Introduction
Oocyte capacity, i.e. the chance of an oocyte to be fertilized and sustain embryo development, is relevant in the discussion of decreasing female fertility, which has been observed over the last years in both human and farm animals (Alviggi et al., Reference Alviggi, Humaidan, Howles, Tredway and Hillier2009; Hansen Reference Hansen2011). Also, oocyte quality is very important to the outcome of assisted reproductive technologies (ART) such as in vitro fertilization (IVF), cytoplasmic transfer (CT) and somatic cell nuclear transfer (SCNT, ‘cloning’). IVF is increasingly used in the treatment of infertility in human (Human Fertilisation and Embryology Authority, 2012; Society for Assisted Reproductive Technologies, 2013) and in cattle, in which the use of genomic selection in breeding programs supports the wish for IVF on oocytes from selected, valuable donors, with subsequent transfer of embryos with high genomic values (Ponsart et al., Reference Ponsart, Le Bourhis, Knijn, Fritz, Guyader-Joly, Otter, Lacaze, Charreauz, Schibler, Dupassieux and Mullaart2014). CT has the purpose to improve oocyte quality from especially older women thereby increasing the chance of pregnancy (Barritt et al., Reference Barritt, Willadsen, Brenner and Cohen2001), whereas SCNT mostly serves a research purpose in particular in relation to the production of transgenic animals (Al-Mashhadi et al., Reference Al-Mashhadi, Sørensennm, Kragh, Christoffersen, Mortensen, Tolbod, Thim, Du, Li, Liu, Moldt, Schmidt, Vajta, Larsen, Purup, Bolund, Nielsen, Callesen, Falk, Mikkelsen and Bentzon2013). Common to all of these techniques is the severe importance of the specific individual donor of the oocytes, making investigations of individual donor oocyte capacity highly interesting and relevant. Such kind of investigations should also include individuals at different stages of the reproductive time span since in cattle, young prepubertal heifers with high breeding values are attractive as oocyte donors while in human, a high percentage of the IVF treatments are performed in women older than 35 years. These examples illustrate the interest and the different needs depending on species. In this study, we used the pig as oocyte donor, since a strong characteristic of Danish slaughterhouse animals is that they are of good health and are well defined in age and weight, making it possible to use a rather homogeneous group of both pre- and postpubertal donors.
Development of a functionally good oocyte with high capacity is a complex process where mitochondria play an important role. In general, mitochondria are central to the cell's energy production, calcium homeostasis and apoptosis (Zick et al., Reference Zick, Rabl and Reichert2009). Mitochondria harbor their own circular genome (mtDNA) consisting of approximately 16,600 base pairs encoding around 37 genes depending on the species (Bernt et al., Reference Bernt, Brabrand, Schierwater and Stadler2013). The number of mitochondria in a cell varies with cell type, cell stage, energy requirements and cell function (Shoubridge & Wai Reference Shoubridge and Wai2007; Eisele et al., Reference Eisele, Schaefer, Randel Nyengaard, Post, Liebetanz, Brüel and Mühlfeld2008). Estimation of mtDNA copy number in oocytes from different animal species is described in the literature and associations are made between oocyte mtDNA quantity and oocyte-related characteristics such as oocyte fertilization (Reynier et al., Reference Reynier, May-Panloup, Chrétien, Morgan, Jean, Savagner, Barrière and Malthièry2001; El Shourbagy et al., Reference El Shourbagy, Spikings, Freitas and St John2006; Zeng et al., Reference Zeng, Ren, Yeung, Shu, Xu, Zhuang and Liang2007) and size of oocyte and follicle (El Shourbagy et al., Reference El Shourbagy, Spikings, Freitas and St John2006; Endo et al., Reference Endo, Kimura, Kuwayama, Monji and Iwata2012; Cotterill et al., Reference Cotterill, Harris, Fernandez, Lu, Huntriss, Campbell and Picton2013). In many of these studies the authors observed a large between-oocyte variation in mtDNA copy number, ranging from a few thousands to more than one million.
Therefore, the aim of this study was to investigate if the oocyte donor has an influence on its oocyte's mtDNA copy number. This was done by measuring the mtDNA copy number in single oocytes from individual porcine donors using quantitative real-time PCR. Furthermore, the relation between oocyte size and mtDNA copy number was investigated using pre- and postpubertal pig oocytes, since such investigations contribute to the basic understanding of mitochondrial dynamics in growing oocytes from animals at different reproductive stages. Preliminary data from this study have been presented earlier (Pedersen et al., Reference Pedersen, Løvendahl, Larsen, Madsen and Callesen2015a).
Materials and methods
Oocyte collection
Ovaries were collected from an abattoir slaughtering 4–6 month old pigs (prepubertal, i.e. ovaries without a corpus luteum) and sows (postpubertal, i.e. ovaries with several corpora lutea) and were transported to the laboratory at 30–33°C. Here, cumulus–oocyte complexes (COCs) from individual donors were aspirated into separate glasses from all visible follicles, with an 18-gauge needle using vacuum suction. COCs were selected for further use, when they had a homogeneous ooplasm and at least one cumulus cell layer. The cumulus cells were removed by vortexing in phosphate buffered saline (PBS) without calcium and magnesium for 8 min in a centrifuge tube, and the removal of all cumulus cells was carefully checked under the microscope. The oocytes were washed three times in T2 (HEPES-buffered M-199, supplemented with 2% serum) and selected for good morphology (smooth and clear cell membrane, homogenous cytoplasm) to discard oocytes known to have a very low developmental competence (Pedersen et al., Reference Pedersen, Liu, Li, Purup, Løvendahl, Holm, Hyttel and Callesen2015b). All oocytes from each donor were transferred to wells (1 oocyte per well) in Porcine Zygote Medium (PZM-3, Yoshioka et al., Reference Yoshioka, Suzuki, Tanaka, Anas and Iwamura2002) for measurement of oocyte diameters in a time-lapse incubator (EmbryoScope®; Unisense FertiliTech A/S, Aarhus, Denmark). Two perpendicular inside-ZP diameter measurements were performed on each oocyte to calculate its average diameter. The aim was to obtain 15 oocytes from each of 10 pre- and 10 postpubertal pigs, with three oocytes in each of the following size intervals (μm): ≤110, 111–114, 115–118, 119–122, ≥123, as defined previously (Pedersen et al., Reference Pedersen, Liu, Li, Purup, Løvendahl, Holm, Hyttel and Callesen2015b). For some of the donors, it was not possible to obtain three oocytes in all intervals resulting in an actual number of six to 15 oocytes from each donor. Selected oocytes were transferred to T2 drops until freezing. Before freezing, each oocyte was washed two times in Embryowater (Sigma) and transferred to DNase-free tubes (Life Technologies) in 10 μl Embryowater. The oocytes were snap-frozen in liquid nitrogen and stored at −80°C.
Mitochondrial DNA quantification by quantitative real-time PCR (Q-PCR)
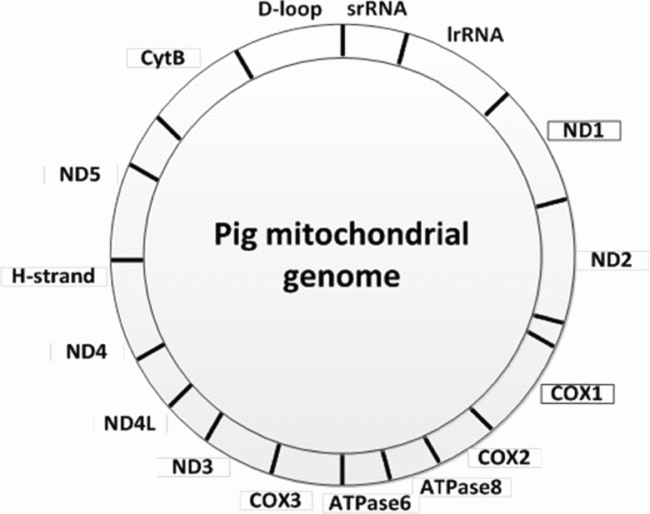
The genes ND1 encoding NADH dehydrogenase subunit 1 and COX1 encoding cytochrome c oxidase subunit 1 (Fig. 1) were used to determine the mitochondrial DNA copy number. Two amplicons were generated by PCR using pig DNA isolated from frontal cortex and primers derived from the Sus scrofa domesticus mitochondrion, complete genome (GenBank ID NC_012095). Oligonucleotide primers used for PCR amplification of a 495-bp ND1 fragment were: ND1-F: 5′-GCATTTACAGCAATTCTCTTCC-3′ and ND1-R: 5′-CTGTTATGATAAGGGTAGTGTAGATAATGGG-3′. A 520-bp COX1 fragment was amplified using the primers: COX1-F: 5′-CAGCCCGGAACCCTACTTGGCGTG-3′ and COX1-R: 5′-TCAGGTTGCGGTCTGTCAGTAGATTAG-3′. The PCR reaction mix contained 100 ng DNA, 1.5 mM MgCl2, 0.2 mM dNTP, 0.5 μM of each primer set and 1 U Phusion DNA polymerase (Finnzymes), in a total volume of 25 μl. The PCR profile was as follows: 98°C for 30 s, 30 cycles of 98°C for 15 s, 70°C/COX1 and 65°C/ND1 for 30 s, 72°C for 30 s and finally an elongation at 72°C for 5 min. The ND1 and COX1 amplicons were visualized and isolated from an ethidium bromide stained 1% agarose gel. The PCR products were cloned into the pCR TOPO 2.1 vector (Invitrogen) and sequenced in both directions. DNA sequencing was performed as previously described (Bjerre et al., Reference Bjerre, Madsen, Bendixen and Larsen2006). These two plasmid preparations were used for preparation of standard curves in the subsequent quantitative PCR analyses. The DNA concentration in the plasmid preparations were measured with a NanoDrop ND-1000 spectrophotometer and serial dilutions were prepared. The number of copies of mtDNA was determined using the DNA copy number calculator (http://cels.uri.edu/gsc/cndna.html). Diluted plasmid solutions ranging from 102−109 which were stored at −20°C. Each oocyte was lysed in 10 μl lysis buffer (0.4 μl Proteinase K (10 mg/ml) and 9.6 μl Proteinase K buffer) and incubated at 45°C for 45 min followed by 99°C for 10 min. The samples were vortexed and centrifuged for 2 min at 11,000 rpm.
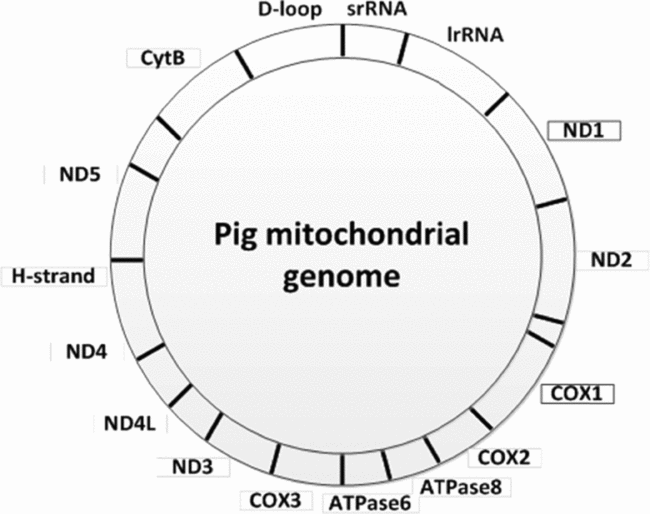
Figure 1 The pig mitochondrial genome showing the position of the genes ND1 and COX1.
Q-PCR reactions using ND1-QF (5′-CCGATACGACCAACTAATACATTTAC-3′), ND1-QR (5′-GCTGTTATAATAGGGAGTGAGATGTG-3′) and probe # 4 (5′-CTTCCTGC-3′, Universal Human Probe library, Roche) for ND1 and COX1-QF (5′-TGAGGATGCCAGAAGTAATAGGA-3′), COX1-QR (5′-GGCCTTTCCACGTATAAACAAC-3′) and probe # 31 (5′-TGGTGGAA-3′, Universal Human Probe library, Roche) were run in triplicate on a ViiA7 Real-Time PCR System (Applied Biosystems). Each 10 μl reaction volume contained 2 μl oocyte lysate, 5 μl TaqMan (Applied Biosystems), 0.15 μl SYBR Green fluorescent probe (Roche), 0.15 μl of each primer (Sigma) and 2.55 μl water. The cycling conditions were as follows: 50°C for 2 min, an initial denaturation at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Statistical analysis
Standard curves showed high correlation coefficients (r 2 = 0.99–1.00) and amplification efficiencies (COX1 91–104%; ND1 84–92%). For the inter-assay control, the standard curves from the six runs were compared. The performance was tested using linear regression models with interaction effects of assay date. This was supported by graphics to illustrate effects. Since primers could have impact on amplification efficiency, tests were carried out for each pair of primer.
Results for the two primers were in good agreement as shown by a high determination coefficient (R 2 = 0.87) for a linear regression of COX on ND both in linear and in log-transformed units. On the log scale COX numbers were equal to 1.34 + 0.705 ND numbers, so that COX was higher than ND at low levels and vice versa at high levels.
The effect of individuals on mtDNA copy number was investigated in an iterative and explorative way driven by (unexpected) findings. At first, distribution of data showed visual indications of deviating from being normally distributed when assessed by QQ-plot, and were therefore log-transformed. However, even after log-transformation bimodal distribution was indicated and supported by cluster analysis (PROC MODECLUS, SAS Institute Inc.), where data could be split into two approximate normal distributions. The data points from the two clusters were at the same time clustering the donors into two groups with high and low mtDNA copy number. When donors and oocytes were assigned to these two groups there was a clear discrimination between them and very little overlap, and thereby satisfying criteria for bi-modality (Schilling et al., Reference Schilling, Watkins and Watkins2002). The effect of oocyte donor was also estimated using ANOVA in a linear mixed model (PROC MIXED, SAS Institute Inc.), by considering donor as random effect. In that model the effect of grouping was investigated as a fixed factor with two levels (high/low), and donor as nested within group. The individuality effect of donor was then expressed as the intra-class correlation coefficient defined as the ratio between donor variance to total variance.
Relationship between oocyte size and mtDNA copy number was analyzed using linear regression. Data analysis was accomplished using procedures in MODECLUS, REG, SGPLOT and MIXED (SAS 9.3, SAS Institute Inc.). A probability of P < 0.05 was considered to be statistically significant.
Results and Discussion
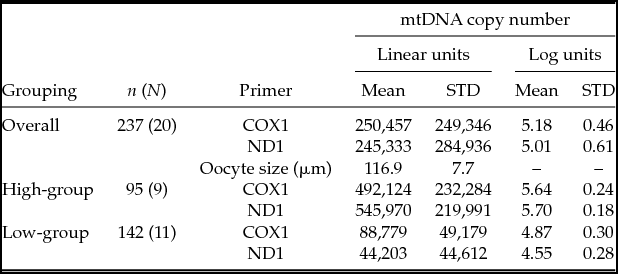
In total, 145 oocytes from 10 prepubertal animals and 93 oocytes from 10 postpubertal animals were analyzed (Fig. 2 and Table 1). The distribution of mtDNA copy number was bimodal, where data points were assigned to groups of high and low oocyte donors. Little overlap between distributions was evident after they were split into two clusters (Fig. 2). Within each group variation was also much smaller than the variance across all donors (Table 1). The intra-class correlation across all donors was 83% and 92% for COX and ND respectively, and dropped to 35% and 39% when the group effect was included in the model. This shows that the group significantly affected the mtDNA copy number, as well as it accounts for a large part of the variation between donors. The variation between donors within group is still large, and of a magnitude comparable with what is observed with other biological traits such as milk yield in dairy cows (e.g. Løvendahl & Chagunda Reference Løvendahl and Chagunda2011). The ANOVA showed no effect of developmental stage (pre- versus postpubertal) on mtDNA copy number. Also, donor had no effect on oocyte size, and oocyte size was not significantly related to mtDNA copy number (Table 1).
Table 1 mtDNA copy number in oocytes from pre- and postpubertal donors, divided into groups of individuals with ‘high’ (≥100,000) or ‘low’ (<100,000) mtDNA copy numbers, n: total number of oocytes from N donors
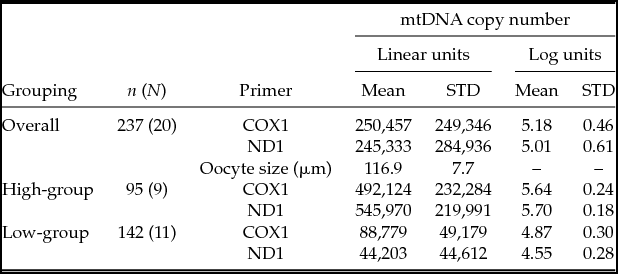

Figure 2 Box and whisker plot of mtDNA copy number in oocytes from each individual prepubertal (P1–P10) and postpubertal (S1–S10) donor, grouped into donors with mean mtDNA copy number above (HI) or below (LO) 100,000. Results are shown for both primers: COX1 (a) and ND1 (b).
This study demonstrates the importance of the oocyte donor in relation to oocyte mtDNA copy number, irrespectively of the donor's puberty status and the oocyte's growth stage. More precisely, we show that individual porcine oocyte donors can be classified as having either a low or high level of oocyte mtDNA copy number (<100,000 or ≥100,000) including both pre- and postpubertal donors. These results confirm previous studies in bovine and porcine oocytes (Iwata et al., Reference Iwata, Goto, Tanaka, Sakaguchi, Kimura, Kuwayama and Monji2011; Sato et al., Reference Sato, Itami, Tasaki, Takeo, Kuwayama and Iwata2014). Several studies have demonstrated an association between oocyte mtDNA copy number and fertilization rate, with a proposed threshold value of 100–200,000 mtDNA copies in the individual oocyte (Reynier et al., Reference Reynier, May-Panloup, Chrétien, Morgan, Jean, Savagner, Barrière and Malthièry2001; May-Panloup et al., Reference May-Panloup, Chretien, Jacques, Vasseur, Malthiery and Reynier2005; El Shourbagy et al., Reference El Shourbagy, Spikings, Freitas and St John2006; Santos et al., Reference Santos, El Shourbagy and St John2006). These results were obtained by comparing the mtDNA copy number in fertilized and non-fertilized oocytes, but without knowing the donor-origin of the oocytes. However, assuming that the donor represents a certain level of mtDNA copies as our experiment indicates, it would be possible to group oocyte donors according to their level of mtDNA copy number. Such kind of experiment has been performed in genetically modified knock-out mice, demonstrating a critical post-implantation (but not pre-implantation) developmental threshold of 40–50,000 mtDNA copies (Wai et al., Reference Wai, Ao, Zhang, Cyr, Dufort and Shoubridge2010). The same principle was used by Iwata et al. (Reference Iwata, Goto, Tanaka, Sakaguchi, Kimura, Kuwayama and Monji2011), demonstrating a positive correlation between mtDNA copy number and abnormal fertilization in older cows. In our study, there were no indications of mtDNA copy number being related to decreased developmental competence of small oocytes/follicles (Marchal et al., Reference Marchal, Vigneron, Perreau, Bali-Papp and Mermillod2002; Bagg et al., Reference Bagg, Nottle, Armstrong and Grupen2007) and prepubertal oocytes (Ikeda & Takahashi Reference Ikeda and Takahashi2003; Li et al., Reference Li, Pedersen, Li, Adamsen, Liu, Schmidt, Purup and Callesen2013; Pedersen et al., Reference Pedersen, Callesen, Løvendahl, Chen, Nyengaard, Nikolaisen, Holm and Hyttel2014). These examples illustrate the need for further research of the importance of mtDNA copy number and the complex mechanisms regulating the mitochondrial complement of an oocyte.
Future investigations should focus on the consequences of mtDNA quantity in relation to oocyte development and on factors determining an individual donor's level of oocyte mtDNA copy number including changes during a lifetime. Such kind of investigations should be performed in cattle to reveal whether oocyte mtDNA copy number could be a way to select young heifers and cows for ovum pick-up and IVF. In the same way, this kind of knowledge would be useful in human fertility treatment to classify women regarding oocyte quality and for possible treatments and prevention.
A necessary basis for the quantification of mitochondria is employment of a reliable quantitative method. The direct approach, i.e. actually counting the mitochondria, is a time-consuming process (e.g. Pedersen et al., Reference Pedersen, Callesen, Løvendahl, Chen, Nyengaard, Nikolaisen, Holm and Hyttel2014; Reader et al., Reference Reader, Cox, Stanton and Juengel2014), hence a commonly used and still plausible method is to assess the mtDNA copy number. In oocytes, the number of mtDNA copies per mitochondrion has been estimated to one to two based on very few studies (Piko & Matsumoto Reference Piko and Matsumoto1976; Reader et al., Reference Reader, Cox, Stanton and Juengel2014). To confirm this, mitochondrial counts and mtDNA copy number estimation could be performed and compared on oocytes from the same individual characterized by a certain mtDNA level. Such kind of results would be valuable in the search for the most precise parameter for estimation of the mitochondrial numbers in oocytes and for selection of the most suitable mitochondrial parameter to characterize oocytes with high developmental competence. Increasing our knowledge on mitochondrial number and function could also be important to studies of mitochondrial-related diseases and the development and recommendations of new technologies, such as cytoplasmic transfer and cloning in different mammalian species.
Acknowledgements
The authors thank M.K. Perera, M. Jeppesen, A.M. Pedersen, J. Adamsen, K. Villemoes and R. Kristensen for invaluable and qualified technical assistance.
Financial support
This work was supported by Department of Animal Science, Aarhus University, Denmark.
Statement of interest
None declared.