Introduction
Oocyte genome cloning is a method by which haploid parthenogenetic embryos can be obtained in such a way that blastomeres from these embryos can be considered as a clone of the original gamete (Surani et al., Reference Surani, Barton and Norris1986; Escribá et al., Reference Escribá and García-Ximénez2001). Parthenogenetic activation can be stimulated in several mammalian species, including humans (Revazova et al., Reference Revazova, Turovets, Kochetkova, Kindarova, Kuzmichev, Janus and Pryzhkova2007), by a wide variety of artificial chemical and physical stimuli that induce oocyte activation. These include electrical stimulation, and treatment with ethanol, thimerosal, strontium, ionomycin (Io) or calcium ionophore (Collas et al., Reference Collas, Fissore, Robl, Sullivan and Barnes1993; Machaty et al., Reference Machaty, Wang, Day and Prather1997; Loi et al., Reference Loi, Ledda, Fulka, Cappai and Moor1998; Rho et al., Reference Rho, Wu, Kawarsky, Leibo and Betteridge1998; Liu et al., Reference Liu, Chen, Cheng and Ju2002; Yi et al., Reference Yi and Park2005; Bhak et al., Reference Bhak, Lee, Ock, Mohana Kumar, Choe and Rho2006; Méo et al., Reference Méo, Yamazaki, Ferreira, Perecin, Saraiva, Leal and Garcia2007). In the bovine, Io in combination with 6-dimethylaminopurine (DMAP; non-specific kinase inhibitors) has been shown to be particularly effective in inducing oocyte activation and subsequent embryo development (Susko-Parrish et al., Reference Susko-Parrish, Leibfried-Rutledge, Northey, Schutzkus and First1994; Wells et al., Reference Wells, Misica and Tervit1999).
Ionomycin in combination with DMAP (Io + DMAP) was used to activate cytoplasts during nuclear transfer that produced a cloned bovine, demonstrating the suitability of this treatment in promoting full-term development (Cibelli et al., Reference Cibelli, Stice, Gluekey, Kane, Jerry, Blackwell, Ponce de León and Robl1998; Salamone et al., Reference Salamone, Barañao, Santos, Bussmann, Artuso, Werning, Prync, Carbonetto, Dabsys and Munar2006). However, if oocytes activated by Io + DMAP are treated with a 3-h interval between Io activation and DMAP (Io + 3h + DMAP), allowing time for the extrusion of the second polar body (2PB), a specifically haploid parthenote is induced (Susko-Parrish et al., Reference Susko-Parrish, Leibfried-Rutledge, Northey, Schutzkus and First1994; Vichera et al., Reference Vichera, Alfonso, Duque, Silvestre, Pereyra-Bonnet, Fernandez-Martín and Salamone2009). If a 3-h interval between Io and DMAP treatments is absent, the 2PB is not extruded in most activated oocytes, resulting in the formation of diploid parthenotes, (Susko-Parrish et al., Reference Susko-Parrish, Leibfried-Rutledge, Northey, Schutzkus and First1994; Wells et al., Reference Wells, Misica and Tervit1999; Cibelli et al., Reference Cibelli, Stice, Gluekey, Kane, Jerry, Blackwell, Ponce de León and Robl1998; Salamone et al., Reference Salamone, Barañao, Santos, Bussmann, Artuso, Werning, Prync, Carbonetto, Dabsys and Munar2006; Vichera et al., Reference Vichera, Alfonso, Duque, Silvestre, Pereyra-Bonnet, Fernandez-Martín and Salamone2009).
Previous reports on haploid parthenogenetic activation have indicated that no difference in cleavage ability was found between haploid and diploid parthenogenetic embryos; however only some of the haploid parthenogenetic embryos developed to the blastocyst stage (Surani et al., Reference Surani, Barton and Norris1986; Escribá et al., Reference Escribá and García-Ximénez1999, Reference Escribá and García-Ximénez2000; Lagutina et al., Reference Lagutina, Lazzari, Duchi and Galli2004). Parthenogenetic embryos are unable to develop to term due to the imprinting process that has an essential role during embryogenesis (MacGrath et al., Reference MacGrath and Solter1983, Reference MacGrath and Solter1984; Surani et al., Reference Surani, Barton and Norris1984, Reference Surani, Barton and Norris1986). Therefore, to obtain viable offspring from haploid parthenogenetic embryos, every single blastomere from these embryos should be combined with the male counterpart (haploid male hemizygotes) in order to restore the normal heteroparental condition. Haploid male hemizygous eggs can be produced by removing the female pronucleus from fertilized eggs (Barton et al., Reference Barton, Surani and Norris1984; McGrath and Solter 1984; Surani et al., Reference Surani, Barton and Norris1984; Kaufman et al., Reference Kaufman, Lee and Speirs1989; Latham and Solter Reference Latham and Solter1991; Hagemann et al., Reference Hagemann and First1992), fertilization of enucleated oocytes (McGrath and Solter 1984; Surani et al., Reference Surani, Barton and Norris1984; Kaufman et al., Reference Kaufman, Lee and Speirs1989; Latham and Solter Reference Latham and Solter1991; Hagemann et al., Reference Hagemann and First1992; Obata et al., Reference Obata, Ono, Akuzawa, Kwon, Yoshizawa and Kono2000) and injection of spermatozoa into oocytes followed by maternal chromosomes removal (Miki et al., Reference Miki, Hirose, Ogonuki, Inoue, Kezuka, Honda, Mekada, Hanaki, Iwafune, Yoshiki, Ishino and Ogura2009).
In this work, we compared the developmental ability of haploid and diploid parthenogenetic bovine embryos. Once obtained, blastomeres of haploid parthenogenetic embryos were used, as female genome donors, to reconstruct biparental embryos by fusion with haploid male hemizygotes. In addition, we generated homogeneous transgene-expressing embryos by fusing parthenogenetic haploid blastomeres that expressed a transgene, with haploid male hemizygotes.
Materials and methods
Reagents
All chemicals were obtained from Sigma Chemical Company (St. Louis, MO, USA), except when otherwise indicated.
Oocyte collection and in vitro maturation
Ovaries were collected at a slaughterhouse and transported to the laboratory. Cumulus–oocyte complexes were aspirated from follicles with a diameter of 2 to 8 mm into Dulbecco's phosphate-buffered saline (PBS; GIBCO BRL, Grand Island, NY, USA) that contained 10% fetal bovine serum (FBS; GIBCO BRL) and 2% antibiotic–antimycotic (ATB; GIBCO BRL). Follicular oocytes covered by at least three layers of granulosa cells and with an evenly granulated cytoplasm were selected for in vitro maturation. The maturation medium was bicarbonate-buffered TCM-199 (GIBCO BRL) that contained 2 mM glutamine, 10% FBS, 2 μg/ml follicle-stimulating hormone (Follitropin®, Bioniche, Belleville, Ontario, Canada), 0.3 mM sodium pyruvate (P2256), 100 μM cysteamine (M9768) and 2% ATB. Oocytes were incubated in 100-μl droplets of medium covered with mineral oil (M8410), in 32 mm Petri dishes. In vitro maturation conditions were 6.5% CO2 in humidified air at 39°C for 22 h.
Production of parthenogenetic haploid and diploid embryos
Matured oocytes vortexed for 2 min in hyaluronidase (1 mg/ml in DPBS) to remove cumulus cells, and washed three times in TALP-H (Bavister et al., Reference Bavister and Yanagimachi1977). Metaphase II (MII) oocytes were identified by first polar body visualization and immediately used for parthenogenetic activation or micromanipulation techniques. Parthenogenetic haploid activation consisted of: (1) incubation with 5 μM Io (Invitrogen, California, USA) for 4 min; and (2) incubation with 5 μM Io for 4 min, then in synthetic oviductal fluid (SOF) for 3 h to permit extrusion of the second polar body (2PB), and finally placed in 1.9 mM 6-DMAP in SOF for 3 h. For parthenogenetic diploid activation, oocytes were placed in 5 μM Io for 4 min, followed by 1.9 mM 6-DMAP in SOF for 3 h.
DNA construction
The plasmid used was pCX–EGFP that contains enhanced green fluorescent protein gene (egfp) under the control of chimeric cytomegalovirus-IE–chicken β-actin enhancer–promoter (Ikawa et al., Reference Ikawa, Kominami, Yoshimura, Tanaka, Nishimune and Okabe1995).
Liposome–DNA coincubation
For the injection experiments, 1 μl of 4 μg/ml DNA in combination with 3 μl of commercial liposome (Fugene; Boehringer-Manheim, Germany) were coincubated for 15 min. The liposome–DNA mixture was diluted to half concentration with 10% polyvinylpyrrolidone (PVP; Irvine Scientific, Santa Ana, CA, USA), resulting in a final DNA concentration of 0.5 μg/ml.
Intracytoplasmic injection of DNA–liposome complexes
After 3 h of haploid parthenogenetic activation, the ooplasm of the activated oocytes was injected with approximately 2 pl of DNA–liposome/PVP mixture, using an injection capillary (0.7 μm in diameter) attached to a Narishige hydraulic micromanipulator (Medical Systems, Great Neck, NY, USA) mounted on a Nikon Eclipse E-300 microscope (Nikon, Melville, NY, USA).
Haploid male hemizygotes production
Haploid male hemizygotes were generated by fertilization of enucleated oocytes. Enucleation was performed as follows: oocytes were held and manipulated in TALP-H supplemented with 3 mg/ml bovine serum albumin (BSA). Denuded MII oocytes were enucleated mechanically using micromanipulators mounted on a Nikon Eclipse E-300 microscope and 20 μm diameter pipettes. Metaphase chromosomes were visualized under UV (< 10 s) after staining with 5 μg/ml Hoechst 33342 for 10 min. Fertilization of enucleated oocytes was performed as follows: bovine frozen semen was thawed in a 37°C water bath for 30 s. Spermatozoa were centrifuged twice (490 g, 5 min) and resuspended in Brackett–Oliphant medium (BO) supplemented with 5 mM caffeine and 20 IU/ml heparin. Spermatozoa were diluted to half concentration with BO that contained 10 mg/ml fatty acid-free BSA (A6003), resulting in a final sperm concentration of 15 × 106/ml. Spermatozoa were coincubated with enucleated oocytes in 100-μl droplets, for 3 h at 39°C in a humidified atmosphere of 5% CO2 in air. After this incubation, hemizygotes were immediately used for biparental embryo reconstruction.
Karyotype analysis
Embryos were cultured in SOF supplemented with 0.05 g/ml demecolcine (D1925) for 6 h at 72 h after IVF (in vitro fertilization) and parthenogenetic activation. Embryos were then exposed to a hypotonic 0.8% sodium citrate solution for 10 min at 37°C. Subsequently, embryos were placed on a clean glass slide and treated with a drop of methanol–acetic acid solution (3:1). Slides were dried and then stained with 5% Giemsa solution (Invitrogen) for 10 min. Chromosome spreads were evaluated at ×400 magnification.
Biparental bovine embryo reconstruction by parthenogenetic haploid blastomere fusion
Parthenogenetic haploid embryos (2–16 cells) that expressed EGFP, or not, were treated with 1.5 mg/ml pronase (Sigma protease) dissolved in TALP-H to remove the zona pellucida (ZP). Gentle pipetting was applied to disaggregate blastomeres from these embryos. Parthenogenetic blastomeres that expressed EGFP were then selected under blue light using an excitation filter at 488 nm and an emission filter at 530 nm. Parthenogenetic blastomeres were fused with haploid male hemizygotes as described below. Haploid male hemizygotes were incubated in 1.5 mg/ml pronase for 5–10 min on a warm plate to remove the ZP. ZP-free haploid male hemizygotes were then transferred individually to a drop of 1 mg/ml phytohemaglutinin dissolved in TCM-199 without serum for a few seconds. Following this step, they were dropped quickly over a single parthenogenetic haploid blastomere resting on the bottom of a 100 μl TALP-H drop. Following attachment, the ZP-free haploid male hemizygote/parthenogenetic haploid blastomere pair was picked up, transferred to fusion medium (0.3 M mannitol, 0.1 mM MgSO4, 0.05 mM CaCl2, 1 mg/ml PVA), for 2–3 min and then to a fusion chamber (BTX Instrument Division; Harvard Apparatus, Holliston, MA, USA) that contained 2 ml of warm fusion medium. Fusion (Fig. 1) was performed with a double direct current (dc) pulse of 65 V, each pulse was for 30 ms, 0.1 s apart. The biparental reconstructed zygotes were then removed carefully and placed in culture.

Figure 1 Schematic diagram showing the production of haploid parthenogenetic embryos and the biparental embryo reconstruction procedures. (See online for a colour version of this figure.)
In vitro culture
Parthenogenetic, IVF and reconstructed embryos were cultured in SOF medium in a system similar to the well of the well (WOW) method (Vajta et al., Reference Vajta, Peura, Holm, Paldi, Greve, Trouson and Callesen2000), whereby microwells were produced using a heated glass capillary slightly pressed to the bottom of a Petri dish and covered with a 100-μl microdrop of culture medium (16 WOW each microdrop, one embryo each WOW). Culture conditions were a humidified atmosphere of 5% O2, 5% CO2 and 90% N2 in air, at 39°C. The medium was supplemented with 10% FBS on day 5 during embryo culture. Cleavage was evaluated on day 2 and blastocysts (Fig. 2) on day 7, post fusion.

Figure 2 Biparental bovine blastocyst produced by fusion of a parthenogenetic haploid blastomere with a haploid male hemizygote and cultured in the well of the well system. Original magnification ×200. (See online for a colour version of this figure.)
Determination of EGFP fluorescence in embryos
Embryos were briefly exposed to blue light during in vitro culture, using an excitation filter at 488 nm and an emission filter at 530 nm, to determine EGFP expression at different stages of development. Embryos were analyzed on days 3 and 7, after parthenogenetic activation or biparental embryo reconstruction.
Determination of blastocyst cell number
Embryos were stained in TCM-199 containing 1 mg/ml Hoechst 33342 (B2261), for 2 min and mounted immediately between coverslips to count total nuclei under an epifluorescence microscope.
Immunocytochemical analysis
Immunocytochemical analysis was performed on bovine blastocysts, obtained as result of biparental embryo reconstruction (n = 2), and on IVF bovine blastocyst controls (n = 3). Briefly, embryos were fixed for 30 min in 4% v/v paraformaldehyde in PBS and permeabilized by 15 min incubation in PBS that contained 0.2% v/v Triton X-100. Non-specific immunoreactions were blocked by incubation with 3% v/v FBS and 0.1% v/v Tween-20 (Promega, H5152) in PBS (blocking buffer) for 30 min. After this pretreatment, affinity-purified primary polyclonal antibody against Oct-4 (Santa Cruz Biotechnology, California, USA) was diluted 1:100 in PBS and applied for 1 h at room temperature. Blastocysts were washed extensively in blocking buffer for 15 min. Then, the samples were incubated with secondary Alexa 488–donkey anti-goat IgG 2 mg/ml (Molecular Probes, Inc. Eugene, USA) diluted 1:1000 for 40 min at room temperature in the dark. After additional washing, the embryos were incubated in PBS that contained propidium iodide for 10 min in the dark. Embryos were mounted on slides in 70% v/v glycerol. Oct-4 negative controls were produced using only the secondary antibody. The embryos were analyzed on a Nikon confocal laser scanning microscope. An excitation wavelength of 488 nm was selected from an argon-ion laser to excite the Alexa-conjugated secondary antibody and a 544 nm wavelength to excite propidium iodide. Images of serial optical sections were recorded every 1.5 to 2 μm along the Z-axis of each embryo. Three-dimensional images were constructed using software EZ-C1 2.20.
Transfer of embryos
Reconstructed embryos were transferred to Aberdeen Angus recipients on day 7 of the oestrous cycle. Reconstructed embryos at the blastocyst stage (day 7) were used for embryo transfer. Each reconstructed embryo was washed several times in TL-HEPES and loaded into a 0.25 ml straw. Fresh embryos were transported to the farm at 35°C within 3 h. Embryos were transferred non-surgically to the uterine horn ipsilateral to the ovary bearing the corpus luteum, using a transverse hole-type transfer device. Each recipient received one embryo. Pregnancies were diagnosed by fetal membrane palpation through rectal inspection at approximately days 60–70 after oestrus.
Experimental design
In the first experiment we compared the developmental ability of parthenogenetic haploid and diploid bovine embryos produced with Io, Io + 3h + DAMP or Io + DMAP. For the best parthenogenetic haploid treatments (Io + 3h + DAMP) we injected a mixture of pCX–EGFP–liposome complexes 3 h following activation, in order to obtain exogenous gene expression to be used as a parthenogenetic cytoplasmic marker. Karyotype analysis was done in order to determine the ploidy of the parthenogenetic embryos and male hemizygotes generated. In the second experiment, haploid parthenogenetic embryos (4–16 cells) either expressing EGFP or not, were disaggregated and the parthenogenetic haploid blastomeres obtained were fused with zona-free haploid male hemizygotes in order to reconstruct biparental embryos. The developmental ability of the reconstructed embryos and the blastocyst cell numbers were evaluated. Additionally, the Oct-4 expression pattern of blastocysts obtained from biparental embryo reconstruction was analyzed by immunocytochemistry. Finally, reconstructed biparental embryos were transferred to recipient cows on day 7 of in vitro development. The procedure is shown in Fig. 1.
Statistical analysis
In vitro embryo development and transgene expression were compared by non-parametric Fisher's exact test. For all statistical analyses, the SAS program was used (SAS Institute, 1989). Differences were considered significant at P < 0.05.
Results
Experiment 1: Development of haploid parthenogenetic embryos, injected or not, with pCX–EGFP–liposome complexes
Development and EGFP-expression rates of haploid parthenogenetic embryos are summarized in Table 1. Statistical differences in cleavage rates were observed between the Io + 3h + DMAP group and the Io group, but not with the Io + 3h + DMAP injected with pCX–EGFP–liposome group. No differences were observed in rates of blastocyst development between the Io + 3h + DMAP and the Io + 3h + DMAP injected with pCX–EGFP–liposome groups, but significant differences were observed between the Io + 3h + DMAP and the Io group. All haploid parthenogenetic groups showed statistical differences in blastocyst rates compared with the diploid parthenogenetic control 40.2% (49/122), but no differences were seen in cleavage rates. The EGFP-expression pattern was evaluated in cleaved embryos (day 3 post activation). Parthenogenetic haploid embryos obtained by Io + 3h + DMAP treatment and then injected with pCX–EGFP–liposome complexes showed an EGFP-expression rate of 54.7% (35/54). Moreover, karyotype analysis of parthenogenetic embryos generated by Io + 3h + DMAP, confirmed that 83.3% (10/12) were indeed haploid. The remaining embryos were mixoploid. Karyotype analysis performed on male hemizygotes confirmed that 80.0% (8/10) of them were indeed haploid. The remaining male hemizygotes were diploid.
Table 1 Parthenogenetic haploid bovine embryo development and transgene expression

a–dValues with different superscripts in a column are significantly different (P < 0.05, Fisher's test). +, injected; –, not injected; DMAP, 6-dimethylaminopurine; EGFP, enhanced green fluorescent protein; Io, ionomycin; NA, not applicable.
Experiment 2: Development and transgene expression of biparental bovine embryos reconstructed by fusion of parthenogenetic haploid blastomeres (EGFP-positive or -negative), with haploid male hemizygotes
Development and EGFP-expression rates of ZP-free biparental embryos reconstructed with parthenogenetic haploid blastomeres that were either positive or negative for transgene expression, are summarized in Table 2. No differences were observed between these groups in the percentages of fusion and development, while biparental IVF controls showed a significantly higher blastocyst rate. All embryos that were reconstructed with EGFP-positive parthenogenetic haploid blastomeres expressed the transgene during development (100%, 29/29) and 96.6% (28/29) of them showed expression in all its blastomeres. Immunocytochemical analysis to determine the Oct-4 expression pattern was positive for the inner cell mass (ICM) and the trophoblast in the blastocysts analyzed (n = 2). A similar result was seen in IVF control embryos. To determine the number of blastocyst cells, biparental reconstructed blastocysts (n = 6) and IVF control blastocysts (n = 8) were stained on day 8, resulting in an average of 72.8 ± 7.0 and 97.0 ± 7.3 cells, respectively.
Table 2 Development and transgene expression (+ EGFP) of bovine embryos reconstructed by parthenogenetic blastomere fusion

a–dValues with different superscripts in a column are significantly different (P < 0.05, Fisher's test). +, injected; –, not injected; DMAP, 6-dimethylaminopurine; EGFP, enhanced green fluorescent protein; Io, ionomycin; IVF, in vitro fertilization; NA, not applicable
Experiment 3: Embryo transfer in recipient cows
Reconstructed embryos (n = 2) were transferred at the blastocyst stage to Aberdeen Angus recipients. One pregnancy was diagnosed by fetal membrane palpation through rectal inspection at approximately 60 days after embryo transfer.
Discussion
Generation of parthenogenetic haploid embryos allows obtaining several blastomeres as identical copies of a single oocyte genome (Surani et al., Reference Surani, Barton and Norris1986; Escribá et al., Reference Escribá and García-Ximénez2001). Initially, we evaluated two methods to generate haploid parthenogenetic embryos, Io and Io + 3h + DMAP treatments. The parthenogenetic embryos produced by activation with only Io exposure, showed low cleavage rates and no blastocysts were obtained. However, parthenogenetic embryos produced by the Io + 3h + DMAP method cleaved and developed successfully regardless or not of whether they were injected with pCX–EGFP–liposome complexes (Table 1). Both these groups showed significant differences only in blastocyst development rates when compared with the diploid parthenogenetic control group. These results agree with previous reports that showed that haploid parthenogenetic embryos are compromised developmentally compared with diploid parthenogenetic embryos in different species such as cow, mouse and pig (Kaufman et al., Reference Kaufman1983; Henery et al., Reference Henery and Kaufman1992; Van De Velde et al., Reference Van De Velde, Liu, Bols, Ysebaert and Yang1999; Lagutina et al., Reference Lagutina, Lazzari, Duchi and Galli2004). Recently, we demonstrated that intracytoplasmic injection of DNA–liposome complexes produces IVF and parthenogenetic embryos with an efficient expression of exogenous genes (Vichera et al., Reference Vichera, Moro and Salamone2010). In the present report, we also showed that haploid parthenogenetic embryos, produced by Io + 3h + DMAP and confirmed by karyotype analysis, cleaved successfully and showed high EGFP expression after injection of pCX–EGFP–liposome complexes (Table 1).
In the second experiment we demonstrated that it is possible to reconstruct biparental bovine embryos using female genome donors obtained from parthenogenetic haploid embryos up to the 16-cell stage (Table 2). Moreover, efficient fusion rates were obtained (90%), regardless of the embryonic stage of the donors (4–16 cells) and these embryos were capable of development to the blastocyst stage (Fig. 2). A previous report in mice observed that nuclear transfer of a maternal genome at the fourth cellular division, or more, in haploid male hemizygotes, severely impaired the developmental ability of the embryos generated (Surani et al., Reference Surani, Barton and Norris1986). This situation could be due to the existence of asynchrony between both parental genomes making a functional integration of both nuclei difficult.
All the biparental embryos reconstructed with EGFP-positive parthenogenetic haploid blastomeres expressed the transgene (100%) and most of them showed transgene expression in all blastomeres (96.6%). Development was not affected in this experiment, which suggested that transgene expression does not compromise in vitro embryonic progression. Expression of EGFP verified the cytoplasmic contribution of parthenogenetic haploid blastomeres in the reconstructed embryos (Fig. 3). On the other hand, positive expression of Oct-4 observed both in the ICM and the trophoblast in the blastocysts analyzed, as well as in the IVF control group embryos, was consistent with appropriate nuclear programming, agreeing with previous reports (Kirchhof et al., Reference Kirchhof, Carnwath, Lemme, Anastassiadis, Schöler and Niemann2000). In this study, statistical differences were found in cell numbers, between reconstructed biparental blastocysts and control IVF blastocysts. This finding indicates that the cell divisions kinetics could be modified by the biparental embryo reconstruction procedure.
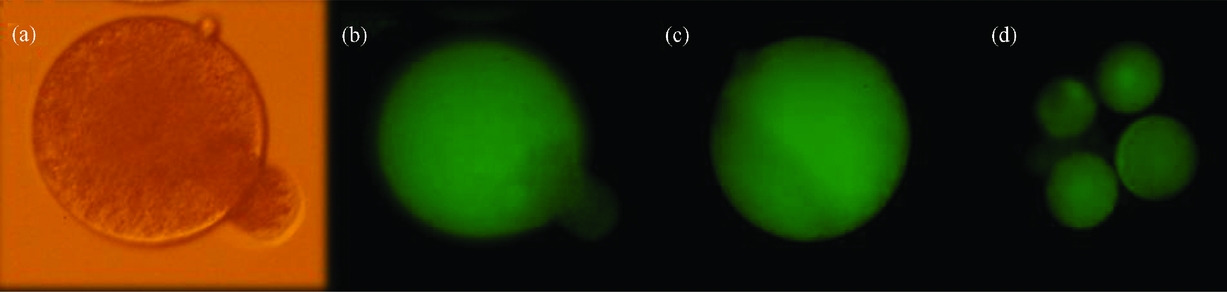
Figure 3 (a) ZP-free haploid male hemizygote fusing to an enhanced green fluorescent protein (+EGFP) parthenogenetic haploid blastomere (using phytohemagglutinin), prior to fusion. (b) The same zona pellucida (ZP)-free haploid male hemizygote fusing to a +EGFP parthenogenetic haploid blastomere under blue light. (c) The same ZP-free haploid male hemizygote completely fused to a +EGFP parthenogenetic haploid blastomere under blue light. (d) Biparental reconstructed ZP-free embryo expressing the transgene in all its blastomeres. The fluorescence was evaluated under blue light (488 nm). Original magnification ×200. (See online for a colour version of this figure.)
Transfer of reconstructed embryos, at the blastocyst stage, to recipient cows, resulted in one pregnancy diagnosed by fetal membrane palpation through rectal inspection at approximately 60 days after embryo transfer. However this pregnancy was subsequently lost. Previous reports in mice showed that viable offspring could be produced by nuclear transfer of haploid parthenogenetic nuclei (Surani et al., Reference Surani, Barton and Norris1986). Offspring produced with this technique were not identical to one another nor to their parents, as variability is given by the parental counterpart. Moreover, live offspring were obtained from chimeras reconstructed from aggregation of parthenogenetic and in vitro fertilized bovine embryos (Boediono et al., Reference Boediono, Suzuki, Li and Godke1999). In the rabbit, oviductal transfer of reconstructed zygotes resulted in 100% pregnancy on day 12, but no pregnancies were diagnosed on day 21 after ovulation (Escribá et al., Reference Escribá and García-Ximénez2001). One year later, the same authors obtained viable offspring when biparental embryos were reconstructed from cryopreserved haploid rabbit parthenotes (García-Ximénez et al., Reference García-Ximénez and Escribá2002). These results indicate that the female gametic endowment can be successfully stored by cryopreservation of parthenogenetic haploid embryos. Germplasm cryopreservation makes the establishment of genetic banks possible for the conservation of biodiversity and contributes to the preservation of endangered species. In the future, the cryopreservation of parthenogenetic haploid embryos could be a very attractive option to maximize the conservation of genetic resources due to its greater resistance compared to oocyte cryopreservation.
In this work, we demonstrated that it is possible to multiply the haploid oocyte genome from a single bovine oocyte and that this haploid oocyte genome replicate can be used to generate biparental bovine embryos. Future research might consider the generation of stable haploid parthenogenetic cell lines as an alternative source of female gametes. The generation of parthenogenetic embryos has been achieved in several mammalian species (Kaufman et al., Reference Kaufman1983; Machaty et al., Reference Machaty, Wang, Day and Prather1997; Loi et al., Reference Loi, Ledda, Fulka, Cappai and Moor1998; Liu et al., Reference Liu, Chen, Cheng and Ju2002; Grabiec et al., Reference Grabiec, Max and Tischner2007; Méo et al., Reference Méo, Yamazaki, Ferreira, Perecin, Saraiva, Leal and Garcia2007; Revazova et al., Reference Revazova, Turovets, Kochetkova, Kindarova, Kuzmichev, Janus and Pryzhkova2007). Previously, karyotypically stable cell lines that maintain a haploid karyotype have been isolated from amphibians and insects (Freed et al., Reference Freed and Mezger-Freed1970; Debec et al., Reference Debec1984). More recently the generation of haploid embryonic stem cells was described in Mekada fish (Yi et al., Reference Yi, Hong and Hong2009). This situation opens the possibility for oocyte genome cloning by multiplying the parthenogenetic haploid line. This would have the potential to generate an unlimited number of biparental embryos by combining these female haploid cells with haploid male hemizygotes to create a new combination of genetic traits from both parents.
In conclusion, we have shown that it is possible to obtain a consistent number of female haploid genome replicates from a single bovine oocyte and, subsequently, to use these replicates to generate biparental embryos. This approach offers enormous potential for livestock production as the use of genetic markers could allow the selection of certain favorable attributes prior to embryo reconstruction. Biparental embryo reconstruction by fusion of haploid oocyte genome replicates also could improve transgenic animal production because, in addition to increasing the number of transgenic embryos produced from a single oocyte, it also generates homogeneous transgene-expressing embryos. The capacity to alter the genome, by the introduction of exogenous genes with high efficiency and homogeneous expression, could increase the generation of transgenic farm animals that are useful in the pharmaceutical industry, in biomedicine or for livestock production.
Acknowledgements
The authors are grateful to CIALE for provision of the biological material. The authors thank Elizabeth Crichton and Lic Lucia Moro for their assistance with English. This work was supported by the Agencia de Promoción Científica y Tecnológica (PICT Redes N° 35142 2005–2009) and by Universidad de Buenos Aires (UBACYT G808).







