigations confirmed this fact. Besides, significant involvement of aspartic cathepsin D was well documented. The possibility of aimed secretion of proteolytic enzymes to the external environment by this ectoparasite of carp gills appears to be purposeless and therefore improbable. From this point of view and based on our results, we believe that cathepsins L, B and D are involved in processing of ingested host's blood in E. nipponicum.
ACKNOWLEDGEMENTS
We are grateful to Dr C. Caffrey and Dr D. Greenbaum, Center for Discovery and Innovation in Parasitic Diseases, UCSF, USA, who kindly provided the probe Green BODIPY-DCG-04. The MS part of the work was carried out with the support of Proteomics Core Facility (‘CEITEC – Central European Institute of Technology’). We also thank Mr Stanislav Sojka, Rybářství Třeboň, Plc., for arrangements concerning collection of parasites.
FINANCIAL SUPPORT
This research was enabled due to the grants of the Czech Science Foundation (L.M., Grant no. P506/12/1258), (Z.Z., Grant no. P206-12-G151); Grant Agency of the Charles University in Prague (L.J., Grant no. 502313); Masaryk University institutional support (M.K., J.I., Grant no. MUNI/A/1484/2014); Charles University institutional/departmental support (M.K., Grant no. UNCE 204017; L.J., H.D., M.K., L.M., Grant no. PRVOUK P41; L.J., H.D., Grant no. SVV 260202/2015); and the European Regional Development Fund (Z.Z., Grant no. CZ.1·05/1·1·00/02·0068).