Introduction
Anatolia (Asia Minor) is located strategically between Asia and Europe and is considered to be the centre of origin of many crop plants, including the cultivated grapevine, Vitis vinifera L. (McGovern, Reference McGovern2004). According to some estimates, grape cultivation in Anatolia and in the neighbouring Transcaucasia began approximately 7000–8000 years ago as the seeds of domesticated grapes, dating to ca. 8000 BP, were found in these regions (This et al., Reference This, Lacombe and Thomas2006). Even today, the wild grape (V. vinifera ssp. sylvestris) continues to thrive in these regions. Wild grapes are also distributed throughout the Near East, ranging from Western Anatolia eastward into southwest Asia, including Afghanistan (Heywood and Zohary, Reference Heywood and Zohary1991). However, the primary grapevine habitats mostly occur in the area known as the Fertile Crescent, which includes various geographical locations: Turkey, Iraq, Lebanon, Syria, Jordan, Israel and Iran. Rather unique biotic and abiotic features of this region have contributed to a wide range of diversity both within and between grape populations (Zohary and Hopf, Reference Zohary and Hopf2000). However, rapid industrialization of the region has recently put an enormous pressure on wild germplasm resources, including wild grape germplasm, which, like wild relatives of any crop plant, is an invaluable genetic resource for grape breeding. Therefore, accurate characterization and preservation of the existing grapevine germplasm of this region is urgently needed to prevent potential genetic erosion and variability loss.
Molecular markers are useful tools for studying genetic diversity. Using molecular markers, genetic analyses of wild grape populations from the Iberian Peninsula (Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted; Lopes et al., Reference Lopes, Mendoça, Rodrigues do Santos, Eiras-Dias and da Camara Machado2009) and France (Di Vechhi-Staraz et al., Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009) have recently been studied. Genetic diversity of V. vinifera ssp. sylvestris from Anatolia has not been studied, even though this region is known to be a wild grape primary habitat. A few earlier reports (Ergül et al., Reference Ergül, Marasalı and Ağaoğlu2002; Ergül et al., Reference Ergül, Kazan, Aras, Çevik, Çelik and Söylemezoğlu2006; Karataş et al., Reference Karataş, Değirmenci, Velasco, Vezzulli, Bodur and Ağaoğlu2007; Şelli et al., Reference Şelli, Bakır, İnan, Aygün, Boz, Yaşasın, Özer, Akman, Söylemezoğlu, Kazan and Ergül2007) have mainly focused only on autochthonous grape cultivars of this region.
In this report we sampled natural grape populations distributed across their ecological range in the Mediterranean and in western and northwestern regions of the Anatolian plateau. Our particular interests were to investigate the existing genetic diversity within and between wild grape populations provide insights into the evolution of grapes and facilitate the conservation of V. vinifera ssp. sylvestris germplasm from these primary habitats. In this paper we also compared the genetic diversity of Anatolian wild and cultivated grape germplasm as well as Anatolian and Iberian wild grapes to draw inferences about the evolutionary history of grapes within the regions neighbouring the Mediterranean basin.
Material and methods
Plant material
Eighty-four wild grape accessions from three Anatolian locations (Anamur, Fethiye and Gökçeada) were used in this analysis (Fig. 1) (Supplementary Table S1, available online only at http://journals.cambridge.org). The environmental conditions in these three locations are typical of wild grapevine habitats, which include lands and forests with a high degree of humidity due to numerous rivers and brooks. These locations are also characterized by abundant tree species such as elms, poplars and oaks, on which grapevines grow as a liana. The plant sampling strategy employed was the same for all populations and designed to prevent potential errors, such as inadvertently collecting individuals from cultivated subspecies (V. vinifera ssp. sativa) and also from rootstocks instead of wild plants. To reduce the likelihood of any sampling error, only dioecious individuals were collected, as cultivated grapes are hermaphrodites while wild subspecies are dioecious. In addition, SSR data from 31 autochthonous Anatolian grape cultivars and Spanish wild grape accessions from Iberian Peninsula (Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted) were used in comparisons referred to in the paper.
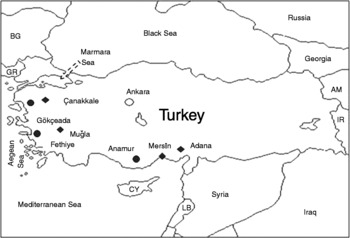
Fig. 1 Collection sites of wild grape populations from Anatolia (closed circles and diamonds indicate the collection sites of wild and cultivated grapes, respectively).
DNA extraction and PCR amplification
Total genomic DNA was extracted from frozen woody shoots of young leaves using DNeasy™ Plant Mini Kit (Qiagen). The DNA extracted was quantified and used as a working DNA solution of 10 ng/μl of the following 15 microsatellite loci well scattered on the genome: VVMD5, VVMD7, VVMD21, VVMD24, VVMD25, VVMD27 (Bowers et al., Reference Bowers, Dangl, Vignani and Meredith1996; Bowers et al., Reference Bowers, Dangl and Meredith1999), VVIN16, VVIP60, VVIH54, VVIB01, VVIV37, VVIN73, VVIP31 (Merdinoglu et al., Reference Merdinoglu, Butterlin, Bevilacqua, Chiquet, Adam-Blondon and Decroocq2005), VVS2 (Thomas and Scott, Reference Thomas and Scott1993) and VMC4f3 (Vitis Microsatellite Consortium).
Amplification reactions were performed in a total volume of 20 μl with 30 ng of DNA template, 0.25–0.50 μM of forward primer labelled either with 6:FAM; 6-carboxyflureiscein, HEX; hexachloro-6-carboxyfluorescein or with NED; 7'8'benzo-5,2,4,7 trichloro-5-carboxyfluorescein, fluorophore; 0.5 μM of unlabelled reverse primer; 150 μM of each dNTP (Boehringer, Mannheim, Germany); 2.5 mM MgCl2+1X buffer AmpliTaq; and 0.8 units AmpliTaq polymerase (PE Applied Biosystems, Foster City, CA, USA). The PCR was done with a GeneAmp PCR system 9700 thermocycler (PE Applied Biosystems). The cycling programme consisted of the following steps: 10 min at 94°C; 35 cycles of 45 s at 92°C, 60 s at 57°C and 90 s at 72°C; and a final extension step of 5 min at 72°C. Labelled amplification products were resolved on an automated 310 ABI PRISM DNA sequencer (PE Applied Biosystems), using a HD400-ROX as an internal size standard. The PCR fragments were detected with the GeneScan analysis software version 3.1, and the alleles were scored using Genotyper DNA fragment analysis software version 2.5.2 (PE Applied Biosystems).
Genetic diversity
Allele sizes and total number of alleles (N a) were determined for each SSR. Putative alleles were indicated by the estimated size in base pair counts. Genetic diversity was estimated using the following statistics: N a, effective number of alleles (N e); information index (I), observed heterozygosity (H o) calculated as the number of heterozygous genotypes over the total genotypes analyzed for each locus and expected heterozygosity (H e) (Nei, Reference Nei1973). All the calculations were tested using GenAlex software version 6.0 (Peakall and Smouse, Reference Peakall and Smouse2006). The Wright's inbreeding coefficient (Fis) was estimated following Weir and Cockerham (Reference Weir and Cookerman1984), and its significance (Fis ≠ 0) was tested after 1000 permutations. A positive value of Fis indicates a deficit in heterozygosity in comparison with the Hardy–Weinberg equilibrium expectations, while a negative value of Fis indicates an excess of heterozygosity.
Cluster analysis
The genetic distances between individuals were calculated based on the proportions of shared alleles, as described by Bowcock et al. (Reference Bowcock, Ruíz-Linares, Tomfohrde, Minch, Kidd and Cavalli-Sforza1994), using the program MICROSAT (Minch et al., Reference Minch, Ruíz-Linares, Goldstein, Feldman, Kidd and Cavalli-Sforza1997). A dendrogram was constructed using the unweighted pair group method with arithmetic mean (UPGMA) cluster analysis along with the use of the program Neighbor of the PHYLIP version 3.57 software (Felsenstein, Reference Felsenstein1989) and constructed using the Tree View program version 1.5 (Page, Reference Page1996).
Genetic differentiation
Analysis of molecular variance (AMOVA, Excoffier et al., Reference Excoffier, Smouse and Quattro1992) was performed to partition the observed genetic variability among and within populations using the GENEALEX program. Fst was estimated over all populations and between each pair of populations using the method of Weir and Cockerham (Reference Weir and Cookerman1984). Because some of the microsatellite markers have imperfect or compound loci and therefore did not follow the stepwise mutation model, we choose to use Fst instead of Rst. The calculations were tested using the FSTAT program (Goudet, Reference Goudet2001). Principal component analysis (PCA) was used to display genetic divergence among samples in a multidimensional space using GENEALEX program version 6.0 (Peakall and Smouse, Reference Peakall and Smouse2006).
Results
Genetic discrimination of V. vinifera ssp. sylvestris from rootstocks and V. vinifera ssp. sativa
In this study, special attention was given to the collection of wild grapes from their natural habitats. However, to further ascertain that the presumed wild accessions are true wild accessions but not naturalized rootstocks or cultivated grapes, we compared the genotype data from these analyses (Supplementary Table S2, available online only at http://journals.cambridge.org) with those obtained from the analysis of rootstocks having the same markers (De Andres et al., Reference De Andres, Cabezas, Ververa, Borrego, Martinez-Zapater and Jouve2007). In these analyses, no individuals identified as rootstocks were found among the wild accessions studied. Moreover, we have compared the wild Anatolian grapevine genotypes with 31 autochthonous grape cultivars from Turkey, using the same 15 microsatellites that we have used for the wild grape accessions (Supplementary Table S3, available online only at http://journals.cambridge.org). The results from these experiments did not indicate the presence of any misclassified cultivated grapevines among our wild grape samples. This indicates that our diversity measurements are not likely to be biased by inadvertently including hybrids in one or the other pool, which would artificially change the genetic diversity values.
Genetic diversity of Anatolian wild grapes
All 15 SSR primer combinations used (see materials and methods) efficiently and reproducibly amplified polymorphic fragments from all wild grape accessions surveyed. The number of scoreable fragments amplified by each SSR primer pair varied from six for VVIN16 and VVIB01 to 21 for VMC4f3.1 with an average of 12.26 per primer combination. A total of 184 scoreable alleles were detected among the 84 genotypes (Table 1). The frequency of these alleles was lower than 25% for most loci, except for the four alleles of the VVMD24, VVIB01, VVIN73 and VVIN16 loci, which had a frequency of higher than 40%, the highest frequency found in this study for wild populations. In addition, there was no correlation between the N a detected and the N e and I values calculated from these analyses. The highest N a appeared in the VMC4f3.1 locus (21 alleles) with N e and I values of 10.523 and 2.6, respectively. The VVMD32 locus had 14 alleles, and the N e and I values were 10.001 and 2.417, respectively. This result suggests that the VVMD32 locus is more informative than the VMC4f3 locus.
Table 1 Total genetic diversity in the 84 wild grape accessions from Anatolia Peninsula

F, fixation index.
The H o values ranged from 0.524 in VVIN16 to 0.881 in VVIP31, with an average value of 0.748, while the H e values ranged from 0.609 in VVIN16 to 0.905 in VMC4f3.1, with an average value of 0.811. A comparison between these two parameters was carried out based on Fis values. For 13 loci, the Fis value was positive, meaning a deficit of heterozygotes, whereas for only two loci (VVMD24 and VVMD25), the Fis value was negative (Table 1). Deviations of genotypic frequency from Hardy–Weinberg equilibrium (HWE) within a single population can be represented by Fis values. In this study, Fis values for 13 out of 15 loci were low and not significantly different from zero, indicating random mating occurring among individuals.
Genetic distance analysis of wild grape germplasm
The genetic distance-based results seen in the neighbour-joining tree divided the wild accessions analyzed into four major groups (Fig. 2) that partially correlated with geographic origin of the accessions. One small cluster called ‘A’ mostly contained wild accessions from the Anamur region. In this cluster, there were also two individuals (G1 and G13) collected from the Gökçeada region. The second cluster called ‘B’ mostly contained Anamur individuals with some Gökçeada individuals (G8, G12, G9, G10, G17, G18, G19, G27 and G30) were also included in this cluster. The third cluster called ‘C’ is divided into two sub-clusters that contained Fethiye and Gökçeada accessions. Finally, the fourth cluster called ‘D’ mainly contained Gökçeada accessions.

Fig. 2 UPGMA analysis of microsatellite diversity based on the proportion of shared alleles. Each branch is colour coded according to the genetic populations (Pop A, B, C and D), G, Gökçeada; F, Fethiye; A, Anamur (A colour version of this figure can be found online at journals.cambridge.org/pgr).
The PCA (Supplementary Fig. S1, available online only at http://journals.cambridge.org) supported the results of clustering by UPGMA. The first principal component (21.77%) of the variation clearly separated the populations into groups (C–A) and (D–B), while the second principal component (18.07%) defined groups (A–B) and (C–D). PCA also indicated that some accessions from the same population were genetically closer to accessions from different populations than those of the same population.
We also analyzed the allelic patterns across four populations (Table 2). The mean allele number (MNA) was higher in cluster B than in the remaining populations. The lowest N a values appeared in cluster A. All the primers showed unique alleles in all the populations except cluster A. The largest average number of unique alleles was detected in cluster B. The mean values of genetic diversity (H e) were calculated for each cluster. All clusters showed high values of genetic diversity, and the highest values found were in cluster B accessions (H e = 0.788).
Table 2 Genetic parameters examined at the four genetic groups of wild individuals

N, sample size.
a Based from 15 SSR loci.
The genetic differentiation of wild grape germplasm using AMOVA analysis showed that most of the genetic diversity was attributable to differences among individuals within populations (92%) and only 8% between populations. The Fst analysis performed revealed a moderate genetic differentiation value (Fst = 0.059, P < 0.0001) between the genetic groups A and D and lower genetic differentiation in the rest of genetic populations (P < 0.0001) (Supplementary Table S4, available online only at http://journals.cambridge.org). The low level of population differentiation is suggestive of high levels of gene migration among populations. The lowest values, on the other hand, were found between the genetic groups A and D.
Comparisons of genetic diversity among Anatolian wild and cultivated grapevines as well as among Anatolian and Iberian wild grapevine populations
The genetic diversity of the wild grape populations around the Mediterranean basin using chlorotype microsatellites has shown that central Mediterranean and Eastern populations have higher genetic diversity values than Western populations as Iberian Peninsula (Arroyo-García et al., Reference Arroyo-García, Ruiz-García, Bolling, Ocete, Lopez, Arnold, Ergul, Söylemezoglu, Uzun, Cabello, Ibáñez, Aradhya, Atanassov, Atanassov, Balint, Cenis, Costantini, Goris-Lavets, Grando, Klein, McGovern, Merdinoglu, Pejic, Pelsy, Primikirios, Risovannaya, Roubelakis-Angelakis, Snoussi, Sotiri, Tamhankar, This, Troshin, Malpica, Lefort and Martinez-Zapater2006). In order to analyze this result using nuclear microsatellites, we have investigated the genetic diversity between V. vinifera ssp. sylvestris from both ends of the Mediterranean basin; the genotype data obtained in this study were compared with genotype data from 192 wild Iberian grape genotypes (Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted).
The genetic diversity values were higher for the wild grapevine from Anatolian Peninsula (H e = 0.811 ± 0.025) than for the wild grapevine from Iberian Peninsula (H e = 0.748 ± 0.007). We also compared the N a at the 15 loci in Anatolian populations with 84 accessions to those found in 192 wild Iberian (Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted) accessions (Table 3). Out of 229 alleles found at 15 loci, 87 (3.3%) were unique (i.e. occurred only in populations from one of the countries). Out of these 92 alleles, 34 occurred in Iberian and 53 in Anatolian populations, and the percentages of unique alleles in populations from each of the countries were 17.9 and 22.36%, respectively. The average, as well as the total N a from all 15 loci, was significantly higher in Anatolian than in Iberian populations. When genetic diversity values between Anatolian and Iberian populations were compared, we found that Iberian populations had an average H e value of 0.65 (Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted) while Anatolian populations had an average H e value of 0.811, indicating higher genetic diversity values at the wild Anatolian grape populations than wild Iberian grape populations.
Table 3 Allelic diversity values between wild grape accessions from IP and AP

IP, Iberian Peninsula; AP, Anatolian Peninsula.
Furthermore, we compared the genetic diversity values of Anatolian wild grapes with those of Anatolian cultivated grapes originated from the same or surrounding locations as the wild grapes used in this study. The SSR data from 31 autochthonous cultivated grapevines showed genetic diversity values (H e = 0.712) that are similar to other cultivated grapevines from the Marmara (H e = 0.742), Aegean (H e = 0.742) and Mediterranean (H e = 0.765) regions (Yüksel, Reference Yüksel2008) (Gök Tangolar et al., Reference Gök Tangolar, Soydam, Bakır, Karaağaç, Tangolar and Ergül2009). This comparison showed that the cultivated grapevine from Turkey has lower genetic diversity than the wild grape accessions.
In order to analyze the genetic relationship between cultivated and wild accessions from Anatolian Peninsula, we have used a PCA based on individual genotypes (Fig. 3). The cultivated grapevine accessions cluster on the left side of the plot, while the wild accessions cluster on the right side of the plot. The first principal component accounts for 33.5% of the total variation, and the second accounts for 16.2% of the variation. A clear separation between cultivated and wild accessions is showed in this analysis.

Fig. 3 PCA of the wild and cultivated grapevine accessions.
Discussion
Wild grapevines have been identified in Anatolia region in a wide range of natural and disturbed habitats (Heywood and Zohary, Reference Heywood and Zohary1991). However, until now, a systematic genetic characterization of the individual plants had not been done to confirm whether they are bona fide wild individuals, naturalized grapevine cultivars or rootstocks, or spontaneous hybrids derived from wild and cultivated forms as described by Di Vecchi-Staraz et al. (Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009). Although their genotypic analysis could not detect the existence of hybrids in the individuals analyzed, we cannot discard the possibility of the existence of a putative hybrid with cultivated grapevine or rootstock. Different studies suggest genetic exchange between cultivated and wild grapevines (Grassi et al., Reference Grassi, Labra, Imazio, Spada, Sgorbati, Scienza and Sala2003; Cunha et al., Reference Cunha, Balerias-Couto, Cunha, Banza, Soveral, Carneiro and Eiras-Dias2007), but we have not identified any putative hybrid formed between the cultivated and wild Anatolian grapevines. This result could be due to the relatively low numbers of the cultivated grapevines analyzed in this study.
The total genetic diversity values found in wild types from Anatolia are higher than those of wild-type accessions from other regions such as those described for the Mediterranean basin (Andrés et al., submitted; Di Vecchi-Staraz et al., Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009; Lopes et al., Reference Lopes, Mendoça, Rodrigues do Santos, Eiras-Dias and da Camara Machado2009; Zinelabidine et al., Reference Zinelabidine, Haddioui, Bravo, Arroyo-Garcia and Martinez-Zapater2010). In general, these values are similar for outcrossing, vegetatively propagated perennial species (Belaj et al., Reference Belaj, Muñoz-Diez, Baldoni, Procedí, Barranco and Satovic2007). The H o is not significantly different (P ≤ 0.01) from H e in the wild Anatolian group, indicating a random mating population. However, reduction in H o has been observed in wild grapevine populations analyzed in Spain, Portugal, France and Italy (Grassi et al., Reference Grassi, Labra, Imazio, Spada, Sgorbati, Scienza and Sala2003; Di Vecchi-Staraz et al., Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009; Lopes et al., Reference Lopes, Mendoça, Rodrigues do Santos, Eiras-Dias and da Camara Machado2009; Andrés et al., submitted), most likely due to the reduction of these populations by human action. As a consequence, these natural populations have a risk of inbreeding depression. In contrast, the Anatolian wild populations showed high genetic diversity and random mating. This result is in agreement with the comparison of the N a at the 15 shared SSR loci between Iberian and Anatolian populations. Of 229 total alleles detected at these loci, 189 were observed only in Iberian Peninsula, while 237 were observed only in Anatolian populations. The number of unique alleles in Anatolian populations was also much higher than those in Iberian populations. This result was expected, as Anatolian populations are located at the primary centre of diversity of this specie and thus are more diverse than in the peripheral populations. In fact, it has been found that the Iberian wild grape population showed lower genetic diversity values and suffer from inbreeding depression (Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted). Similarly, French and Portuguese wild populations have been shown to have lower genetic diversity and putative inbreeding depression (Di Vecchi-Staraz et al., Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009; Lopes et al., Reference Lopes, Mendoça, Rodrigues do Santos, Eiras-Dias and da Camara Machado2009). Furthermore, analysis of wild grapes from Eastern countries such as Iran or Georgia, the presumed centre of primo domestication, will be fundamental because this might help to elucidate the genetic diversity genetic pool involved in grape domestication.
The UPGMA clustering method revealed the existence of four genetic groups in wild grapevines from Anatolia, which partially correlated with the geographic origin of these genotypes. The highest genetic diversity (H e) values in wild grapes were found in the genetic group B, which predominantly contains Anamur accessions. When considering the four different genetic groups identified by the UPGMA analysis, the genetic group B showed higher genetic diversity than the rest of the groups. The relatively lower genetic variation in the A, C and D groups might be due to a sampling bias, as the lowest N a was detected in the population with the smallest size, and a positive correlation of N a and sample size can be generally observed (Riahi et al., Reference Riahi, Soghlami, El-Heir, Laucou, Cunff, Boursiquot, Lacombe, Mliki, Ghorbel and This2010).
The results indicated that the genetic diversity in the Anatolian wild grape germplasm is randomly distributed, and putative gene flow occurs between populations. As shown in the clusters given in Fig. 2, the accessions from Fethiye and Anamur mostly clustered with other accessions from the same two regions, while Gökçeda accessions frequently clustered together with the accessions from the other two regions. These results agree with the moderate genetic differentiation values that we have observed between genetic groups. The gene flow may have occurred among the populations of these three regions. However, the distances between these regions are much further than the possible gene flow distance (~2 km) estimated for wind-pollinated tree species such as pine and oak (Streiff et al., Reference Streiff, Ducousso, Lexer, Steinkellner, Gloessl and Kremer1999; Schuster and Mitton, Reference Schuster and Mitton2000), suggesting that wind pollination may not be a factor responsible for this gene flow. Another possibility that may have contributed to the relocation of wild grapes from one environment to another may be seed dispersal. Once new plants are introduced into a new environment through seed dispersal, further breeding is expected to occur between the introduced plant and the plants from local populations, leading to the gene flow from one population to other. Finally, we cannot discard the possibility that part of the moderate genetic differentiation between the genetic groups could result from the different history of their relationship with the cultivated group.
The AMOVA analyses, which showed partitioning of the genetic variability by means of gene diversity statistics (Nei, Reference Nei1973), indicated that, on average, 92% of SSR diversity was distributed within the populations and only 8% between populations. This is consistent with findings from other studies conducted on woody plants that considerable genetic diversity is partitioned within, rather than between populations (Turpeinen et al., Reference Turpeinen, Tehola, Manninen, Nevo and Nissila2001; Belaj et al., Reference Belaj, Muñoz-Diez, Baldoni, Procedí, Barranco and Satovic2007).
The comparison of the genetic diversity values with the autochthonous grape cultivars from Anatolia Peninsula indicated that diversity is greater in the wild grapes than in the cultivated ones. These results are contrary to the results found in other studies (Lopes et al., Reference Lopes, Mendoça, Rodrigues do Santos, Eiras-Dias and da Camara Machado2009; Riahi et al., Reference Riahi, Soghlami, El-Heir, Laucou, Cunff, Boursiquot, Lacombe, Mliki, Ghorbel and This2010). These results suggest that the Anatolian wild populations are not suffering inbreeding depression as observed in the other wild populations in the Mediterranean basin (Lopes et al., Reference Lopes, Mendoça, Rodrigues do Santos, Eiras-Dias and da Camara Machado2009; Di Vecchi-Staraz et al., Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009; Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted).
The genetic relationship between cultivated and wild accessions indicated that the cultivated accessions do not derive directly from local wild populations. However, the possibility that some cultivars derived from ancestral events of local domestication or cross hybridization with native wild plants could not be ruled out. In fact, this has been described in some putative hybrids between wild and cultivated compartment (Di Vecchi-Staraz et al., Reference Di Vechhi-Staraz, Laucou, Bruno, Lacombe, Gerber, Bourse, Boselli and This2009; Andres et al., Reference Andres, Benito, Perez-Rivera, Ocete, Lopez, Gaforio, Muñoz, Cabello, Martinez-Zapater and Arroyo-Garciasubmitted). In conclusion, the present study suggests that there is no immediate reason for concern about any demographic bottlenecks facing the wild grape populations of this region, and the presence of high number of rare alleles in populations investigated here is clear evidence for this finding. For the future, in situ conservation of the populations in the primary centre of diversity should be advanced by a dynamic approach to keep the level and composition of genetic diversity as high as possible for safeguarding these precious genetic resources for crop improvement.
Acknowledgements
G. P.-R. was the recipient of a technical fellowship from INIA. This research was supported by grants CGL-2005-06821-C02-01 and RTA2008-00032-C02-01.








