With the advent of powerful genome sequencing tools, and consequently an extensive number of sequenced genomes, new opportunities have arisen for dissecting gene regulation and molecular evolution (Rijnkels et al., Reference Rijnkels, Elnitski, Miller and Rosen2003). Comparative genomics is one such opportunity which allows the discovery of new genes and aids in the identification of functional components. In comparative genomics two or more genomes are compared in a large-scale holistic approach to discover the differences and similarities between the individual genomes (Wei et al., Reference Wei, Liu and Dubchak2002). Over 17 000 fully sequenced and draft sequences of archaa, bacteria, and eukaryote genomes with a wealth of sequence data are freely available for public use (https://www.ncbi.nlm.nih.gov/genome). With comparative genomics there is a potential to gain major scientific insights about gene gain or loss, species origins, mammalian orders and survival (Wei et al., Reference Wei, Liu and Dubchak2002).
Casein genes (αs1-, αs2-, β- and κ-casein) have evolved from members of a group of secreted calcium phosphate binding phosphoprotein genes, specifically the ODAM gene (Kawasaki et al., Reference Kawasaki, Lafont and Sire2011). In milk, caseins and large amounts of colloidal calcium and phosphate form aggregates that are known as casein micelles (Walstra and Jenness, Reference Walstra and Jenness1984). The formation of casein micelles is very critical to the effective transport of high concentrations of calcium and phosphate from the lactating mother to the neonate via milk (Holt et al., Reference Holt, Carver, Ecroyd and Thorn2013). However, it appears that some mammalian species are devoid of some casein types. African elephant milk, for example, lacks α-caseins, but nevertheless contains casein micelles just as observed in the milk of all mammalian species studied so far (Martin et al., Reference Martin, Cebo, Miranda, McSweeney and Fox2013; Madende et al., Reference Madende, Osthoff and Patterton2015).
This observation has prompted an investigation into the distribution of casein genes across several mammalian species by comparative genomics. Several comparative studies have been done on caseins in the past, albeit mostly at protein level (Ginger and Grigor, Reference Ginger and Grigor1999; Holt, Reference Holt2015). Comparison of casein genes (presence/absence and gene sequences) may shed light into the possible functional aspects of casein genes and their gene products, particularly with reference to their importance in the formation of casein micelles in milk, especially in those species where some of the caseins are absent.
Materials and methods
Ensembl genome browser (https://www.ensembl.org) tool of comparative genomics was utilized in the comparison of casein genes across mammalian species (Herrero et al., Reference Herrero, Muffato and Beal2016; Aken et al., Reference Aken, Achuthan and Akanni2017). In summary, Ensembl provides comprehensive evidence-based annotation of all supported genome sequences. The gene annotations across all species provided by Ensembl gene build are automatically integrated. Gene trees were constructed from all casein genes available and the data were used to extract homologs (orthologs and paralogs). Using LastZ and its predecessor BlastZ tools, the synteny mappings from pair-wise alignments of species whose mammalian genomes are not too fragmented were derived. For this study, casein gene comparison was focused on eutherian (placental) mammals.
Results
αs1-casein
Alpha s1 casein is one of the least represented genes across eutherian mammalian species. In total, there are only 22 homologs of the αs1-casein (CSN1S1) gene, of which 11 of the homologs are primates and rodents, whereas 3 of the homologs are Laurasiatheria mammals (super order of Laurasia originating mammals). The rest of the homologs, 8 in total, are eutherian mammals, which include cow, human and sheep as the notable examples. Noted as the species of interest in this study, African elephant does not have the CSN1S1 gene, although its closest relative, the hyrax, does. The gene sequence alignment representation of the 22 αs1-casein homologs is depicted in Fig. 1a. The alignment shows several gaps in the sequences. Most of the gap positions are consistent with each mammalian group or sub-tree, for example, the primates have 3 large gaps (indicated in white), that are consistent amongst the gene sequences.
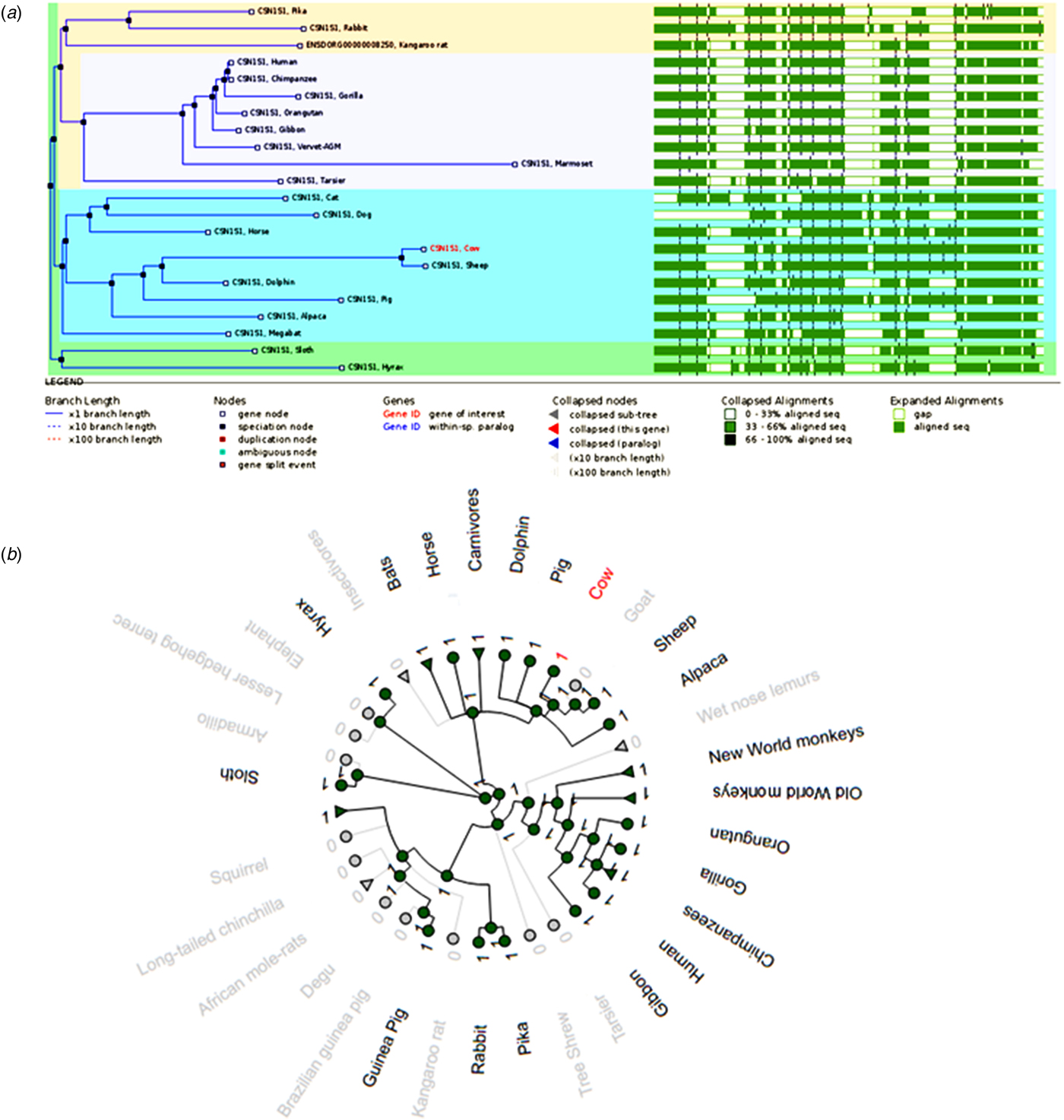
Fig. 1. A comparison of αs1-casein genes. (a) Gene tree of the relationship between 22 αs1-casein gene sequences and their sequence alignment. (b) The radial gene gain or loss representation of αs1-casein gene amongst several mammalian species. The green circular nodes indicate the presence of the gene whereas the gray nodes indicate its loss or absence.
Figure 1a also underlines the disparate nature of CSN1S1 gene sequences, most of the regions in the sequence alignment share only 33–66% sequence homology and none of the regions have over 66–100% sequence homology. The αs1-casein sequences also vary in length from one to the other. As an example, the cow gene is translated into a 214 amino acid long protein, whereas the pig gene is translated into a 206 amino acid long protein. All the 22 homologs of the αs1-casein gene are orthologous, meaning they all have a common ancestral gene and were separated through a speciation event. Orthologous gene products often retain the same function in the new species (Koonin, Reference Koonin2005).
Figure 1b depicts the CSN1S1 gene gain or loss in a radial view. The gene gain or loss figure shows that the CSN1S1 gene has been lost in a number of ancient mammalian species such as elephant and armadillo. Interestingly, this gene, which had been predicted as the oldest of the casein genes, has been lost even between the closely related species. The classical example is between the vervet monkey, olive baboon and the macaque. Although these 3 primates branched from the same αs1-casein ancestral gene, only the vervet monkey retained the αs1-casein gene. This aforementioned pattern between closely related species is observed consistently throughout the gene gain or loss tree. However, it must be mentioned that only genome sequences that are not too fragmented, were considered for this study. In the case of the absence of CSN1S1 gene in elephant and armadillo, several genome databases have been consulted, and in all the cases, the CSN1S1 gene was absent (see online Supplementary Fig. S1).
αs2-casein
Like the CSN1S1 gene, the αs2-casein (CSN1S2) gene is also minimally represented. Only 13 homologs can be observed in Fig. 2a. The placental or eutherian mammals are the most represented with 9 homologs whereas the rodents and rabbits family only have four representing members. Interestingly the rat and mouse are unusual in possessing a CSN1S2-like casein gene copy which is represented in the alignment as csn1s2b. Unlike the CSN1S1 gene, the CSN1S2 gene family has paralogs present in addition to the orthologs. In cats specifically, the CSN1S2 gene has undergone a duplication event through the course of evolution, resulting in a CSN1S2 gene that shares a common ancestor with other αs2-casein homologs (see online Supplementary Fig. S2). In most cases, paralogous gene products usually perform different functions in the same species. Interestingly, African elephant lacks both α-casein genes, its close relatives, such as the hyrax, only lack the CSN1S2-like casein gene, which developed later than all the other casein genes. The hyrax retained the CSN1S1 gene, which is the oldest of the casein genes. The absence of both α-casein genes appears to be unique to African elephant, although the gene loss or gain plots show absence of both α-casein genes in the squirrel, this is not exactly accurate. The squirrel does have the CSN1S1 gene but lacks the CSN1S2-casein gene, these data have not been taken into account on the gene gain or loss plots due to the high fragmentation of the squirrel gene sequence database.

Fig. 2. A comparison of αs2-casein genes. (a) Gene tree of the relationship between 13 αs2-casein gene sequences and their sequence alignment. (b) The radial gene gain or loss representation of αs2-casein gene amongst several mammalian species. The green circular nodes indicate the presence of the gene whereas the gray nodes indicate its loss or absence.
There are also several gaps in the sequences as shown by the sequence alignment. Sequence comparison across all the 13 species indicates that there is between 33–66% homology between the sequences. Figure 2b further shows that there is increased homology among CSN1S2 gene sequences that are in the same subgroup. For example, when the placental mammals' subgroup is considered, the sequence homology increases to between 66–100% (as indicated by dark green shaded areas on the alignment). Furthermore, Fig. 2b also highlights the high homology and conserved nature of the signal peptide, which is located at the N-terminus region. The sequence length also varies considerably from one species to the other, as highlighted by the gaps in the sequences. This further increases variability among orthologous gene products. Some gaps are much larger, making the sequences shorter (armadillo), whereas other gaps in the sequence are relatively smaller, making the sequences longer (sheep).
β-casein
The β-casein (CSN2) encoding gene is much more common among mammalian species compared to both αs1- and αs2-casein encoding genes. The gene tree and alignment in Fig. 3a shows 40 homologs of the CSN2 gene. The primates and rodents are the most represented species with up to 19 β-casein homologous genes. Interestingly, the squirrel and microbat genomes show the presence of paralogs of the β-casein gene, as depicted by duplication nodes (colored in red) in both Figs 3a and 3b. Paralogs are a consequence of evolution through gene duplication, resulting in two active sets of genes, whose products usually assume different functions although having a common ancestral gene (Koonin, Reference Koonin2005).
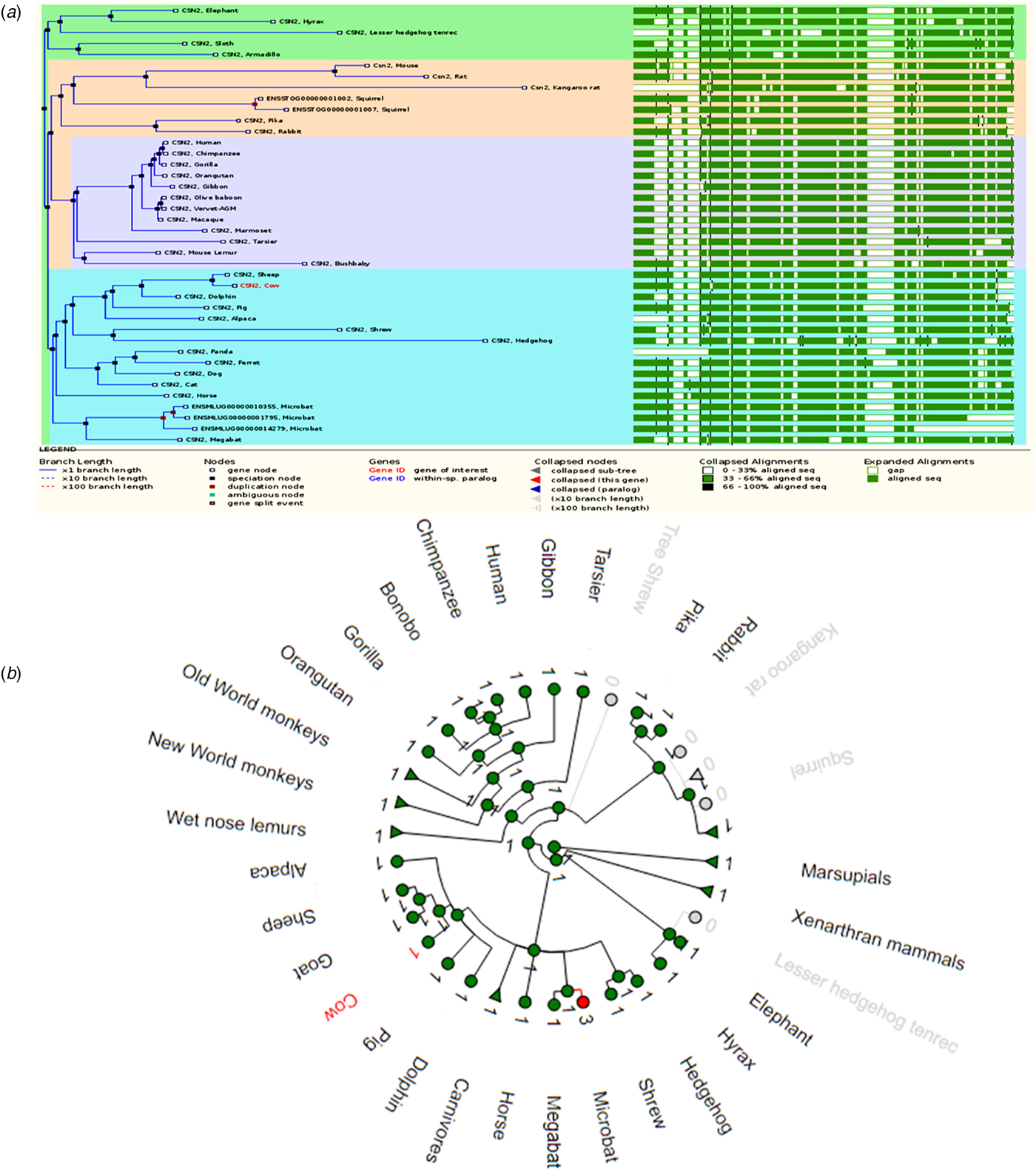
Fig. 3. A comparison of β-casein genes. (a) Gene tree of the relationship between 40 β-casein gene sequences and their sequence alignment. (b) The radial gene gain or loss representation of β-casein gene amongst several mammalian species. The green circular nodes indicate the presence of the gene whereas the gray nodes indicate its loss or absence.
Like CSN1S1 and CSN1S2 genes, the sequence alignment across all the species shows a higher degree of divergence with between 33–66% sequence homology. However, the opposite is true for a sequence alignment of species that are in the same subgroup, where a much higher sequence homology of between 66–100% is observed (Fig. 3b). Several gaps also exist in the sequence alignment, with most gaps consistent throughout the alignment. As mentioned before, such gaps increase the heterogeneity of casein genes and their products. The CSN2 gene is more conserved among the closely related species. In addition to retaining the CSN2 gene, it appears that more genes have been gained through duplication events over the course of evolution, as illustrated by red nodes. The expanded gene tree, showing the distribution of CSN2 gene among mammals and non-mammalian species, is supplied as online Supplementary Fig. S3.
κ-casein
The κ-casein (CSN3) gene is the most studied casein gene, and as a result, this gene presents interesting comparative genomics. Unlike αs1-, αs2- and β-casein encoding gene products, that are calcium sensitive, the gene product of κ-casein is soluble in calcium (Ginger and Grigor, Reference Ginger and Grigor1999). Figure 4a shows a CSN3 gene tree relationship between 35 mammalian species. All homologs presented on the gene tree are also orthologs, meaning they are the result of a speciation event rather than a duplication event. The gene tree members are dominated by primates and rodents with 18 members, bats are the least represented with only 2 sequences.
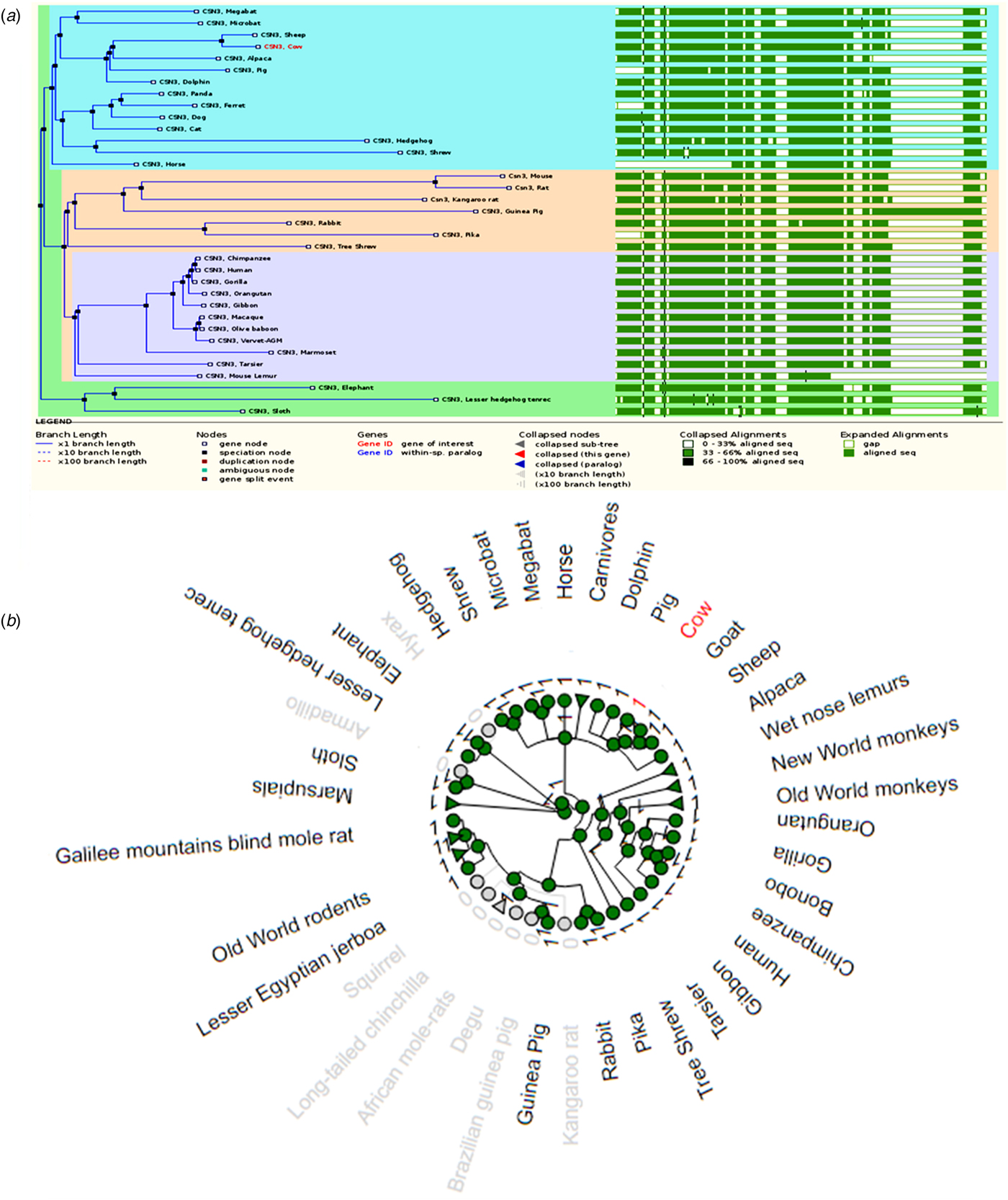
Fig. 4. A comparison of κ-casein genes. (a) Gene tree of the relationship between 35 κ-casein gene sequences and their sequence alignment. (b) The radial gene gain or loss representation of κ-casein gene amongst several mammalian species. The green circular nodes indicate the presence of the gene whereas the gray nodes indicate its loss or absence.
A look at these homologous gene sequences shows a great deal of divergence. As was noted for the CSN1S1, CSN1S2 and CSN2 genes, the sequences mostly share between 33–66% (light green color) homology when compared together. Increased sequence homology is observed when sequences are compared within a subgroup. Gaps in the sequences are also common amongst CSN3 gene sequences, although these gaps are much more consistent throughout the sequences. In addition, the sequence comparison also reveals the differences in length of each gene. The horse CSN3 gene appears to be the shortest of the 35 genes. It is important to note that the length of genes do not necessarily reflect the length of its gene products. Events such as exon skipping often result in short length gene products that are shorter than their full length counterparts.
Figure 4b shows the gene gain or loss relationship of CSN3 genes from a variety of species. The figure also suggests that some mammalian species such as the wallaby and squirrel have lost the κ-casein gene, this is not entirely correct. The CSN3 gene is present in these species, but the sequences are highly fragmented, and therefore were omitted from the gene gain or loss plot. The genome sequence of the squirrel is complete but the sequence is of very low quality and therefore several mistakes are expected from the genome. Nevertheless, it is clear from the figure that most mammalian species have retained the CSN3 gene. The same observation was noted for CSN2 gene tree. The expanded gene tree showing the distribution of CSN3 gene among mammals and non-mammalian species is supplied as online Supplementary Fig. S4.
Discussion
A number of comparative studies of caseins have been previously conducted at protein level with the most recent study conducted by Holt (Reference Holt2015). Because no single organism can adequately describe the functionality of the other, comparative studies are of paramount importance if more light is to be shed on debatable concepts and models. Caseins are rapidly evolving genes and this was evident in our study of comparative genomics of casein genes across all mammalian species whose sequence data is available and is not too fragmented. Of the 4 casein genes, the α-casein genes are the less represented group. In contrast, most mammalian species do possess CSN2 and CSN3 genes.
With regards to alignment of casein genes sequences, it is clear that casein gene sequences (αs1-, αs2-, β- and κ-caseins) are very diverse, although sequences of very closely related species show increased homology. Apart from the signal peptide sequence that is highly conserved, the rest of the mature peptide sequence is very diverse. Moreover, gaps in the sequences, that are introduced to maximize the multiple alignment, also contribute to the diverse nature of casein gene sequences, since they highlight the conserved and non-conserved sequence regions. The non-homologous nature of casein gene sequences could be related to the rather less specific function of their gene products in the casein micelle assembly. In addition to gene sequence differences, further divergence and variability of caseins is introduced by events such as exon skipping, which occur during processing of primary transcripts (Martin et al., Reference Martin, Cebo, Miranda, McSweeney and Fox2013).
In addition to the presence of orthologs, paralogs are also a common feature amongst casein genes, specifically the CSN1S2 and CSN2 gene family. Paralogs occur due to duplication events, leading to the same species having more than one pair of the particular casein gene. The gene gain or loss figures also highlight the absence of α-casein encoding genes in most ancient mammalian species, such as the African elephant and armadillo, whereas in most modern mammals, such as cow and horse, the gene is present. It is interesting to note that, according to the casein micelle models, all four caseins have a role to play in the formation of a bovine casein micelle (Horne, Reference Horne1998). Bovine casein micelle models generally fall into mainly three categories: coat-core models, sub-micelle models and internal structure models (Phadungath, Reference Phadungath2005). Based on experimental evidence, models that fall into the coat-core category proposed that a casein micelle structure is composed of a layer of κ-casein that forms a coat around a core polymer of both αs1- and β-caseins that contain charged phosphorylated loops (Wong, Reference Wong1988). Unlike the coat-core models, the sub-micelle models proposed that casein micelles are formed by smaller sub-micelles that are in turn bound together with colloidal calcium phosphate, and stabilized by hydrophobic interactions and calcium caseinate bridges (Rollema, Reference Rollema and Fox1992). The individual subunits are composed of a combination of αs1-, β- and κ-caseins. Lastly, the internal structure models are based on the properties of all the casein types directing the formation of the internal structure of casein micelles (Wong, Reference Wong1988). These models generally cement the role of colloidal calcium phosphate and the outside location of the hairy layer of κ-casein (Smyth et al., Reference Smyth, Clegg and Holt2004). Furthermore, experimental data suggested that casein micelles are stabilized by negative charges of protruding κ-casein, as well as a zeta potential of approximately −20 mV at pH 6.7.
It appears from the comparative genomics of casein genes that not all four caseins are present in all the mammals that were investigated, however both ancient and modern mammalian species have CSN2 and CSN3 genes as a common feature, therefore suggesting that these two genes and their gene products may have a much bigger and important role to play in casein micelle formation. Beta and κ-caseins are important for provision of hydrophobic interactions and electrostatic repulsion, respectively, and their importance was consistently highlighted in all the three categories of casein micelle structure described above (Horne, Reference Horne1998). Data from the current study support this observation, although we also propose that casein micelles do not strictly require all four casein types for their formation. Recent data from proteomics analysis of African elephant milk, which is devoid of αs-caseins (Madende et al., Reference Madende, Osthoff and Patterton2015), shows that this milk contains casein micelles (unpublished). Moreover, its β-casein, which is highly abundant, also displays a very different phosphorylation profile compared to that of cow β-casein, and could potentially take up the role that αs-caseins play in the formation of casein micelles (Madende et al., Reference Madende, Kemp, Stoychev and Osthoff2018). Human milk lacks αs2-casein, and it may be possible that its role in casein micelle formation could be shifted to αs1-casein, which is capable of forming disulfide-linked heteromultimers with κ-casein (Martin et al., Reference Martin, Cebo, Miranda, McSweeney and Fox2013). This demonstrates that caseins can be multifunctional with regards to micelle formation, and as a result, the presence of all four (sometimes five) caseins may not be a prerequisite for casein micelle formation in milk.
The gene comparison data show a lot of differences with regards to the presence or absence of casein genes among mammalian species. Some of these differences are rather extreme, for example, the gene gain or loss tree shows that the squirrel only has β-casein, while the rest of the caseins are absent. It is of paramount importance to note that comparative genomics data is only as good as the quality of the genome databases (Muller et al., Reference Muller, Naumann and Freytag2003). Sequencing errors that are carried over to the actual genome database may be misleading and therefore result in inaccurate interpretation of results. In addition, gene data is also dependent on databases and the quality of sequencing, and therefore some of the genes are shown to be absent from the gene tree because of incompleteness of the genome database or an omission error. In the case of the squirrel, the draft genome of the thirteen-lined squirrel has been sequenced in full, but has not been assembled into chromosomes, and the sequence data is not of the highest quality (Di Palma et al., Reference Di Palma, Alfoldi, Johnson, Berlin, Gnerre, Jaffe, MacCallum, Young, Walker and Lindblad-Toh2011). Recently, the Wellcome Sanger Institute and its collaborators have sequenced high quality full genomes of 25 species, including the red squirrel and the gray squirrel, for the first time (http://www.sanger.ac.uk/science/collaboration/25-genomes-25-years). Currently the aforementioned genomes are being assembled before they are made available for public use and may provide further insight into the casein genes of the squirrel.
The evolution of casein genes follows the order: CSN1S1, CSN2, CSN1S2 and CSN3 (Martin et al., Reference Martin, Cebo, Miranda, McSweeney and Fox2013), the development of CSN1S2 gene occurred less than 147.7 million years ago (MYA). Interestingly, the sloth and armadillo, which are ancient mammals, lack αs2- and αs1-casein encoding genes respectively. Assuming that their genome sequences are complete and without errors, then the sloth clearly did not develop the CSN1S2 gene and the armadillo lost the CSN1S1 gene during evolution.
The above highlights the rapidly evolving nature of casein genes, which may be linked to the specific nutritional requirements and adaptability of mammals. The African elephant is also an ancient mammal and its genome database shows that it lacks both αs2- and αs1-casein encoding genes. It appears that the CSN1S2 gene developed between 147.7 and 91 MYA and some species did not develop it (for example, the sloth). The CSN1S1 gene has also been lost, for example in the elephant and armadillo. It appears that the β-casein gene is the more conserved of the ancient genes, whereas the κ-casein gene (the last casein gene to develop) has been acquired by most, if not all, mammalian species. This highlights the importance that these genes may have in casein micelle formation and maintenance.
In conclusion, casein gene sequences are very diverse from each other. The diverse gene sequences and absence of some casein genes in a number of mammalian species that contain casein micelles, suggest different mechanisms of casein micelle formation, opposed to those described for bovine casein micelles, where all four caseins are present. The genes encoding α-caseins are absent in most mammalian species, in contrast, genes encoding β- and κ-caseins are widely distributed amongst mammals, and this suggests that the latter gene products have a more significant role to play in milk, particularly in the assembly and mineral (calcium and phosphate) sequestration of the casein micelle.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0022029919000414.
Acknowledgements
This study was supported by grants from the National Research Foundation (grant number 85939) and the University of the Free State of South Africa (grant number 27241).






