Introduction
Parmotrema pseudotinctorum (des Abb.) Hale is a rosette-forming foliose lichen which colonizes large areas on volcanic rock surfaces in arid or very dry and warm areas in the Canary Islands, mainly in the infra- and thermo-Mediterranean bioclimatic belts, surrounded by crassicaule or xeric Mediterranean vegetation (Molins et al. Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013). In some localities, even under extreme environmental conditions, this lichen can grow abundantly giving a distinctive appearance to the general landscape.
Recently, Roca-Valiente et al. (Reference Roca-Valiente, Divakar, Ohmura, Hawksworth and Crespo2013) studied the mycobiont using molecular phylogeny combined with morphological features, resolving the taxonomic identity of this species with regard to the closely related Parmotrema tinctorum (Delise ex Nyl.) Hale, which was occasionally considered to be a synonym. Parmotrema pseudotinctorum seems to be restricted to the Canary Islands, Africa, the Cape Verde Islands and India, the only place where it grows as an epiphyte on bark.
Thallus anatomy and ultrastructure were described by Molins et al. (Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013), who detected the presence of a characteristic biofilm of bacteria that covers the underside of the thallus. In populations from Tenerife and Lanzarote, a collection of bacterial strains isolated from this lichen have been characterized physiologically and metabolically (Gimeno et al. Reference Gimeno Molina, Barreno and Biosca2016). The results revealed that most bacterial strains were capable of producing proteases and nucleases as well as solubilizing polysaccharides and phosphate. In addition, siderophores and more than 75% of fixed nitrogen were produced as well as some auxins and they formed biofilms.
These activities may have an important role in the functioning of the symbiosis and undoubtedly have biotechnological applications.
Over the last ten years, several studies have focused on the phycobiont inventory of the closely related P. tinctorum and P. pseudotinctorum lichens. These investigations revealed the existence of considerable phycobiont diversity within the Trebouxia clade G sensu Helms (Reference Helms2003). First, Ohmura et al. (Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006) detected a total of six phycobiont lineages within P. tinctorum thalli sampled in the Shimizu district of Japan. Later, Mansournia et al. (Reference Mansournia, Wu, Matsushita and Hogetsu2012) investigated the population heterogeneity of phycobionts growing within P. tinctorum in the Chiba Prefecture, Japan. They revealed the presence of four phycobiont lineages, two of them being identical to the lineages already detected by Ohmura et al. (Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006). Finally, Molins et al. (Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013) identified three phycobiont lineages in their investigation of P. pseudotinctorum lichens growing on the Canary Islands, Spain. Interestingly, none of these lineages were identical to any of the previously sequenced P. tinctorum phycobionts (Ohmura et al. Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006; Mansouria et al. Reference Mansournia, Wu, Matsushita and Hogetsu2012). In addition, all three previously-mentioned studies detected the presence of multiple phycobiont lineages within single Parmotrema thalli.
In general, none of the previously identified lineages of P. tinctorum/P. pseudotinctorum phycobionts could be assigned to previously described Trebouxia species, emphasizing our poor knowledge of the diversity, evolution and taxonomy of the phycobiont genus Trebouxia. However, such knowledge is essential for our understanding of the functioning and complexity of lichen symbiotic interactions. In this paper, we therefore aimed to obtain a more complete picture of the intrathalline phycobiont diversity in P. pseudotinctorum, and to morphologically characterize and formally describe the most abundant phycobiont associated with this lichen. Here, we describe this phycobiont as Trebouxia crespoana sp. nov.
Materials and Methods
Lichen material
Five representative populations of Parmotrema pseudotinctorum collected from the Canary Islands were included in the analyses. Samples from San Sebastián (La Gomera, LG) were collected on old basalt rocks. The samples from Los Cancajos and Breña Baja (La Palma, LP) were collected on basalts from the flow of the “Cumbre Vieja” volcano. Lichen thalli from Jameos del Agua (Lanzarote, LZ) were collected on subalkaline basalt rocks originating from the sub-recent flow of the La Corona volcano, and samples from Buenavista (Tenerife, TF) were collected on old basalts. The samples were dried and stored at −20 °C until processing. Each lichen specimen used in this study was encoded as indicated in Supplementary Table S1 (available online).
Sample handling
Lichen thalli were examined under a stereomicroscope to remove surface contamination (e.g. bark, sand, mosses, fragments of other lichen species, or infection by lichenicolous fungi). Lichen thalli were sterilized by sequential immersion in 96% ethanol (10s), 0·5% NaOCl (2 min) and 70% ethanol (2 min) (Arnold et al. Reference Arnold, Miadlikowska, Higgins, Sarvate, Gugger, Way, Hofstetter, Kauff and Lutzoni2009). Fragments from different parts of the thallus (centre, mid-point and edge) were randomly excised and pooled together. The mycobiont and the primary phycobiont were identified by Sanger sequencing, and the Los Cancajos PAL 4 sample was also analysed by 454-pyrosequencing. The ultrastructure of the phycobionts was characterized by transmission electron microscopy (TEM) for the Breña Baja 2 and 5 samples. The Los Cancajos 10 sample was used to isolate the phycobiont in the culture for its investigation by conventional light (LM) and confocal microscopy (CM).
DNA extraction, amplification and sequencing
Total genomic DNA was isolated and purified using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Two algal loci were amplified; a region of the chloroplast LSU rDNA gene using the algal specific primers 23SU1 and 23SU2 (del Campo et al. Reference del Campo, Casano, Gasulla and Barreno2010), and the ITS rDNA using the primer pair Parm_ITS_Phyco_F (5´- TGA TTC TAT CGT GCC AAC ACC G -3´) and Parm_ITS_Phyco_R (5´- GAT ATG CTT AAG TTC AGC GGG TG -3´) designed in our laboratory based on the previous ITS1-ITS2 alignments obtained in Molins et al. (Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013). Fungal ITS rDNA was amplified using the primer pair Parm_ITS_Myco_F (5´- TGA GAG AGG GGC TTC GCG CTC C -3´) and Parm_ITS_Myco_ R (5´- ATC CGA GGT CAA TAT TGG AAG CA- 3´) designed for this study. PCR reactions were performed in 50 µl using EmeraldAmp GT PCR Master Mix (Takara, Shiga, Japan), which required the addition of the template DNA, specific primers and water. The PCR program for amplification consisted of an initial denaturation at 94 °C for 2 min and 30 cycles at 94 °C for 30 s, 56 °C for 45 s and 72 °C for 1 min, followed by a final elongation at 72 °C for 5 min. Amplifications were carried out on a 96-well SensoQuest Labcycler (Progen Scientific Ltd., South Yorkshire, UK). The PCR products were visualized on 2% agarose gels and purified using the Gel Band Purification Kit (GE Healthcare Life Science, Buckinghamshire, UK). The amplified PCR products were sequenced with an ABI 3100 Genetic Analyzer using the ABI BigDyeTerminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA).
Sequence analyses
The newly determined algal nuclear ITS rDNA and chloroplast LSU rDNA sequences were aligned with the selection of Trebouxia sequences from the GenBank database, with emphasis given to the authentic strains of Trebouxia species. The alignment was then supplemented by a number of sequences belonging to Trebouxia clade G, including all new lineages identified by Ohmura et al. (Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006), Mansournia et al. (Reference Mansournia, Wu, Matsushita and Hogetsu2012) and Molins et al. (Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013). The sequences were aligned using MAFFT v.6 software (Katoh et al. Reference Katoh, Misawa, Kuma and Miyata2002) under the Q-INS-I strategy, N treated as a wildcard and checked for obvious sequencing errors. The alignment of ITS rDNA sequences was improved by eliminating the ambiguously aligned regions using the program Gblocks v.0.91b (Castresana Reference Castresana2000). The two loci were concatenated, yielding an alignment of 1477 characters. The identical sequences were merged, resulting in the final matrix containing 54 ITS rDNA and 26 LSU rDNA sequences. For each locus, the most appropriate substitution model was estimated using the Bayesian information criterion (BIC) as implemented in jModelTest 2.1.4 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012). This BIC-based model selection procedure selected the GTR+I+Γ and TPM3uf+I+Γ models for the ITS and LSU rDNA regions, respectively.
The phylogenetic trees were inferred by Bayesian inference (BI) using MrBayes v.3.2.6 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Hohna, Larget, Liu, Suchard and Huelsenbeck2012), carried out on partitioned datasets using the GTR+I+Γ substitution models for both partitions (the TPM3uf model was not implemented). Two parallel MCMC runs were carried out for six million generations, each with one cold and three heated chains. Trees and parameters were sampled every 100 generations. Convergence of the two cold chains was assessed during the run by calculating the average standard deviation of split frequencies (SDSF). The SDSF value between simultaneous runs was 0·0024. Finally, the burn-in values were determined using the ‘sump’ command. Bootstrap analyses were performed by maximum likelihood (ML) using GARLI v.2.01 (Zwickl Reference Zwickl2006). ML analysis consisted of rapid heuristic searches (100 pseudoreplicates) using automatic termination (genthreshfortopoterm command set to 100 000). The analysis was performed on partitioned datasets using the different substitution models selected by jModelTest 2.1.4. All analyses were run on the CIPRES Science Gateway v.3.3 web portal (Miller et al. Reference Miller, Pfeiffer and Schwartz2010).
454-pyrosequencing and phylogenetic analyses
A first RT-PCR (RT-PCR I) and a first PCR (PCR I) were performed using the genomic DNA from the Los Cancajos PAL 4 sample as a template and nr-SSU-1780/5.8S 2R primers (Moya et al. Reference Moya, Molins, Martínez-Alberola, Muggia and Barreno2017). The number of cycles of PCR I (19 cycles) was determined by the average Ct (cycle threshold) of the RT-PCR I. We then performed a second RT-PCR (RT-PCR II) and a second PCR (PCR II) using 1 μl of the PCR I as template and the fusion primers designed following the GS Junior System Guidelines for Amplicon Experimental Design (Roche, Branford, USA). The specific cycle number for the PCR II (7 cycles) was determined by the average Ct from the RT-PCR II. The RT-PCRs and PCRs were performed and purified as previously described in Moya et al. (Reference Moya, Molins, Martínez-Alberola, Muggia and Barreno2017). Algal ITS rDNA sequences were determined using a GS Junior 454 system (Roche 454 Life Sciences, Branford, CT, USA) following the Roche Amplicon Lib-L protocol at the Genomics Core Facility at the University of Valencia (Spain). Reads were processed as described in Moya et al. (Reference Moya, Molins, Martínez-Alberola, Muggia and Barreno2017) and clustered based on 99% score coverage threshold and 90% length coverage threshold (-S 99 -L 0·9) criteria. The consensus sequences of the OTUs were identified using the BLAST tool in the GenBank database (Altschul et al. Reference Altschul, Gish, Miller, Myers and Lipman1990).
Ultrastructural examinations
Transmission electron microscopy (TEM) examinations were performed on samples 2 and 5 from Breña Baja. For TEM, the cells were fixed in 2% Karnovsky fixative for 12 h at 4°C and washed three times for 15 min with 0·01 M PBS (pH 7·4) then post-fixed with 2% OsO4 in 0·01 M PBS (pH 7·4) for 2 h at room temperature. Thereafter, they were washed three times in 0·01 M PBS (pH 7·4) for 15 min and then dehydrated at room temperature in a graded series of ethanol solutions, starting at 50% and increasing to 70%, 95% and 100% for no less than 20–30 min at each step (Molins et al. Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013; Moya et al. Reference Moya, Škaloud, Chiva, García-Breijo, Reig-Armiñana, Vančurová and Barreno2015). The fixed and dehydrated samples were embedded in Spurr’s resin according to the manufacturer’s instructions (http://www.emsdiasum.com/microscopy/technical/datasheet/14300.aspx). Sections (90 nm) were cut with a diamond knife (DIATOME Ultra 45°) using an ultramicrotome (Reichert Ultracut E), mounted on oval hole copper grids coated with formvar and post-stained with 2% (w/v) aqueous uranyl acetate and 2% lead citrate, using the “SynapTek Grid Staining Kit” (http://www.ems-diasum.com/microscopy/technical/datasheet/71175.aspx). The sections were observed with a JEOL JEM-1010 (80 kV) electron microscope equipped with a MegaView III digital camera and “AnalySIS” image acquisition software. TEM examinations were carried out at the SCSIE Service of the University of Valencia.
Isolation and propagation of microalgae
Phycobionts were isolated from sample no. 10 from Los Cancajos using the micromethod described by Gasulla et al. (Reference Gasulla, Guéra and Barreno2010). Samples were homogenized with a mortar and pestle in an isotonic buffer (0·3 M sorbitol, 50 mM HEPES, pH 7·5) and filtered through muslin. Isolation was carried out by a gradient centrifugation method using Percoll®. The algal suspension was diluted with sterile water and 10 μl was streaked onto sterile 1·5% agar Bold’s Basal Media Petri dishes (BBM) (Bold Reference Bold1949; Bischoff & Bold Reference Bischoff and Bold1963). The isolated algae were maintained at 15 μmol m−2 s−1 photosynthetic photon flux density (PPFD) with a 12 h photoperiod at 21°C. To obtain unialgal cultures, small populations of phycobionts were transferred onto the fresh BBM agar slants and incubated accordingly.
Microscopy
The morphology of the isolated phycobiont strain was investigated by both conventional light (LM) and confocal (CM) microscopy. LM observations were performed using an Olympus BX51 microscope equipped with differential interference contrast. Micrographs were taken with an attached Canon EOS 700D camera. For CM, a Leica TCS SP2 laser scanning confocal microscope equipped with an argon-krypton laser was used. We applied a 488 nm excitation line and an AOBS filter-free system collecting emitted light between 498 and 700 nm. The autofluorescence of chlorophyll was exploited for visualization of the chloroplast structure. A series of optical sections through chloroplasts was captured and used for 3-dimensional reconstruction of their morphology. The chloroplast reconstructions were produced by the ImageJ v.1.34p program (Abramoff et al. Reference Abramoff, Magalhaes and Ram2004), using the “Volume viewer” plugin. Zoospore formation was induced by transferring the culture in different ontogenetic stages to distilled water.
Results
Trebouxia crespoana Barreno, Molins, Moya & Škaloud sp. nov.
AlgaeBase ID: Aea4fa8abaec4fb8a
Differing from other Trebouxia species by the formation of pyriform cells bearing local cell wall thickening and by the order of the nucleotides in ITS rDNA sequences.
Type: Spain, Canary Islands, La Palma, Breña Baja, flow of “Cumbre Vieja” volcano rocks, 28°38′33″N, 17°45′30″W, phycobiont in Parmotrema pseudotinctorum, 11 December 2016, A. Santos-Guerra (MAF-Lich 21323—holotype; MA-Lich 19165, VAL_Lich 30719—isotypes).
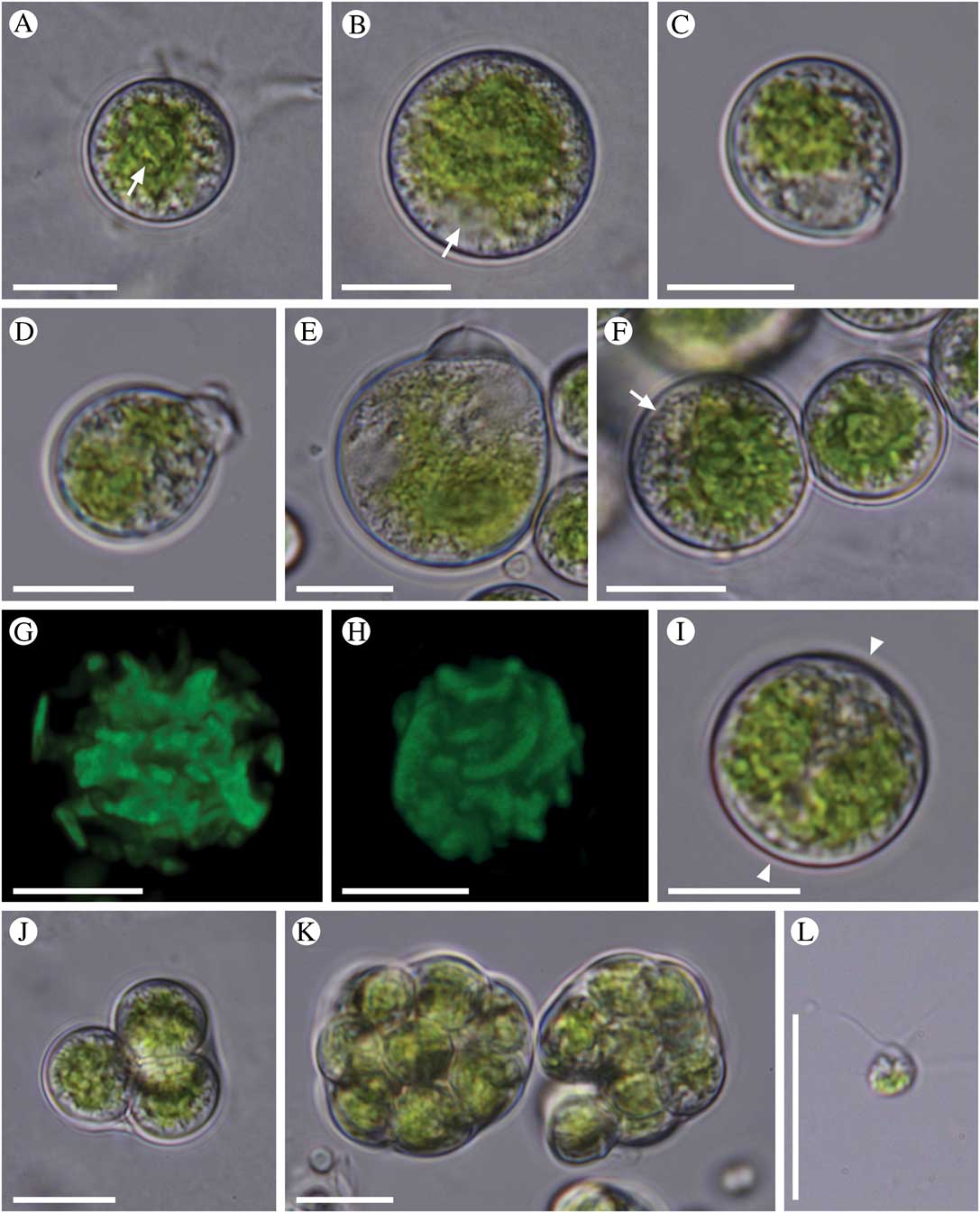
Fig. 1 Morphology of Trebouxia crespoana sp. nov. A, young cell with a prominent pyrenoid (arrow); B, mature vegetative cell (nucleus labelled with an arrow); C, a pyriform cell with papilla-like cell wall thickening; D, cell with a prominent cell wall thickening; E, old cell with a distinct cell wall bulge; F, cells with a central, crenulate chloroplast (peripheral vesicles labelled with an arrow); G, reconstruction of chloroplast in a mature cell; H, reconstruction of a chloroplast surface, note the elongated ends of chloroplast lobes; I, a cell with two adjacent chloroplasts (space between the chloroplasts is marked with arrowheads); J, small autosporangium with 4 autospores; K, irregularly-shaped autosporangia; L, a zoospore. Scales=10 µm.
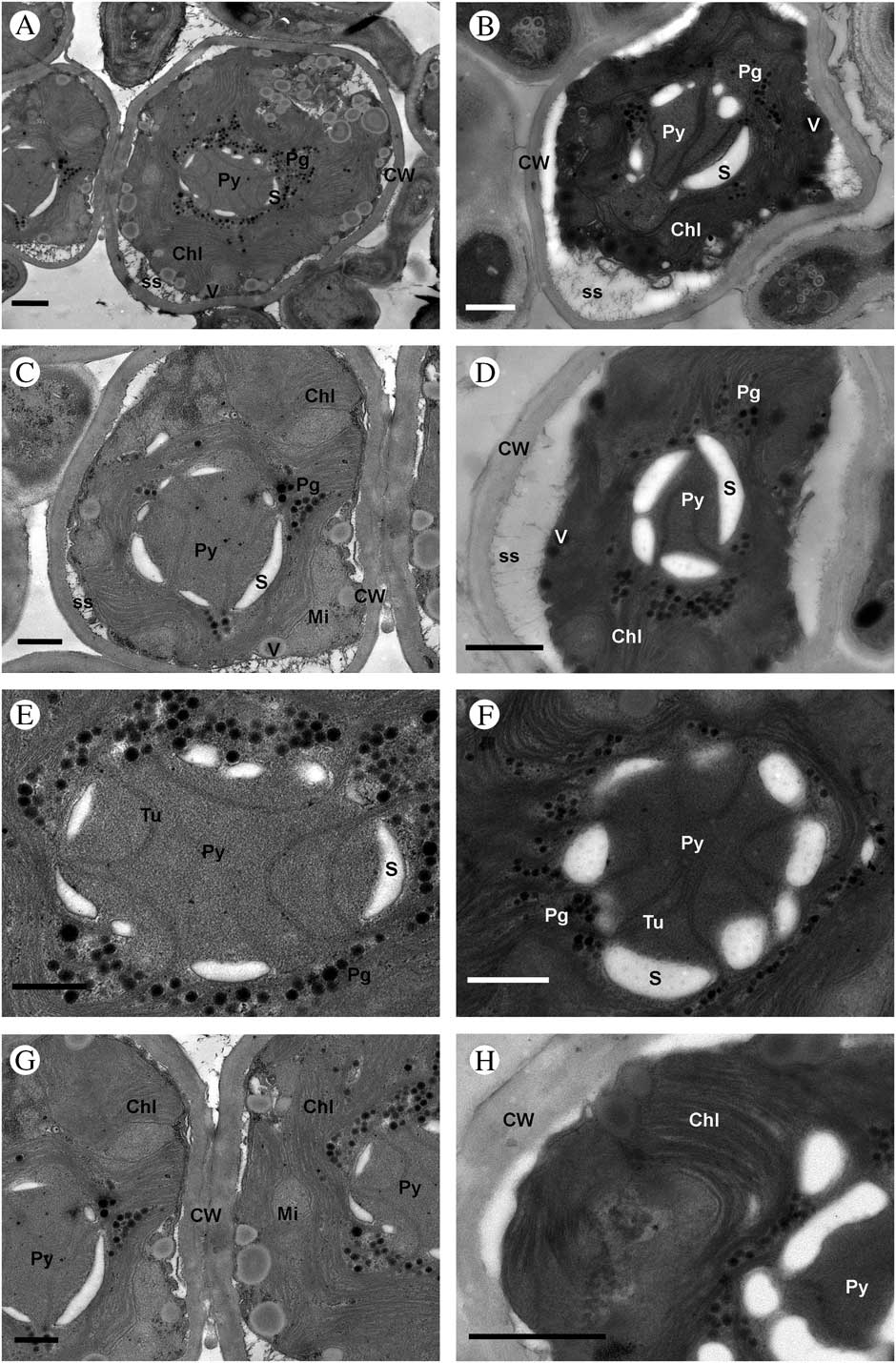
Fig. 2 Ultrastructure of Trebouxia crespoana sp. nov. A–D, T. crespoana cells within thallus; E & F, details of pyrenoid; G & H, detail of the cell wall and thylakoid arrangement. Abbreviations: Chl (chloroplast), CW (cell wall), Mi (mitochondrion), Pg (pyrenoglobuli), Py (pyrenoid), S (starch grain), SS (secretory space), Tu (tubules) and V (vesicles). Scales: A & H=1 μm; B, D & F=800 nm; C, E & G=600 nm.
Cells are solitary, usually spherical, rarely oval and pyriform, 11–21(–26) µm diam. Cell wall is usually thin. Sometimes a characteristic cap-like cell wall thickening is produced on one side of vegetative cells. The cells possess a central crenulate chloroplast with several lobes elongated at their ends; only rarely are two crenulate chloroplasts formed within a single cell. The chloroplast contains one spherical or irregularly elongated pyrenoid covered by a layer of a few large, arched starch grains. Pyrenoid is corticola-type with very thin, unbranched tubules of curved profile, and pyrenoglobuli developed in the closest parts of the chloroplast stroma. In mature and old cells, the pyrenoid is usually indistinct. Asexual reproduction involves the formation of autospores and zoospores. The autosporangia of irregular shape usually contain 4–16 autospores, tightly appressed to each other. Zoospores are biflagellate, drop-shaped or oval, 5·5–6·0 µm long and 4·0–4·5 µm wide, with posterior chloroplast. Sexual reproduction was not observed.
Reference strains. CAUP H 1019 (Culture Collection of Algae of Charles University in Prague), Strain 132 (E. Barreno’s personal collection at the University of Valencia).
Etymology. The specific epithet refers to the Canarian Professor of Botany Dr Ana Crespo. She has developed lichen studies in Spain over the last quarter of the 20th century and pioneered studies of phylogenetic systematics with molecular markers; especially innovative were her analyses of Parmeliaceae s. a.
Ecology and distribution. Trebouxia crespoana was found only in symbiosis with the foliose lichen Parmotrema pseudotinctorum, growing as saxicolous on diverse types of volcanic rock on several islands of the Canarian archipelago. This rosette-forming lichen is prevalent and yields large biomass in the infra-Mediterranean arid bioclimatic belt but can reach the thermo-Mediterranean subhumid belt in the driest areas of the laurel forests. So far, T. crespoana is known only from the Canary Islands.
Morphology of cultured Trebouxia crespoana
Young cells were spherical, containing one axial chloroplast with several lobes and a distinct pyrenoid (Fig. 1A). Mature vegetative cells were up to 21(–26) µm in diameter, usually spherical in shape (Fig. 1B) but oval and pyriform cells were also observed. The cell wall was usually thin; however, in some cells a local cell wall thickening had developed on one side of the vegetative cells. First, a small, papilla-like cell wall thickening occurred, followed by the formation of a distinct cell wall bulge (Fig. 1C). We did not observe any cell wall ruptures or other processes following the cell wall bulging; instead, the cell wall thickening was retained in mature and old cells (Fig. 1D & E). The thickenings measured 0·5–3·5 µm. T. crespoana possesses a central crenulate chloroplast with several lobes spreading toward the cell periphery (Fig. 1F & G). The lobes were notably elongated at their ends (Fig. 1H). Rarely, two adjacent crenulate chloroplasts were observed (Fig. 1I). The chloroplast contained one centrally positioned pyrenoid (Fig. 1A) surrounded by starch grains (Fig. 1F). The pyrenoid was clearly visible in young cells but indistinct in mature and old cells. The cells were uninucleate, with a lateral nucleus positioned between the chloroplast lobes (Fig. 1B). Usually a large number of small peripheral vesicles were observed underneath the cell wall (Fig. 1F). The cells reproduced only asexually, by the formation of autospores and zoospores. The autosporangia were first spherical, later irregularly-shaped and deformed by the growing autospores, up to 24 µm in length, usually containing 4–16 spherical autospores (Fig. 1J & K). When released, they measured 5·5–9·0 µm in diameter. Zoosporangia were spherical, containing an unspecified number of zoospores. Zoospores were biflagellate, teardrop-shaped or oval, 5·5–6·0 µm long and 4·0–4·5 µm wide, with posterior chloroplast (Fig. 1L).
Ultrastructure of lichenized Trebouxia crespoana (Fig. 2)
Lichenized cells were mostly oval to rounded, up to 7·8 µm in diameter (Figs 2A–C). The cell wall was thin, 0·18–0·33 µm wide (Figs 2C & D). The secretory space was evenly distributed except in the zones in contact with mycobiont haustoria (Fig. 2D). The interaction of phycobionts and the mycobiont was frequently made through type 2 haustoria sensu Honegger (Reference Honegger1986). The centrally positioned chloroplast extends into dividing lobes showing a regular and dense distribution of the thylakoid membranes (Figs 2C & H) with some pyrenoglobules. The central pyrenoid was spherical or irregularly elongated, of the corticola-type sensu Friedl (Reference Friedl1989), with very thin, curved, unbranched tubules (Figs 2E & F). Pyrenoglobuli developed primarily in the innermost parts of the chloroplast stroma (Fig. 2E). Starch grains were closely associated with the pyrenoid matrix, forming a starch sheath made up of a few large, arched plates adjacent to the pyrenoid (Figs 2E & F). The cells possess a few mitochondria, and in the peripheral zones numerous vesicles with a central electrodense granular content in a non-electrodense matrix (Figs 2C & G). These vesicles were clearly visible in cultured cells observed under light microscopy (Fig. 1F).
Mycobiont identification and the diversity of primary phycobionts
Morphological identification of investigated lichens was verified by BLAST searches of mycobiont ITS rDNA sequences. All 39 mycobiont sequences were closely related or identical to P. pseudotinctorum isolates molecularly characterized by Roca-Valiente et al. (Reference Roca-Valiente, Divakar, Ohmura, Hawksworth and Crespo2013).
Diversity of primary phycobionts was much higher in comparison to the mycobiont partners. All algal sequences were inferred within the Trebouxia clade G sensu Helms (Reference Helms2003). The majority of lichen thalli contained the new phycobiont lineage described here as T. crespoana sp. nov. (Fig. 3). This species was found in all lichen populations investigated and was previously reported by Molins et al. (Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013) and Leavitt et al. (Reference Leavitt, Kraichak, Nelsen, Altermann, Divakar, Alors, Esslinger, Crespo and Lumbsch2015). Seven lichen thalli collected in the Buenavista and Los Cancajos populations contained a different algal lineage, sister to T. crespoana and Trebouxia OTU G04 sensu Leavitt et al. (Reference Leavitt, Kraichak, Nelsen, Altermann, Divakar, Alors, Esslinger, Crespo and Lumbsch2015). This lineage was previously reported in many Japanese P. tinctorum lichens collected by Ohmura et al. (Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006) and Mansournia et al. (Reference Mansournia, Wu, Matsushita and Hogetsu2012), and labelled as clade I and T. corticola, respectively. Finally, the primary phycobionts of three lichens collected in the Jameos del Agua population were inferred in a lineage related to another P. tinctorum phycobiont labelled as clade V and Trebouxia RG1 by Ohmura et al. (Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006) and Mansournia et al. (Reference Mansournia, Wu, Matsushita and Hogetsu2012), respectively.

Fig. 3 Phylogeny of the genus Trebouxia obtained by Bayesian inference of the concatenated nuclear ITS rDNA and chloroplast LSU rDNA sequences. Values at the nodes indicate statistical support estimated by MrBayes posterior-node probability (left of slash) and maximum-likelihood bootstrap (right of slash). Thick branches represent nodes receiving the highest posterior probability support (1·00). Newly sequenced strains and newly generated sequences are marked in bold. Identical sequences obtained from the same localities are shown in one row, along with the sampling codes. Clade affiliation sensu Ohmura et al. (Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006) and Mansournia et al. (Reference Mansournia, Wu, Matsushita and Hogetsu2012) is indicated next to the sequences by black circles and white rectangles, respectively. Four main Trebouxia clades (A, I, S, G) are indicated by letters on the backbone of the tree. Scale bar represents the expected number of substitutions per site.
Phycobiont diversity by 454-pyrosequencing
Sequencing of ITS rDNA amplicons produced 5096 sequence reads for Los Cancajos PAL 4. Singleton reads (26) were filtered out. By clustering with a 99% similarity cut-off, eight OTUs were recognized: OTU1–OTU8 (Table 1). These OTUs were representative of three phycobiont genera: Trebouxia, Asterochloris and Myrmecia. The OTUs 1 and 7 matched with T. crespoana and with the primary phycobiont detected by Sanger sequencing in Los Cancajos PAL 4. OTU 3 fitted with T. asymmetrica. OTU 5 was related to T. impressa and OTUs 2 and 4 to T. decolorans. Significant matches were obtained with Asterochloris mediterranea (AF345435) and Myrmecia sp. (KF907687) for OTU 8 and OTU 6, respectively.
Table 1 Taxonomic identification of the eight OTUs detected according to BLAST matches in GenBank and numbers of the corresponding sequences present in each OTU

Discussion
Distinctness of Trebouxia crespoana sp. nov.
Trebouxia represents an extremely diverse and probably the most common genus of lichen phycobionts, occurring worldwide in thalli of numerous lichen fungal genera (Tschermak-Woess Reference Tschermak-Woess1988; Miadlikowska et al. Reference Miadlikowska, Kauff, Högnabba, Oliver, Molnár, Fraker, Gaya, Hafellner, Hofstetter and Gueidan2014). Traditionally, some Trebouxia species were exclusively delimited based on morphological features such as cell size and shape, chloroplast morphology, number and morphology of pyrenoids, dissociation of autospores, and zoospore morphology (Ettl & Gärtner Reference Ettl and Gärtner1995). However, our knowledge concerning overall Trebouxia species diversity has dramatically changed over the last two decades, since a number of molecular phylogenetic investigations revealed substantial hidden diversity within the genus (Kroken & Taylor Reference Kroken and Taylor2000; Dahlkild et al. Reference Dahlkild, Källersjö, Lohtander and Tehler2001; Blaha et al. Reference Blaha, Baloch and Grube2006; Muggia et al. Reference Muggia, Zellnig, Rabensteiner and Grube2010, Reference Muggia, Pérez-Ortega, Kopun, Zellnig and Grube2014; Sadowska-Des et al. Reference Sadowska-Deś, Dal Grande, Lumbsch, Beck, Otte, Hur, Kim and Schmitt2014; Leavitt et al. Reference Leavitt, Kraichak, Vondrak, Nelsen, Sohrabi, Pérez-Ortega, Clair and Lumbsch2016). Despite this, only a very small fraction of newly recognized lineages have been formally described as new species (Beck Reference Beck2002; Voytsekhovich & Beck Reference Voytsekhovich and Beck2016). With the discrepancy between observed diversity and described species in mind, we are here proposing a new species for the most common lineage of photobionts we detected during our investigation of Parmotrema pseudotinctorum thalli. Though T. crespoana represents a distinct and statistically well-supported lineage within Trebouxia clade G, a morphological comparison to all previously described Trebouxia species is necessary to avoid describing a junior synonym of an already described yet molecularly uncharacterized species. Morphologically, T. crespoana is well recognized by the formation of pyriform cells bearing local cell wall thickening (Fig. 1C–E). Such a feature has been observed only in T. asymmetrica, a symbiont described and isolated by Friedl & Gärtner (Reference Friedl and Gärtner1989) from the lichen Diploschistes albescens. However, the cells of this species are much smaller and more ovoid than those of T. crespoana. In addition, the authentic strain of T. asymmetrica (UTEX 2507) has been sequenced by Piercey-Normore & DePriest (Reference Piercey-Normore and DePriest2001), showing its phylogenetic position within the Trebouxia clade A sensu Helms (Reference Helms2003), unrelated to all P. pseudotinctorum phycobionts.
Trebouxia crespoana matches with Trebouxia OTU G01, forms a well-supported clade together with Trebouxia OTU G04 sensu Leavitt et al. (Reference Leavitt, Kraichak, Nelsen, Altermann, Divakar, Alors, Esslinger, Crespo and Lumbsch2015) and a lineage comprising several phycobionts found in lichen thalli collected in the Buenavista (samples 1, 2, 5) and Los Cancajos (samples 1, 4, 5, PAL1) populations. Although the majority of these phycobionts were detected in Parmotrema lichens sampled in the Canary Islands, it is highly likely that both Trebouxia OTU G04 and the Buenavista-Los Cancajos lineage represent two distinct yet undescribed Trebouxia species. The genetic differences among these lineages and T. crespoana are almost as great as those between any sister Trebouxia species (Fig. 3). Indeed, T. crespoana and Trebouxia OTU G04 were already recognized as different species by Leavitt et al. (Reference Leavitt, Kraichak, Nelsen, Altermann, Divakar, Alors, Esslinger, Crespo and Lumbsch2015) using the ABGD delimitation method. In addition, several published studies demonstrated the strong ecological differentiation of closely related green-algal lineages, including lichen phycobionts (Peksa & Škaloud Reference Peksa and Škaloud2011; Moniz et al. Reference Moniz, Rindi, Novis, Broady and Guiry2012; Škaloud & Rindi Reference Škaloud and Rindi2013; Sadowska-Des et al. Reference Sadowska-Deś, Dal Grande, Lumbsch, Beck, Otte, Hur, Kim and Schmitt2014; Malavasi et al. Reference Malavasi, Škaloud, Rindi, Tempesta, Paoletti and Pasqualetti2016; Ryšánek et al. Reference Ryšánek, Holzinger and Škaloud2016), indicating these lineages represent distinct cryptic species. Although we did not collect any ecological data with the samples collected, we are quite sure that applying a broad species concept, grouping all Canary Island phycobionts into a single taxon T. crespoana, would lead to masking the real species diversity within the genus Trebouxia. Our data rather demonstrate that the vast majority of aerophytic and symbiotic green algae still remain undescribed, even though they may represent the most common and ecologically important organisms in certain areas. The Canary Islands are a suitable example of such a phenomenon (Molins et al. Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013; Vančurová et al. Reference Vančurová, Peksa, Němcová and Škaloud2015).
Investigation of intrathalline phycobiont diversity
Although 454-pyrosequencing has already been surpassed by other high throughput sequencing (HTS) approaches, it still represents a powerful and complementary approach to the traditional Sanger sequencing. HTS approaches supply information to evaluate species diversity at diverse taxonomic levels and provide much higher resolution to reveal the multiplicity of microalgae associated with the lichen thallus. The coexistence of multiple microalgae inside a single lichen thallus has been reported in some lichen species (Blaha et al. Reference Blaha, Baloch and Grube2006; Ohmura et al. Reference Ohmura, Kawachi, Kasai, Watanabe and Takeshita2006; Piercey-Normore Reference Piercey-Normore2006; Muggia et al. Reference Muggia, Zellnig, Rabensteiner and Grube2010, Reference Muggia, Pérez-Ortega, Kopun, Zellnig and Grube2014; Schmull et al. Reference Schmull, Miadlikowska, Pelzer, Stocker-Wörgötter, Hofstetter, Fraker, Hodkinson, Reeb, Kukwa, Lumbsch and Kauff2011; Molins et al. Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013, Reference Molins, Moya, García-Breijo, Reig-Armiñana and Barreno2018; Leavitt et al. Reference Leavitt, Kraichak, Nelsen, Altermann, Divakar, Alors, Esslinger, Crespo and Lumbsch2015; Moya et al. Reference Moya, Molins, Martínez-Alberola, Muggia and Barreno2017), whereas other lichens can be considered as highly specific, accepting only one single algal lineage in their thallus as a partner (Piercey-Normore & DePriest Reference Piercey-Normore and DePriest2001; Beck et al. Reference Beck, Kasalicky and Rambold2002; Yahr et al. Reference Yahr, Vilgalys and Depriest2004; Doering & Piercey-Normore Reference Doering and Piercey-Normore2009; Peksa & Škaloud Reference Peksa and Škaloud2011; Piercey-Normore & Deduke Reference Piercey-Normore and Deduke2011; Molins et al. Reference Molins, Moya, García-Breijo, Reig-Armiñana and Barreno2018). The sample of P. pseudotinctorum analyzed by 454-pyrosequencing in the present work showed only one Trebouxia sp. associated with the thallus (Trebouxia sp. OTU G01: 5044 reads). The remaining seven OTUs detected represent between 0·03 and–0·1% of the sequence reads and cannot be considered as strictly intrathalline.
These huge differences in species abundances raise questions about the levels of specificity and the influence of ecological settings in the association of lichenized fungi with certain algae available in the substratum pool (Peksa & Škaloud Reference Peksa and Škaloud2011). Sterilization of lichen material is essential in avoiding surface contamination, but OTUs representing<0·5% can be considered as the possible epithalline algal fraction (Moya et al. Reference Moya, Molins, Martínez-Alberola, Muggia and Barreno2017).
Ultrastructural investigations
In this study, TEM was used to characterize the ultrastructure of T. crespoana cells isolated from P. pseudotinctorum (Fig. 2). The observations were made on different samples and no differences were found between them. Cells showed a characteristic corticola-type pyrenoid (Friedl Reference Friedl1989) with the absence of pyrenoglobuli associated with the matrix thylakoids, and starch grains forming a starch sheath. Besides molecular techniques, phycobiont identity should be validated through microscopic examination including TEM (Muggia et al. Reference Muggia, Zellnig, Rabensteiner and Grube2010; Casano et al. Reference Casano, del Campo, García-Breijo, Reig-Armiñana, Gasulla, del Hoyo, Guéra and Barreno2011; Molins et al. Reference Molins, García-Breijo, Reig-Armiñana, del Campo, Casano and Barreno2013, Reference Molins, Moya, García-Breijo, Reig-Armiñana and Barreno2018; Moya et al. Reference Moya, Škaloud, Chiva, García-Breijo, Reig-Armiñana, Vančurová and Barreno2015; Catalá et al. Reference Catalá, del Campo, Barreno, García-Breijo, Reig-Armiñana and Casano2016). Although T. crespoana represents a distinct and well-supported lineage within Trebouxia clade G, the cells showed the same pyrenoid type as T. corticola, T. galapagensis, T. higginsiae and T. usneae (Friedl Reference Friedl1989). Ultrastructural traits of pyrenoids from cultured phycobionts have been traditionally used to characterize Trebouxia species (Friedl Reference Friedl1989). However, our up-to-date knowledge of Trebouxia diversity has expanded and the original classification proposed by Friedl (Reference Friedl1989) is in need of revision. This will allow ongoing and future studies to better delimit heterogeneous lineages in Trebouxia.
This project was supported by the Ministerio de Economía y Competitividad (MINECO, Spain) (CGL2016-79158-P), Excellence in Research (Generalitat Valenciana, Spain) (PROMETEOII/2013/021) and the Primus Research Programme of Charles University SCI/13. We would like to thank the technicians (Teresa Mínguez and Nuria Cebrián) at the Servicio de Microscopía Electrónica, SCSIE and Jardí Botànic (Universitat de Valencia) for their help with the TEM process. Daniel Sheerin revised the English. We dedicate this article to Ana Crespo in honour of her retirement.
Supplementary Material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0024282918000208






