Introduction
Pou5f1 encodes the POU class V family (POU-V) of transcription factors that are key molecules for the maintenance of pluripotent cell ability and cell specification of vertebrate embryos. In mice, Pou5f1 is expressed in the inner cell mass (ICM), epiblast and germ cell lineage (Yeom et al., Reference Yeom, Fuhrmann, Ovitt, Brehm, Ohbo, Gross, Hubner and Schöler1996; Niwa et al., Reference Niwa, Miyazaki and Smith2000). In the absence of Pou5f1, mouse embryos cannot establish pluripotency in the ICM and are lethal during preimplantation stages (Nichols et al., Reference Nichols, Zevnik, Anastassiadis, Niwa, Klewe-Nebenius, Chambers, Schöler and Smith1998). Pou5f1 is one of the four factors essential for reprogramming somatic cells to pluripotent stem cells (iPSCs) in mouse and human (Takahashi and Yamanaka, Reference Takahashi and Yamanaka2006; Takahashi et al., Reference Takahashi, Tanabe, Ohnuki, Narita, Ichisaka, Tomoda and Yamanaka2007). A conditional knockout of mouse Pou5f1 in the primordial germ cells (PGCs) induced apoptosis of PGCs because of reduction of viability of PGC (Kehler et al., Reference Kehler, Tolkunova, Koschorz, Pesce, Gentile, Boiani, Lomeli, Nagy, McLaughlin, Schöler and Tomilin2004), indicating that the role of Pou5f1 was not exclusively restricted to maintenance of pluripotent cell ability.
Since the initial characterization of mouse Pou5f1, many homologues have been identified in vertebrates. Although there have been several discussions about the orthologous relationship among vertebrate POU-V homologues, these POU-V family genes have been classified into the Pou5f1 group and its paralogous Pou5f3 group (Frankenberg et al., Reference Frankenberg, Frank, Harland, Johnson, Nichols, Niwa, Schöler, Tanaka, Wylie and Brickman2014). Comparative genomics indicated that Pou5f1 and Pou5f3 might come from gene duplication of a common ancestral gene (Frankenberg and Renfree, Reference Frankenberg and Renfree2013). According to these scenarios, eutherians have lost Pou5f3 and retained Pou5f1, whereas teleost fishes and anurans have retained only Pou5f3. As noteworthy evidence, marsupials and monotremes have retained both Pou5f1 and Pou5f3 (Frankenberg and Renfree, Reference Frankenberg and Renfree2013). Another important example is the axolotl (Ambystoma mexicanum) that has retained both Pou5f1 and Pou5f3 in the genome (Tapia et al., Reference Tapia, Reinhardt, Duemmler, Wu, Arauzo-Bravo, Esch, Greber, Cojocaru, Rascon, Tazaki, Kump, Voss, Tanaka and Schöler2012). In other urodele examples, a partial sequence with Pou5f1 characteristics has been reported in eastern newt (Notophthalmus viridescens) (Maki et al., Reference Maki, Martinson, Nishimura, Tarui, Meller, Tsonis and Agata2010) and Iberian ribbed newt (Pleurodeles waltl) (Wallacieds et al., unpublished). Because of poor genome information, the characterization of POU-V genes in urodeles still remains to be resolved.
Amphibian larva and adult are well known as lively animals having a high ability to regenerate (Brockes, Reference Brockes1997). Especially in newts, this high regeneration capability has been shown in eye, jaw, limb and tail, even in an aged adult (Sanchez Alvarado and Tsonis, Reference Sanchez Alvarado and Tsonis2006; Eguchi et al., Reference Eguchi, Eguchi, Nakamura, Yadav, Millan and Tsonis2011). Based on their regenerative capability, urodele limbs and eyes have been used to understand the cellular and molecular mechanisms of tissue regeneration (Chaar and Tsilfidis, Reference Chaar and Tsilfidis2006; Kragl et al., Reference Kragl, Knapp, Nacu, Khattak, Schnapp, Epperlein and Tanaka2008; Simon and Tanaka, Reference Simon and Tanaka2013). However, trigger molecules for tissue regeneration remain unclear. To identify the essential molecules working in the initial phase of newt regeneration, a more detailed investigation should be performed for cell-to-cell signalling and transcriptional regulation. The POU class V family transcription factor is one of the most important regulators in reprogramming of somatic cells and maintenance of cellular pluripotency. In the present study, we identified two cDNA clones named CpPou5f1 and CpPou5f3 that encode POU-V proteins in Cynops pyrrhogaster. To understand the difference between the two paralogues, their gene expression was examined during the periods of oogenesis, embryogenesis and limb regeneration. Based on their molecular structure and gene expression profiles, we discuss the functional differences between the two paralogous POU class V genes working in one urodele species.
Materials and Methods
Experimental animals and embryos
We purchased sexually mature Japanese red bellied newts, Cynops pyrrhogaster, from Ohuchi Shop (Saitama, Japan), and kept females together with males in the same plastic containers at 12°C. To obtain fertilized eggs, 50 μl (30 U) of gonadotropic hormone (Gestron, Kyoritsu Seiyaku, Japan), was injected subcutaneously into the abdomen of females. Preparation of embryos was performed according to the modified method of Casco-Robles et al. (Reference Casco-Robles, Yamada, Miura and Chiba2010). Embryos were cultured in 0.1× MBS (modified Barth saline; Gurdon, Reference Gurdon1977) at 18°C until the start of the study. The eggs and embryos were dejellied with 2% l-cysteine in 0.1× MBS (pH 9.2–9.5) and rinsed with 0.1× MBS. The developmental stages of the embryos were determined according to Okada and Ichikawa (Reference Okada and Ichikawa1947). For isolation of oocytes, sexually matured females were anaesthetised with 0.1% ethyl 3-amino-benzoate solution (MS-222, Sigma), and their ovaries were excised. The ovaries were dissected in 0.1× MBS and oocytes adhering to the ovarian membrane were collected using tweezers. The isolated oocytes were divided into three stages, previtellogenic, vitellogenic and mature oocytes, according to Scheer et al. (Reference Scheer, Trendelenburg and Franke1976).
Regeneration of limbs
Animals were anaesthetised by soaking in 0.1% MS-222 at st.57 larval stage, at 1 month and at the 1-year-old adult stage. To understand limb development, the full length of the forelimbs was dissected and used for morphological and histological analyses. For regeneration experiments, larvae at st.57 were anaesthetised with 0.1% MS222, and the tip of the forelimb was amputated by cutting with razor blade around the wrist area. Forelimbs immediately after amputation were used as 0 day post-amputation. After amputating the tip of forelimbs, the larvae were reared in 0.1× MBS containing 0.02% kanamycin at 18°C. The larvae were reared in tap water and fed from 1 week after amputation.
Gene cloning
Total RNA was isolated from the ovary of sexually matured female using RNAiso Plus (TaKaRa, Japan). Oligo-dT-primed cDNAs were synthesized using GoScript™ reverse transcriptase (Promega) according to manufacturer’s instruction. To obtain cDNAs encoding POU class V family proteins, PCR was performed using the ovary cDNAs as template. First, we amplified partial cDNAs encoding the POU domain using degenerate primers for the conserved regions in the POU domain (Table 1). Thereafter, 5′ Rapid amplification of cDNA ends (RACE) and 3′RACE were performed to obtain full-length cDNAs. PCR conditions were as follows: pre-heating at 94°C for 2 min and 35 cycles of 94°C/30 s for melting, 55°C/30 s for annealing, and 72°C/2 min for extension. PCR products were inserted into pBluescript II SK(+) using the TA cloning method.
Table 1. Primers used for gene cloning and RT-PCR analyses

* Mix primers containing IUPAC ambiguity codes.
Protein comparison and phylogenetic analysis
Sequence searches were performed using the BLAST search engine on the NCBI website (http://www.ncbi.nih.gov/). Amino acid sequences were aligned using Genetyx software (ver.9, Genetyx Corp., Japan). For phylogenetic analysis, all amino acid sequences were analyzed using neighbour-joining methods through Clustal Omega on the EMBL-EBI website (https://www.ebi.ac.uk/).
RT-PCR analysis
Total RNAs were extracted from oocytes, embryos and larvae at the desired stages or from dissected adult organs using RNAiso Plus (Takara, Japan). For RT-PCR analysis in forelimbs, RNAs were extracted from full length of forelimbs in control animals, or from regenerating tips of amputated forelimbs in operated animals. Single-stranded cDNAs were synthesized with GoScript™ Reverse Transcriptase (Promega) and an oligodT primer, and used as templates for PCR. Semi-quantitative RT-PCR was performed using primers for specific sites in CpPou5f1 and CpPou5f3 (Table 1). The primers for β-tubulin and EF1α were used as an internal control for an amplification standard. RT-PCR conditions were as follows: pre-heating at 94°C for 2 min and 20 cycles (embryos and larvae) or 30 cycles (adult organs and limb regeneration) of 94°C/30 s for melting, 55°C/30 s for annealing, and 72°C/30 s for extension. PCR products were examined by electrophoresis on 2% agarose gels and ethidium bromide-stained bands were captured using a Gel Doc EZ imager (Bio-Rad). The densities of the electrophoretic bands were quantified using NIH Image software (https://imagej.nih.gov/ij/), and a relative value was calculated by normalizing against EF-1α.
Visualization of skeletons and histology
To visualize the skeletal structure of the forelimb, dissected forelimbs were fixed with 10% formalin at 4°C overnight. After washing with 1× PBS, all skin and muscle were removed from the limbs using tweezers. The isolated skeletons were decolorized with 10% aqueous hydrogen peroxide overnight. After washing with water, the cartilage tissues were stained with 15% alcian blue solution (glacial acetic acid 10 ml, alcian blue 8GX 7.5 mg, 95% ethanol 40 ml) for 24 h. After washing twice with 95% ethanol, the forelimb skeletons were stained with 0.025% alizarin red solution (0.3% alizarin red S 10 ml, 0.5% KOH 10 ml, distilled water (DW) 100 ml) overnight. After decolorizing in 20% glycerol containing 1% KOH for more than 3 days, photographs were taken under a dissection microscope (Olympus SZX16). For histological examination, dissected forelimbs were fixed with 4% paraformaldehyde for 12 h at 4°C. The fixed forelimbs were incubated in decalcifying solution (sodium citrate 10 g, formic acid 22.5 g, DW 100 ml) at 4°C overnight for demineralization. After washing with 1× PBS, the forelimbs were dehydrated through a graded series of ethanol, cleared in xylene and embedded in TissuePrep (Fisher Scientific). The tissues were sectioned serially at 10-μm thickness and stained with haematoxylin and eosin.
Results
Identification of CpPou5f1 and CpPou5f3
To obtain POU class V family genes from adult ovary cDNAs of Cynops pyrrhogaster, we first amplified short PCR fragments using degenerate primers for the conserved amino acid sequences in the POU domain. Next, 5′-end and 3′-end of the PCR fragments were elongated RACE, and two clones were isolated as full-length cDNAs. The shorter clone was a 1648-bp cDNA encoding one open reading frame (ORF) containing 390 amino acid residues, and the longer clone was a 1868-bp cDNA encoding one ORF containing 435 amino acid residues. As shown in Fig. 1, the predicted amino acid sequence showed the highly conserved POU-specific domain (red line) and the POU-homeodomain (blue line). To characterize these two clones, the amino acid sequences were compared with those of POU class V family proteins from axolotl, coelacanth, platypus, wallaby, mouse and zebrafish. In the shorter clone, the characteristic MAGH sequence was recognized at the N-terminus of the predicted amino acid sequence, but not in the longer clone (Fig. 2A, arrow 1). The last amino acid in the POU-specific domain was aspartic acid in both clones (Fig. 2A, arrow 2). Deletion of arginine at the fifth position of the POU-homeodomain was recognized in the shorter clone, but not in the longer clone (Fig. 2A, arrow 3). These results indicated that the shorter clone and the longer clone encoded Pou5f1 and Pou5f3 family proteins, respectively. Based on these characteristics, we named the two genes, CpPou5f1 and CpPou5f3. To verify the classification of these two genes, a phylogenetic analysis of POU class V family proteins was performed using the neighbour-joining method and Clustal Omega software. As shown in Fig. 2B, CpPou5f1 and CpPou5f3 were classified into the clade of the POU5F1 and POU5F3 family, respectively.

Figure 1. Predicted amino acid sequence of CpPou5f1 and CpPou5f3. Full-length cDNA clones encoding POU class V transcription factors were isolated from Japanese red bellied newt and named CpPou5f1 and CpPou5f3. The predicted amino acid sequences are represented using capitalized one-letter codes. Black shading amino acids indicate common in two proteins. POU-specific domain and POU-homeodomain are indicated by red and blue line, respectively.
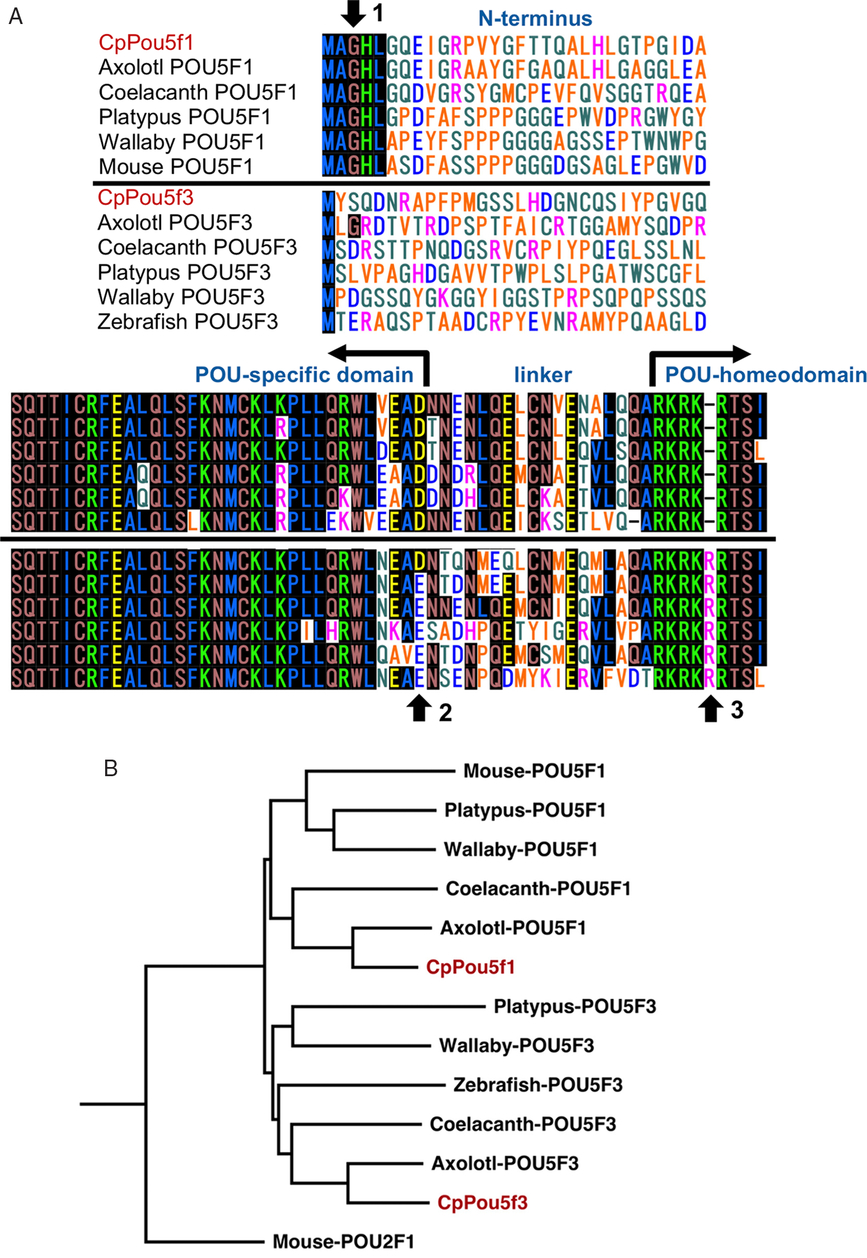
Figure 2. Phylogenetic analysis of CpPou5f1 and CpPou5f3. (A) Sequence comparison among POU class V family proteins. N-terminal (upper panel) and POU-domain sequence (lower panel) of CpPou5f1 (LC215873) and CpPou5f3(LC438345) were compared with those of axolotl (AY542376, AGN30963), coelacanth (XP005998216, XP005994419), platypus (NM001242727, CP007654084), wallaby (FJ998419, XP00529150), mouse (NM013633) and zebrafish (NM131112). Pou5f1 and Pou5f3 family proteins are aligned above and below the central solid line, respectively. Black shading amino acids indicate common in more than six species. Arrows 1–3 show the amino acid sequences which are used to distinguish two families of Pou5f1 and Pou5f3. (B) Phylogenetic tree of POU class V homologues from different species. The branches are shown with their calculated relative similarity. Mouse POU2F1 was used as the outgroup to emphasize the split of Pou5f1 and Pou5f3.
Gene expression in oogenesis and early development
To clarify gene expression of CpPou5f1 and CpPou5f3 during oogenesis, oocytes were isolated from sexually matured female ovaries, and classified into three stages: previtellogenic, vitellogenic and mature oocyte, according to Scheer et al. (Reference Scheer, Trendelenburg and Franke1976). In RT-PCR analysis of transcripts extracted from oocytes, marked gene expression of CpPou5f1 was detected from vitellogenic and mature oocytes, whereas weak gene expression of CpPou5f3 was recognized from previtellogenic to mature oocytes (Fig. 3A). Next, the expression profiles of CpPou5f1 and CpPou5f3 were examined during early development. As shown in Fig. 3B, maternal transcripts of CpPou5f1 were detected from unfertilized eggs. The same level of transcripts was recognized in the cleavage-stage embryos. However, transcripts of CpPou5f1 were increased from gastrula (st.12) to neurula (st.16), suggesting that zygotic expression of CpPou5f1 occurred during gastrulation. In CpPou5f3, the maternal transcript was not observed in the cleavage-stage embryos, and a low level of zygotic expression was detected in blastula, gastrula and neurula stages (Fig. 3B). Gene expression of CpPou5f1 and CpPou5f3 was not detected from embryos after the tail-bud stage (st.30).

Figure 3. CpPou5f1 and CpPou5f3 expression in oogenesis and early development. (A) Gene expression of CpPou5f1 and CpPou5f3 during oogenesis. Isolated oocytes were classified into three stages, previtelogenic (PT), viterogenic (VT) and mature oocytes (MT), and gene expression of POU family genes was examined by RT-PCR. EF1α is the internal marker of gene expression. RT-PCR products without reverse transcriptase reaction. (B) Gene expression of CpPou5f1 and CpPou5f3 during early development. β-Tubulin is the internal marker of gene expression. RT-PCR products without reverse transcriptase reaction. Developmental stages of embryos were determined according to Okada and Ichikawa (Reference Okada and Ichikawa1947). UE, unfertilized eggs.
To examine a functional difference between CpPou5f1 and CpPou5f3, gene expression of CpPou51 and CpPou5f3 was examined in adult organs. As shown in Fig. 4, gene expression of CpPou5f1 was detected only in the ovary among the nine organs tested. In contrast, gene expression of CpPou5f3 was detected in various organs such as testis, ovary, liver, spleen, stomach and intestine.

Figure 4. CpPou5f1 and CpPou5f3 expression in adult organs. Gene expression of CpPou5f1 and CpPou5f3 was examined in adult organs by RT-PCR. EF1α is the internal control of gene expression. RT-, PCR products without reverse transcriptase reaction.
Forelimb formation and POU family genes
Before the study on gene expression in forelimb, morphological change and skeletal maturation were examined in forelimbs from the larval stage to the 1-year-old adult. At st.57, the final stage of the larvae, complete formation of four fingers was observed in the forelimb (Fig. 5Aa). However, skeletons in the forelimbs were made only from the alcian blue-stained cartilage, which was confirmed by histological examination (Fig. 5Ab, c). In the 1-year-old newt, the forelimb grew up four times larger in size, and ossification of skeletons was detected by staining with alizarin red (Fig. 5Ad, e). Histological analysis showed that bone marrow formation occurred in the 1-year-old forelimb (Fig. 5Af). These results suggested that endochondral ossification occurred in forelimbs for the 1 year after metamorphosis. Next, gene expression of CpPou51 and CpPou5f3 was examined during forelimb development after metamorphosis. As shown in Fig. 5B, gene expression of CpPou5f1 was not detected at all stages, whereas weak gene expression was detected in CpPou5f3 from st.57 to the 1-year-old newt stage.

Figure 5. Forelimb development and gene expression of CpPou5f1 and CpPou5f3. (A) Comparison of forelimb between larval stage (a–c, st.57) and adulthood (d–f, 1 year-old). Outer morphology (a, d), skeletons stained with alcian blue and alizarin red (b, e) and histological section stained with haematoxylin and eosin (c, f) of forelimbs were compared between two stages. Asterisks indicate bone marrow cavities. (B) Gene expression of CpPou5f1 and CpPou5f3 during forelimb development. RNAs were extracted from forelimbs at st.57, 1 month after metamorphosis (1M) or 1-year-old newt (1Y), and gene expression of CpPou5f1 and CpPou5f3 were examined by RT-PCR. EF1α is the internal control of gene expression. RT-, PCR products without reverse transcriptase reaction.
Regeneration of forelimb and gene expression of CpPou5f1 and CpPou5f3
To examine the gene expression of CpPou5f1 and CpPou5f3 during regeneration of forelimbs, tips of the forelimb were amputated from st.57 larvae by cutting the wrist, and morphological change of the amputated tips was observed during 4 weeks. Wound healing of the cut edge was recognized at 3 days post-amputation, and the transparent blastema was observed on the tip of forelimb at 1 week post-amputation (Fig. 6Aa–d). Finger rudiments were recognized at 2 weeks post-amputation, and regeneration of the forelimb was completed until 4 weeks post-amputation (Fig. 6Ae, f). During the 4 weeks after amputation, the stump region of the amputated forelimb was isolated and gene expression was examined by RT-PCR. Gene expression of CpPou5f1 was not recognized in the stump just after amputation. However, weak and transient gene expression was detected at 3 days post-amputation (Fig. 6B, C). Thereafter, gene expression of CpPou5f1 was suppressed and not detected in the amputated forelimb at 1 week post-amputation. Whereas, low level of CpPou5f3 expression was recognized in normal forelimb of st.57 larvae, and gradually upregulated after amputation (Fig. 6B, C). CpPou5f3 showed a peak expression at 1 week post-amputation, and thereafter drastically decreased in expression level (Fig. 6B, C).

Figure 6. Gene expression of CpPou5f1 and CpPou5f3 during regeneration of forelimb. (A) Morphological change of regenerating forelimb. Tips of forelimbs of st.57 larva (a) was amputated (b), and morphological change was examined at 3 days (c), 1 week (d), 2 weeks (e) and 4 weeks post-amputation (f). Scale bars indicate 500 μm. (B) Gene expression of CpPou5f1 and CpPou5f3. Tip region of the amputated forelimbs were isolated and gene expression was examined by RT-PCR. EF1α is the internal control of gene expression. RT-, PCR products without reverse transcriptase reaction. (C) Quantitative analysis of gene expression. Expression level of CpPou5f1 and CpPou5f3 were shown with a relative value normalized by gene expression of EF-1α.
Discussion
Mouse Pou5f1 was the first gene identified as POU class V member (Schöler et al., Reference Schöler, Hatzopoulos, Balling, Suzuki and Gruss1989; Okamoto et al., Reference Okamoto, Okazawa, Okuda, Sakai, Muramatsu and Hamada1990; Rosner et al., Reference Rosner, Vigano, Ozato, Timmons, Poirier, Rigby and Staudt1990). Thereafter, many POU class V genes have been identified from vertebrates. After several confusions in nomenclature, these genes were categorized into three paralogues, Pou5f1, Pou5f2 and Pou5f3, based on phylogenetical characteristics (Frankenberg et al., Reference Frankenberg, Frank, Harland, Johnson, Nichols, Niwa, Schöler, Tanaka, Wylie and Brickman2014). In the present study, we identified two POU-V homologues in Cynops pyrrhogaster, and named them CpPou5f1 and CpPou5f3. The predicted amino acid sequence showed that the POU-specific domain and the POU-homeodomain are highly conserved in the two proteins. Frankenberg and his colleague have reported that the Pou5f1 family proteins have a typical MAGH sequence at the N-terminus of the protein, which is absent in Pou5f3 family proteins (Frankenberg et al., Reference Frankenberg, Pask and Renfree2010). Therefore, the N-terminal MAGH sequence is a clear marker to distinguish members of Pou5f1 from those of Pou5f3. In the present study, CpPou5f1 has the N-terminal MAGH in the predicted amino acid sequence, but not in CpPou5f3. In addition, the deletion of arginine at the fifth position of POU-homeodomain is recognized in CpPou5f1 but not in CpPou5f3, which is another remarkable characteristic of Pou5f1 family proteins as indicated previously (Frankenberg et al., Reference Frankenberg, Pask and Renfree2010). These results suggested that two cDNAs newly isolated from Cynops pyrrhogaster encodes Pou5f1 and Pou5f3 proteins, respectively. This classification was verified by phylogenetic analysis using Clustal Omega software.
In the quantitative analysis of gene expression in newt POU-V genes, maternal expression of CpPou5f1 was observed in oocytes and early embryos. This finding is consistent with the expression profiles of axolotl AxPou5f1 (Bachvarova et al., Reference Bachvarova, Masi, Drum, Parker, Mason, Patient and Johnson2004; Tapia et al., Reference Tapia, Reinhardt, Duemmler, Wu, Arauzo-Bravo, Esch, Greber, Cojocaru, Rascon, Tazaki, Kump, Voss, Tanaka and Schöler2012) and mouse Pou5f1 (Schöler et al., Reference Schöler, Dressler, Balling, Rohdewohld and Gruss1990). CpPou5f1 is probably expressed as a maternal transcript and used for the maintenance of pluripotency in the embryonic cells. Gene expression in oocytes and early embryos was detected also in CpPou5f3, which is similar to zebrafish Pou5f3 (previously pou2) (Takeda et al., Reference Takeda, Matsuzaki, Oki, Miyagawa and Amanuma1994). Zebrafish spiel-ohne-grenzen (spg) mutants lacking Pou5f3 caused the gastrulation defects and abnormal neurogenesis (Burgess et al., Reference Burgess, Reim, Chen, Hopkins and Brand2002), indicating the essential role of Pou5f3 in the early development. In Xenopus, triplicated Pou5f3.1, Pou5f3.2 and Pou5f3.3 (previously Oct91, Oct25, Oct60) showed the diversified gene expression (Frank and Harland, Reference Frank and Harland1992; Hinkley et al., Reference Hinkley, Martin, Leibham and Perry1992; Whitfield et al., Reference Whitfield, Heasman and Wylie1993, Reference Whitfield, Heasman and Wylie1995; Watanabe et al., Reference Watanabe, Yasuoka, Mawaribuchi, Kuretani, Ito, Kondo, Ochi, Ogino, Fukui, Taira and Kinoshita2017). These genes have the ability to rescue the loss of self-renewal in Pou5f1-deleted mouse ES cells, demonstrating that Xenopus Pou5f3s are functional homologues of mouse Pou5f1 (Morrison and Brickman, Reference Morrison and Brickman2006). Among three Pou5f3s in Xenopus, only Pou5f3.3 is maternally expressed, which is different from the zygotic expression of Pou5f3.1 and Pou5f3.2 (Hinkley et al., Reference Hinkley, Martin, Leibham and Perry1992). In marsupials and monotremes, Pou5f1 and Pou5f3 are retained in their genome. In the tammar wallaby, Pou5f1 is maternally expressed during oogenesis, whereas Pou5f3 is expressed in a broad range of adult organs (Frankenberg et al., Reference Frankenberg, Pask and Renfree2010). In Cynops pyrrhogaster, CpPou5f1 showed maternal expression in oocytes and ovary, while gene expression of CpPou5f3 was detected in various adult organs. This expression profile is quite similar to the expression pattern of POU-V genes in the tammar wallaby (Frankenberg et al., Reference Frankenberg, Pask and Renfree2010). The Pou5f1 and Pou5f3 genes might divide their role in organogenesis after gene duplication.
In the present study, CpPou5f1 and CpPou5f3 showed a different expression profile at early phases of limb regeneration. After amputating the tip of forelimbs, transient gene expression of CpPou5f1 was induced at 3 days post-amputation, until wound healing was completed. Thereafter, blastema formation was observed on the tip of forelimbs at 1 week post-amputation, when upregulation of CpPou5f3 was detected. It has been known in regeneration of newt forelimbs that the amputated stumps are covered by a wound epithelium, followed by dedifferentiation of the somatic cells and blastema formation (Chaar and Tsilfidis, Reference Chaar and Tsilfidis2006; Simon and Tanaka, Reference Simon and Tanaka2013). Transcriptional profiling using axolotl limb regeneration showed the three distinguishable phase of gene expression among wound healing, blastema establishment and re-development of limb (Knapp et al., Reference Knapp, Schulz, Rascon, Volkmer, Scholz, Nacu, Le, Novozhilov, Tazaki, Protze, Jacob, Hubner, Habermann and Tanaka2013). In the previous reports, it has been shown that the limb regeneration reactivates some developmental genes in order for the blastema cells to grow and differentiate (Simon and Tanaka, Reference Simon and Tanaka2013). Taking together, the temporary upregulation of CpPou5f3 after amputation might be involved in growth and differentiation of blastema cells. In the present study, a temporal difference of gene activation was observed between CpPou5f1 and CpPou5f3, which suggests a functional difference of two POU-V genes in limb regeneration. Limb amputation-induced gene activation in CpPou5f1 and CpPou5f3 is inconsistent with a previous report that gene expression of the Oct4 homologue was not detected in limb regeneration of eastern newt, N. viridescens (Maki et al., Reference Maki, Martinson, Nishimura, Tarui, Meller, Tsonis and Agata2010). In the experiment using two axolotl POU-V genes, both AxPou5f1 and AxPou5f3 could induce pluripotency in mouse and human fibroblasts, suggesting that two axolotl POU-V genes have the ability to reprogramme the somatic cells (Tapia et al., Reference Tapia, Reinhardt, Duemmler, Wu, Arauzo-Bravo, Esch, Greber, Cojocaru, Rascon, Tazaki, Kump, Voss, Tanaka and Schöler2012). Interestingly, it has been reported that ectopic expression of mouse Oct4, Sox2 and Klf4 could induce transdifferentiation of Xenopus tadpole muscles (Vivien et al., Reference Vivien, Scerbo, Girardot, Le Blay, Demeneix and Coen2012). Taking together, it is likely that CpPou5f1 and CpPou5f3 might play a role in the production of pluripotent cells during regeneration of newt forelimb.
Recent advances in genome research has drawn a scheme indicating the origin of POU family class V genes (Frankenberg and Renfree, Reference Frankenberg and Renfree2013). Comparative analyses of POU-domain sequences from available animal genomes have shown that the first original POU-V gene might have been separated from the class III POU genes at branching of the vertebrate lineage (Gold et al., Reference Gold, Gates and Jacobs2014). In comparison with gene alignment among vertebrate genomes, gene synteny is conserved around POU-V genes, suggesting that gene duplication must produce Pou5f1 and Pou5f3 genes from the common ancestral POU-V gene (Frankenberg and Renfree, Reference Frankenberg and Renfree2013). In existing animals, marsupial and monotremes have kept both Pou5f1 and Pou5f3 in their genome (Frankenberg et al., Reference Frankenberg, Pask and Renfree2010). Human and mouse have lost Pou5f3 and retained Pou5f1, while teleost fish and frog have lost Pou5f1 and retained Pou5f3. In contrast with only Pou5f3 in anuran frog, urodele axolotl has kept two POU-V genes, AxPou5f1 and AxPou5f3 (Tapia et al., Reference Tapia, Reinhardt, Duemmler, Wu, Arauzo-Bravo, Esch, Greber, Cojocaru, Rascon, Tazaki, Kump, Voss, Tanaka and Schöler2012). In the present study, we succeeded in cloning two POU-V genes, CpPou5f1 and CpPou5f3 from Cynops pyrrhogaster. Present results demonstrated that Japanese red bellied newts have two POU-V genes similar to axolotl, suggesting a common evolutional feature of urodeles POU class V genes. Urodeles have a high ability for regeneration (Brockes, Reference Brockes1997), and are used as model animals for understanding cellular and molecular mechanism of tissue regeneration (Chaar and Tsilfidis, Reference Chaar and Tsilfidis2006; Simon and Tanaka, Reference Simon and Tanaka2013). Expression of the duplicated POU-V genes in wound repair might support the high ability of regeneration in urodeles.
Financial support
This work was supported by Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science (grant number 24240062), and by the Strategic Research Foundation Grant-aided Project for Private Universities (grant number S1201003) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Ethical standards
Not applicable
Conflicts of interest
None









