Introduction
Primordial germ cells (PGCs) are the precursors of sperm and egg cells and derived from a population of pluripotent epiblast cells during early embryonic development. After separation from the embryonic lineage, PGCs migrate from the base of the allantois and reach the genital ridge of the fetal gonad at around embryonic (E) days E10.5 to E11.5. After colonizing the genital ridge, PGCs differentiate into male or female germ cells at around E12.0 (Suzuki et al., Reference Suzuki, Saba, Sada and Saga2010). Several conserved germline genes, such as vasa, dazl and dead end (dnd), are required for specifying and/or maintaining PGC development (Xu et al., Reference Xu, Gui and Hong2005; Peng et al., Reference Peng, Xie, Zhou, Hong and Gui2009; Liu et al., Reference Liu, Li, Li, Wang, Li, Zhong, Zhang, Zhang, Zhou and Gui2015). Nanos, an evolutionarily conserved family of RNA-binding proteins, was first identified as a determinant of abdomen formation in Drosophila (Subramaniam and Seydoux, Reference Subramaniam and Seydoux1999; Mochizuki et al., Reference Mochizuki, Sano, Kobayashi, Nishimiya-Fujisawa and Fujisawa2000; Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003; Kato et al., Reference Kato, Katsuki, Kokubo, Masuda and Saga2016) and expressed specifically in PGCs from both invertebrate and vertebrate animals (Asaoka-Taguchi et al., Reference Asaoka-Taguchi, Yamada, Nakamura, Hanyu and Kobayashi1999; Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003). Ablation of Nanos and its orthologues results in the loss of PGCs in all species examined including Drosophila (Forbes and Lehmann, Reference Forbes and Lehmann1998), Caenorhabditis elegans (Kraemer et al., Reference Kraemer, Critenden, Gallegos, Moulder, Barstead, Kimble and Wickens1999), zebrafish (Beer and Draper, Reference Beer and Draper2013), sea urchin (Oulhen and Gary, Reference Oulhen and Gary2016), Xenopus (Lai et al., Reference Lai, Singh and King2012), and mouse (Haraguchi et al., Reference Haraguchi, Tsuda, Kitajima, Sasaoka, Nomura-Kitabayashid, Kurokawa and Saga2003). Furthermore, in humans, mutation in Nanos genes is associated with loss of PGCs and this may lead to infertility (Kusz-Zamelczyk et al., Reference Kusz-Zamelczyk, Sajek, Spik, Glazar, Jedrzecjczak, Latos-Bielenska, Kotecki, Pawelczyk and Jaruzelsaka2013). These findings suggest an evolutionarily conserved and indispensable role for Nanos protein during PGCs development.
In vertebrates, three Nanos genes have been identified that have differential expression patterns, functions and regulatory mechanisms among different species. For example, in human, Nanos1 is expressed ubiquitously, while Nanos2 and Nanos3 are expressed in ovary, testis and fetal brain (Julaton and Pera, Reference Julaton and Pera2011). In mouse, the presence of Nanos1 is demonstrated in the central nervous system (Haraguchi et al., Reference Haraguchi, Tsuda, Kitajima, Sasaoka, Nomura-Kitabayashid, Kurokawa and Saga2003), while Nanos3 was found in PGCs as early as embryonic day 9.5 (E9.5) to E15.5. Nanos2 was restricted in the developing male PGCs at E13.5 and then expressed predominantly in male germ cells (Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003; Kato et al., Reference Kato, Katsuki, Kokubo, Masuda and Saga2016). Nanos2 mRNA was first detected in germ cells that colonized the male embryonic gonad at around E13. Expression of Nanos2 gene in PGCs and gonocytes is transient during embryogenesis and neonatal development (Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003; Kato et al., Reference Kato, Katsuki, Kokubo, Masuda and Saga2016). Importantly, inactivation of Nanos2 gene in mice resulted in loss of germ cells only in the male embryos around E15.5, indicating that Nanos2 gene expression is restricted in male PGCs. Furthermore, Nanos2 expression in the mouse starts in male PGCs just after their determination in primary testes, then decreased shortly before birth. It is maintained in the adult, but only in a limited number of spermatogonia (Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003). Nanos2 protein function in the mouse cannot be replaced by Nanos3, however, but the function of Nanos3 can be restored by Nanos2 (Suzuki et al., Reference Suzuki, Tsuda and Saga2007). Therefore, it seems that Nanos2 is a unique and highly specialized protein that has an indispensable role for male germ-cell development. The expression pattern of Nanos2 in PGCs during embryonic development, as well as in spermatogonial stem cells (SSCs) in rat testes, has only been rarely investigated. In the present study we investigated the expression pattern of Nanos2 gene in PGCs from different embryonic developmental days (prenatal period), as well as in SSCs (postnatal period) in a rat animal model.
Materials and methods
Chemicals
All chemicals used in this study were procured from Sigma Chemical Co. (St. Louis, MO, USA) unless stated otherwise.
Experimental animal
Sexually mature female and male rats (120 ± 5 g body weight) Charles-Foster strain were purchased from the Central Animal House, Institute of Medical Sciences, Banaras Hindu University, Varanasi and housed in air-conditioned, light-controlled rooms, with food and water ad libitum. To obtain pregnant females, 12 proestrus stage rats were divided into two groups (six for immunostaining and six for protein and RNA isolation) and kept for mating with proven fertile male rat as per the standard rat laboratory breeding protocol. The following day, all female rats were examined for vaginal plug formation and this was considered as embryonic day 1 (E1). The developing embryos (n=24) were dissected out and 12 embryos (four embryos from each embryonic day E8.5, E10.5 and E11.5) were directly placed in fixative solution for immunostaining. The remaining 12 embryos from different embryonic days were used for protein and RNA isolation. To isolate SSCs, male rat (~100 g; n=6) were killed by cervical dislocation and their testes were removed. From each experimental animal, one testis was directly fixed in 10% normal buffered saline for histological studies and other testis used for isolation of SSCs.
Whole-mount embryo immunostaining
Embryos from different embryonic developmental days E8.5, E10.5 and E11.5 (three embryos from each embryonic day) were excised and fixed overnight in Bouin’s solution for immunocytochemistry. Embryos from different embryonic days were mounted over gelatin-coated (0.1% gelatin to increase adherence of the embryos) glass slides. To localize Nanos2 protein, the mounted embryos were washed three times in PBS containing 0.1% Triton X-100 (PBS-T) and incubated overnight with a blocking solution (PBS-T containing 5% fetal calf serum + 0.1% Triton X-100). The embryos were then incubated with mouse monoclonal Nanos2 antibodies (1:200 in PBS-T; Santa Cruz Biotechnology Inc. cat no. sc-393794) at 4°C for overnight. To confirm PGCs, embryos from different days were also incubated with monoclonal stage-specific embryonic antigen-1 (SSEA-1) antibodies (1:200 in PBS-T; Santa Cruz Biotechnology Inc. cat no. sc-21702). After three washes in PBS-T, the embryos were incubated with FITC and Texas red conjugated mouse IgG (1:1000 dilution in PBS-T; Santa Cruz Biotechnology Inc.) for SSEA-1 and Nanos2 respectively at 37°C for 2 h. After three successive washing with PBS-T, immunological signals were detected using fluorescence microscope (EVOS 4300 colour imaging system, Life Technologies).
Western blot
To confirm the immunocytochemistry results, SSCs (1×106 cells) and embryos from different developmental days (E8.5, E10.5, E11.5) were lysed in 1× cell lysis buffer (Cell Signalling Technology) containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin. For isolation of protein, the manufacture’s protocol was followed with some modifications. Total homogenate was centrifuged at 10,000 g for 20 min and the pellet was resuspended in a small volume of medium A. Protein concentration was determined using the bicinchoninic acid method and bovine serum albumin (BSA) as a standard. Protein (60 µg) from SSCs and embryos from different embryonic days was separated using 15% SDS-PAGE and then transferred onto polyvinylidene fluoride membranes (Millipore Corporation, Bedford, MA, USA). The membranes were blocked in 5% non-fat milk in Tris-buffered saline at room temperature for 2 h and then incubated with monoclonal mouse anti-Nanos2 antibodies diluted 1:500 in TBS at room temperature for 3 h. The membranes were washed in TBST (0.1% Tween-20 in TBS) three times at room temperature for 15 min, and then incubated with horseradish peroxidase (HRP)-conjugated IgG (Santa Cruz Biotechnology) for 1 h at room temperature. After washes with TBST, membranes were processed using an enhanced chemiluminescence system according to the manufacturer’s protocol. These experiments were repeated at least three times.
RT-PCR
Total RNA from embryos (each embryonic days) and SSCs (1×106 cells) was extracted using TRIzol reagent and treated with DNase I to avoid genomic DNA contamination. Total RNA was reverse transcribed using the SuperScript First Strand cDNA Synthesis kit (Invitrogen) according to the manufacturer’s instructions. Primer sequences used for PCR were as follows: Nanos2 forward AGTGCCATGGACCTACCGCCCTTT and Nanos2 reverse, TCTCAATTATCGCTTGACTCTGC; GAPDH forward, GGGTGGTCCAGGGTTTCTTACT and GAPDH reverse, AGGTTGTCTCCTGCGACTCA. Semi-quantitative RT-PCR, 35 cycles, was performed to amplify Nanos2 and GAPDH from embryos and SSCs.
Isolation and culture of spermatogonial stem cells
SSCs from rat testes were isolated by a two-step enzymatic digestion procedure. In brief, seminiferous tissue was dissociated and digested using an enzymatic solution of collagenase (5 mg/ml; type IV, Sigma Chemical Co., St Louis, MO, USA) and trypsin (1 mg/ml, Sigma) and was incubated at 37°C for 30 min with continuous shaking in a water bath. After three washes in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium and removal of most of the interstitial cells, seminiferous cord fragments were further incubated in medium containing 10 mg/ml DNase I and 1 mg/ml hyaluronidase (Sigma-Aldrich) and trypsin (1 mg/ml, Sigma) at 37°C for 20 min. Enrichment of SSCs was performed by differential plating to eliminate any contaminating somatic cells (myoid and Sertoli cells). In brief, dissociated cell suspensions were cultured over 0.1% gelatin-coated plate in selection medium composed of DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% FBS, 10 mM 2-mercaptoethanol (Sigma) and 1% non-essential amino acids (Sigma) at 37°C under humidified conditions of 5% CO2 in air. After 6 h of incubation, a mixed population of single type A SSCs (As) and paired type A SSCs (Apr) was collected from the cell suspension and cultured in DMEM supplemented with 10% FBS, 10 mM 2-mercaptoethanol, 1% non-essential amino acids, 10 ng/ml recombinant human basic fibroblast growth factor, 100 IU/ml penicillin/streptomycin and 40 μg/ml gentamicin at 37°C and 5% CO2 in air. Cultured SSCs were dissociated using Accutase solution (Sigma) and re-plated every 5 to 6 days. SSCs colonies were maintained over a feeder layer of Sertoli cells at 37°C in 5% CO2 and the medium was changed every 48 h. The viability of isolated SSCs was checked using 0.4% trypan blue staining and cell viability percentage was calculated as per standard procedures.
Flow cytometric analysis
Flow cytometric analyses were performed using a standard laboratory protocol. SSCs colonies were dissociated using cell dissociation reagent Accutase (Sigma) and approximately 1×106 cells were suspended in 0.1 ml of PBS containing 2.5% fetal bovine serum and incubated with primary antibodies (anti-mouse THY-1 and Nanos2; Santa Cruz Biotechnology). Primary antibodies were then detected using 5 mg/ml of FITC-conjugated or Alexa488-conjugated secondary antibodies (Santa Cruz Biotechnology). Control cells were also processed in same way except incubation with primary antibodies. The cells were kept in the dark in ice and cell sorting was performed using a Becton Dickinson fluorescence-activated cell sorter (FACS) IV Calibur (Becton Dickinson, San Jose, CA, USA). A minimum of 5000 or 10,000 events were acquired for each sample and analyzed using CFlow Plus software.
Immunocytochemistry
For immunocytochemistry, a mixed population of As, Apr and other undifferentiated SSCs was fixed over gelatin-coated (0.1%, to increase adherence of the cells) glass slides. Rat testicular sections (5 μm thickness) were prepared using a microtome (Leica Instruments, Nussloch, Germany). After a PBS wash, the cells and testicular sections were fixed in 4% paraformaldehyde for 10 min at room temperature. Non-specific background staining was blocked using 10% normal mouse serum at room temperature for 30 min. Cells and section from all groups were then incubated with mouse anti-Nanos2 and THY1 antibodies mouse anti-Nanos2 (1:200, Santa Cruz Biotechnology) for 1 h at 37°C. After washing with PBS, cells and tissue sections were incubated further with FITC-linked secondary antibodies (1:500, Santa Cruz Biotechnology). Immunofluorescence signals were analysed using fluorescence microscopy (EVOS 4300 colour imaging system, Life Technologies). For the negative control, the first antibody was omitted, using the same amount of mouse IgG. The same experiments were performed at least three times.
Statistical analysis
All the experiments were performed at least three times and at least in triplicate for each sample.
Results
Expression of Nanos2 in PGCs during different embryonic developmental days
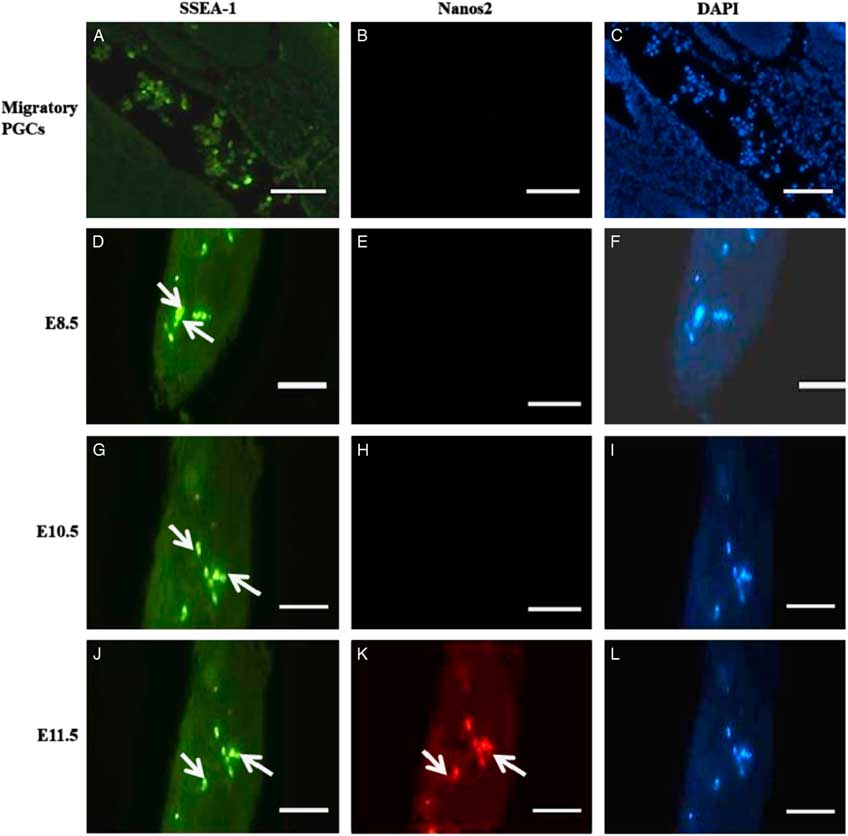
As shown in Fig. 1(K), Nanos2 protein was predominantly localized in migrating PGCs in E11.5 day embryos. Nanos2 expression was exhibited in the cytoplasm of developing PGCs. By contrast, no Nanos2 immunoreactivity was found in E8.5 and E10.5 day embryos, indicating that PGCs do not express Nanos2 protein on early embryonic E8.5 and E10.5 days (Fig. 1E, H). SSEA-1 expression was found in migrating PGCs from different embryo days, indicating that the migrating cells are PGCs (Fig.1A, D, G, J).
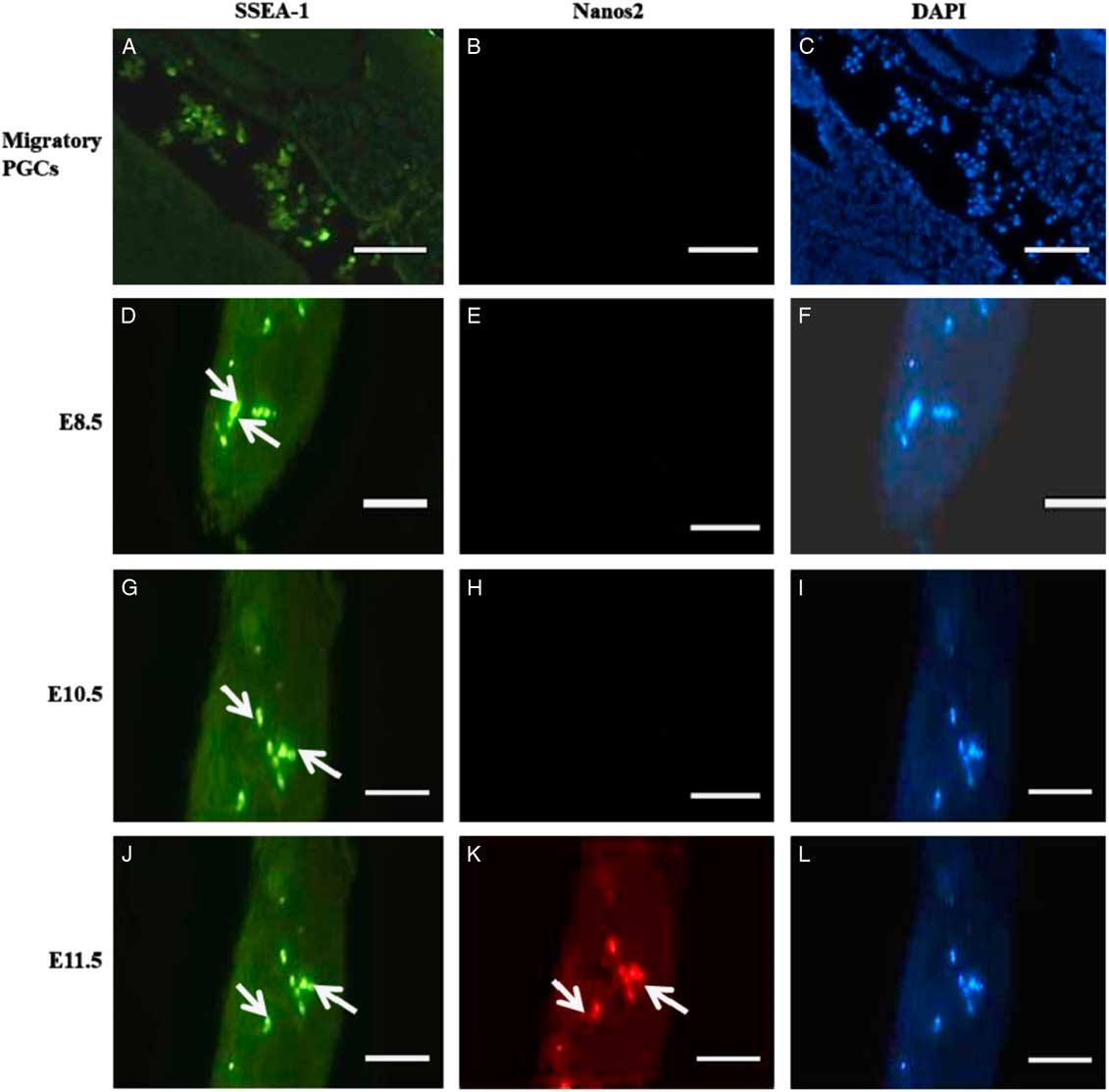
Figure 1 Localization of Nanos2 protein in PGCs on different embryonic developmental days. (A, D, G, J) Fluorescence images showing expression of SSEA-1 in migrating PGCs towards the genital ridge of all embryonic days (green signal; white arrow). (B, E, H) E8.5 and E10.5 day embryos staining negative with anti-Nanos2 antibody. (K) E11.5 day embryo staining positive for Nanos2 protein (red signal; white arrow). White asterisks represent specific signals for Nanos2 protein in PGCs. Scale bars represent 50 µm.
Western blot
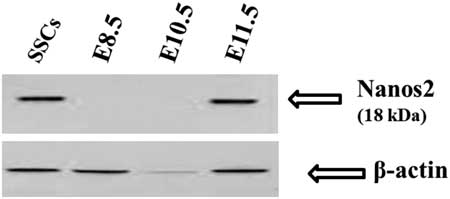
To further verify our immunocytochemistry results, western blotting using Nanos2-specific antibodies was performed. In western blot analysis a single 18 kDa Nanos2 protein band was detected in SSCs and E11.5 day embryos. However, no band was seen from E8.5 and E10.5 day embryos (Fig. 2); there was no cross-reactivity of Nanos2 antibodies with other proteins.
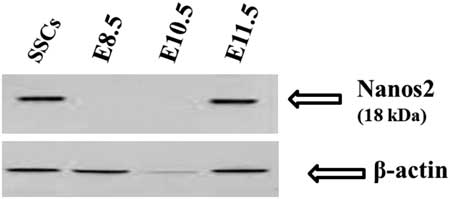
Figure 2 Western blot analysis of E8.5, E10.5, E11.5 embryos and spermatogonial stem cells (SSCs). Nanos2-specific antibodies recognized a specific protein as a single band with a molecular mass of 18 kDa (arrows) in SSCs and E11.5 embryos. The same amount of protein was loaded from all samples and blotted for Nanos2 (top) and β-actin expression (bottom).
mRNA expression
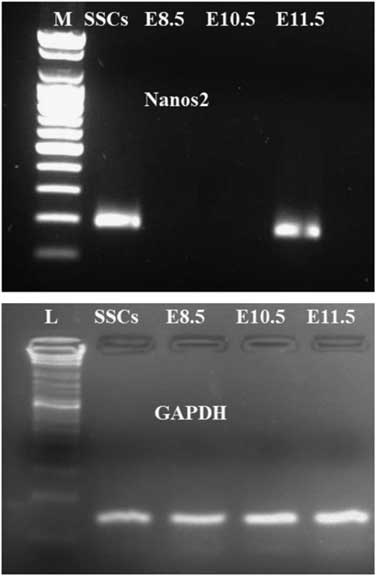
Endogenous Nanos2 mRNA expression in SSCs and embryos from different days (E8.5, E10.5 and E11.5) was examined by semi-quantitative RT-PCR. Nanos2 transcripts (185 bp) were detected in SSCs as well as in E11.5 day embryo, however no expression was found in E8.5 and E10.5 day embryos (Fig. 3), confirming other results.
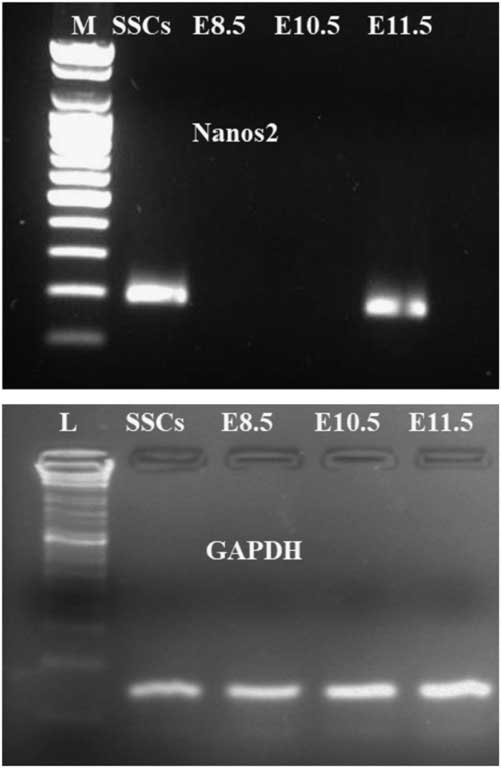
Figure 3 Expression analysis of Nanos2 transcripts for different (E8.5, E10.5 and E11.5) day embryos and in SSCs. PCR amplification showing a 185-bp Nanos2 gene fragment only in E11.5 and SSCs samples; no amplification was found for E8.5 and E10.5 samples. Lane 1: M: molecular weight marker; lane 2: SSCs; lane 3: E8.5; lane 4: E11.5; lane 5: E10.5. The same amount of cDNA from all samples was amplified for GAPDH gene expression.
Expression of Nanos2 protein in spermatogonial stem cells
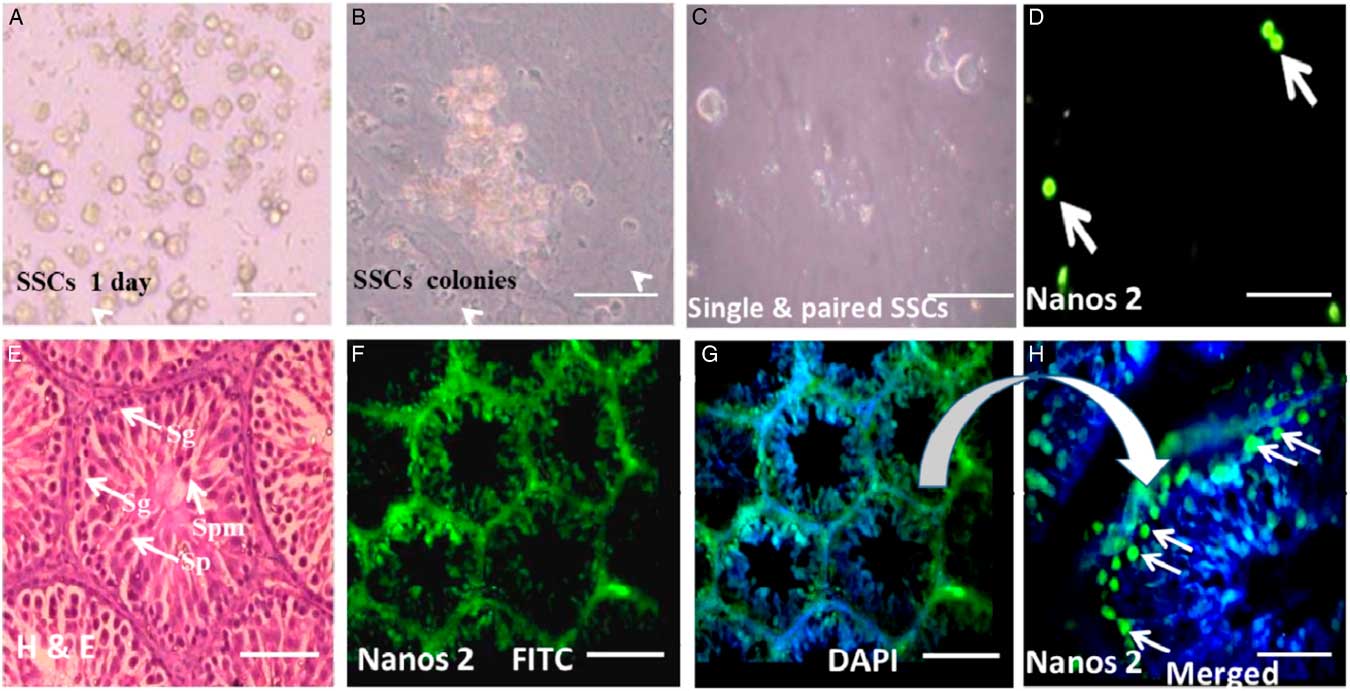
SSCs colonies were cultured and maintained over feeder layers of Sertoli cells (Fig. 4A, B). To examine expression of Nanos2 protein, a mixed population of SSCs was used to localize Nanos2 protein as in the standard protocol. As and Apr undifferentiated spermatogonial cells (Fig. 4C) were immunopositive for Nanos2 (Fig. 4D arrowhead). These observations support the hypothesis that undifferentiated SSCs expressing Nanos2 protein may belong to precursors of the A1 type of spermatogonial cells.
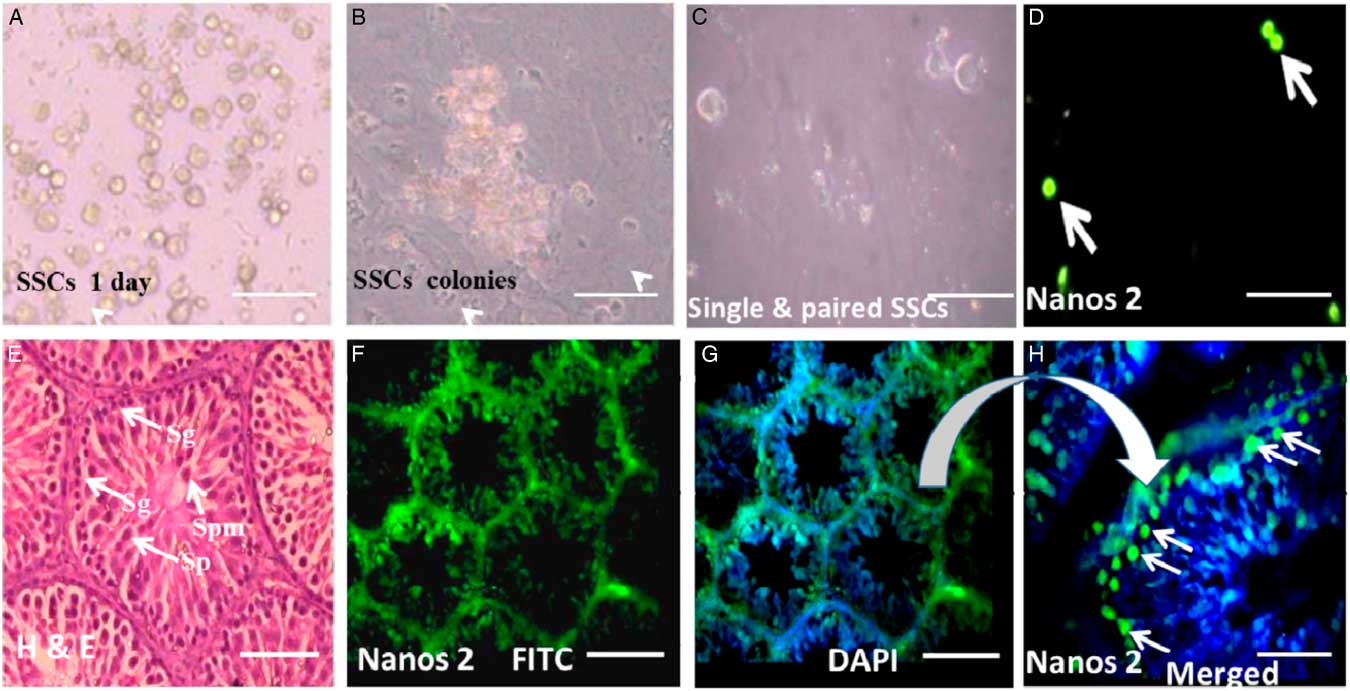
Figure 4 Expression of Nanos2 in spermatogonial stem cells (SSCs). (A) Isolated and cultured SSCs. (B) SSC colonies over a feeder layer. (C) Population of single and paired SSCs. (D) Positive immunoreactivity for Nanos2 protein (green signal, white arrow). (E) Hematoxylin and eosin staining showing a clear picture of different testicular cells (white arrow). (F) Nanos2 immunoreactivity in whole testicular tissue. (G) (H) Positive stain for Nanos2 in type A SSCs found in the periphery of testicular section. Sg: spermatogonial germ, Sp: spermatid, Spm: spermatozoa. Scale bars represent 100 µm.
To verify our immunocytochemistry results, Nanos2 protein localization was also determined in rat testes sections. Expression of Nanos2 protein was detected in spermatogonial germ (Sg) cells only (Fig. 4H, arrowhead). The spermatogonial germ cells (Sg) showed intense cytoplasmic staining for Nanos2 compared with other cell types from the adult testis. The control section representing spermatogenesis showed negative staining for Nanos2. The normal section of testis representing different stages of spermatogenesis was counterstained with classical haemotoxin and eosin (H&E) stain (Fig. 4E).
Flow cytometric analysis
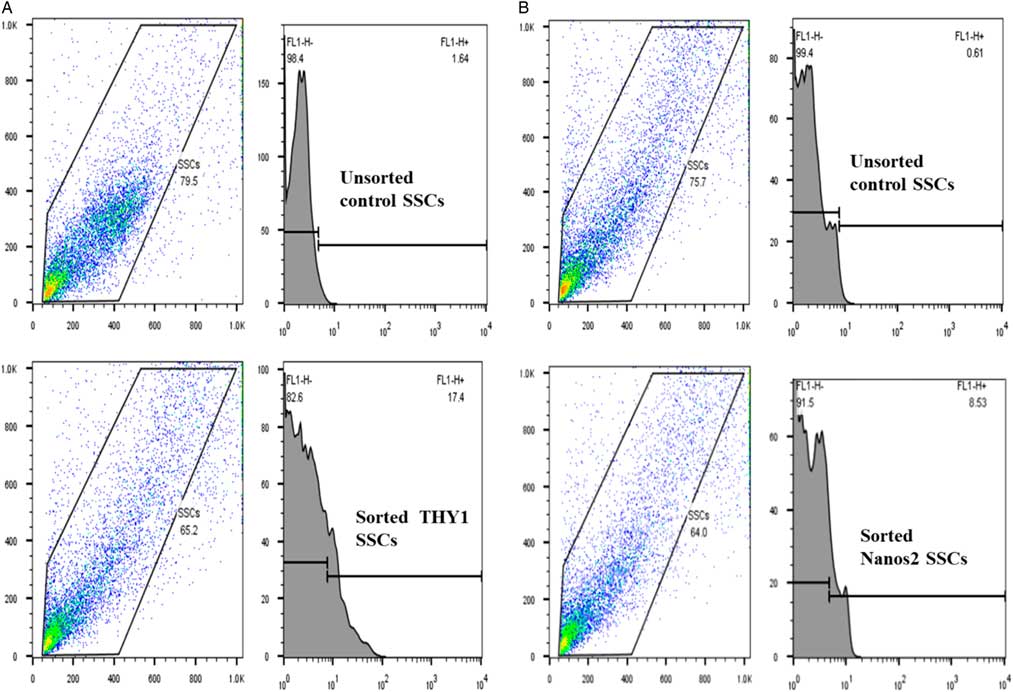
THY1 is a marker of SSCs, whereas Nanos2 is a specific marker for germ cells. Therefore, analysis of the expression of both markers in a sorted SSC population would help to determine the cell type. As shown in Fig. 5(A), flow cytometry analysis revealed that 17.4% of sorted rat testicular cells expressed the spermatogonial stem cell marker THY1, while 8.53% of cells expressed Nanos2 (Fig. 5B). The THY1-positive fraction was found to contain Nanos2-expressing spermatogonial cells, although there was a low population of Nanos2-expressing cells. The negative gate was established based on staining with isotype control IgG.
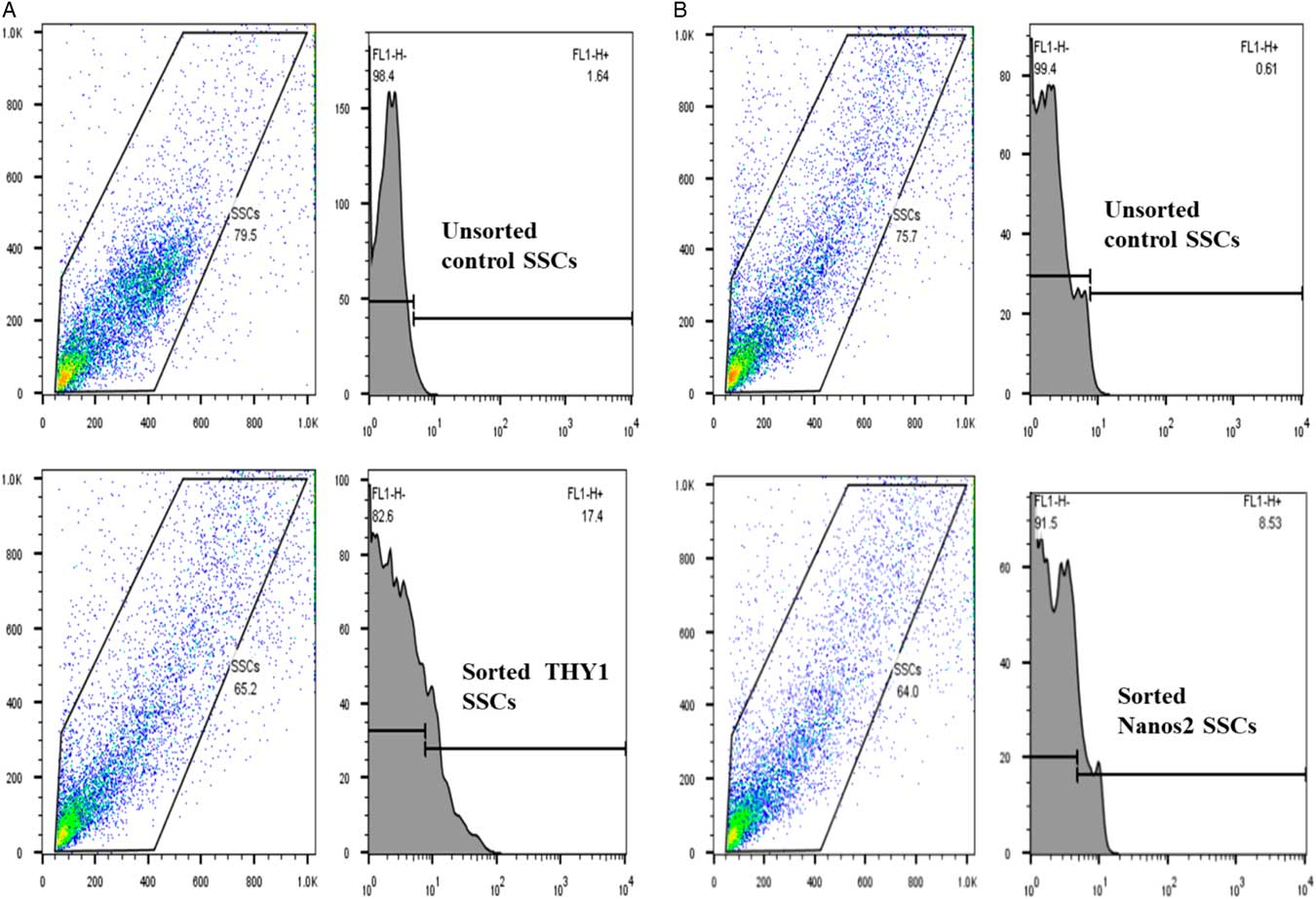
Figure 5 Fluorescence-activated cell sorting (FACS) of rat spermatogonial stem cells (SSCs) stained for THY1 or Nanos2. Rat SSCs were FACS sorted into negative (no stain for THY1 and Nanos2) and positive (stain for THY1 and Nanos2) fractions. (A) Unsorted population and sorted populations stained for THY1 (17.4%). (B) Unsorted population and sorted populations stained for Nanos2 (8.53%). Experiments were repeated at least three times.
Discussion
Nanos2 is a highly conserved RNA-binding protein that is involved in controlling germ-cell sexual differentiation during fetal development in many mammalian species (Suzuki et al., Reference Suzuki, Saba, Sada and Saga2010; Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003). It has also been thought to act as an anti-meiotic gene, because its expression in developing PGCs decides the fate of PGCs to become either male or female PGCs during fetal development (Suzuki and Saga, Reference Suzuki and Saga2008). Nanos2 plays a crucial role in translational regulation and sexual differentiation in PGCs as disruption of Nanos2 causes male infertility (Tsuda et al., Reference Tsuda, Sasaoka, Kiso, Abe, Haraguchi, Kobayashi and Saga2003; Kato et al., Reference Kato, Katsuki, Kokubo, Masuda and Saga2016). As very limited information is available about the Nanos2 gene during germ-cell development, especially in rat, expression pattern of Nanos2 in developing PGCs during different embryonic days (E8.5, E10.5 and E11.5), SSCs and testes was examined and its different expression patterns were found. In this study, there was no expression of Nanos2 around 8.5 and 10.5 days post coitum (d.p.c.), but its expression at 11.5 d.p.c. suggest that Nanos2 is a stage-specific protein during the prenatal period and may also be involved in the suppression of translation in somatic cells, because genetic studies suggest that Nanos2 acts as a suppressor of meiosis in male fetal germ cells (Suzuki and Saga, Reference Suzuki and Saga2008; Kato et al., Reference Kato, Katsuki, Kokubo, Masuda and Saga2016).
SSCs were sorted using flow cytometry and spermatogonial stem/progenitor cell markers: sorted cells were positive for both Nanos2 and THY1. These observations further strengthened our previous data that undifferentiated spermatogonial cells from mature rat testes were SSCs. By analysis of the flow cytometry data, we found that Nanos2-expressing cells were present only within the THY1-positive spermatogonial cell sub-populations and indicated that Nanos2 is a gene that is expressed in SSCs. Studies have also shown that loss of Nanos2 in SSCs by conditional deletion led to the loss of spermatogonia stem cells (Sada et al., Reference Sada, Suzuki, Suzuki and Saga2009; Suzuki et al., Reference Suzuki, Tsuda and Saga2007). Sub-populations of SSCs do not enter into meiosis and retain stem cell features, as demonstrated by their ability to complete spermatogenesis when transplanted into adult testes (Ohta et al., Reference Ohta, Wakayama and Nishimune2004). It has also been suggested that modulation of Nanos2 and Stra8 is essential for meiotic entry of differentiating spermatogonia in vitro (Pellegrini et al., Reference Pellegrini, Filipponi, Gori, Barrios, Lolicato, Grimaldi, Rossi, Jannini, Geremia and Dolci2008). In addition, it has been demonstrated that AtRA, a derivative of RA, was also able to downregulate Nanos2 at both the mRNA and protein levels in male PGCs (12.5 d.p.c.) as well as in undifferentiated spermatogonia (Pellegrini et al., Reference Pellegrini, Filipponi, Gori, Barrios, Lolicato, Grimaldi, Rossi, Jannini, Geremia and Dolci2008; Bowles et al., Reference Bowles, Knight, Smith, Wilhelm, Richman, Mamiya, Yahiro, Chawengsaksophak, Wilson, Rossant, Hamada and Koopman2006). Hence, it can hypothesized that Nanos2 expresses in proliferating PGCs (onwards E11.5) and when the meiotic window is active, then it is re-expressed in the male only postnatally in undifferentiating SSCs (As and Apr) for maintaining the stemness of SSCs.
In western blot analysis, a band of 18 kDa of Nanos2 protein was identified only in fetal gonad (E11.5) and SSCs samples, while there was no expression in E8.5 and E10.5 embryos. To further verify this result, we checked for expression of stem cell-specific marker antibody THY1 in all samples and found that THY1 is only expressed in SSCs samples. This expression pattern indicated that Nanos2 protein expression is embryonic stage specific and is also restricted to a small number of spermatogonial cells (As and Apr). Immunohistochemical and immunocytofluorescence results showed that the expression pattern of Nanos2 protein in rat SSCs diverged from that of germ cells. Immunocytofluorescence staining patterns showed intense cytoplasmic staining of Nanos2 in PGCs at E11.5 and in a small number of SSCs (As and Apr). These data suggested abundant expression of Nanos2 gene during PGCs development, especially when extended to the genital ridge and that its expression continued in a subpopulation of SSCs in matured rat testes, indicating their role in PGCs development and in SSCs. These observations were also supported by RT-PCR, from which a similar expression pattern for Nanos2 was seen in PGCs (E11.5) as well as in SSCs. Consistent with previous reports showing Nanos2 expression at E12.5 and its role in repressing meiotic fate (Suzuki and Saga, Reference Suzuki and Saga2008; Saba et al., Reference Saba, Kato and Saga2014).
In conclusion, the present study revealed the expression pattern of Nanos2 gene in PGCs during different embryonic developmental days (E8.5, E10.5 and E11.5) and in SSCs. The expression profile of Nanos2 gene in PGCs strongly suggested that Nanos2 expression is stage specific as its expression started around E11.5 and continued up to SSCs (As and Apr). Our study indicated that Nanos2 may potentially be involved in the development and differentiation of PGCs during embryonic developmental days may also play a role in self-renewal and meiotic commitment to SSCs.
Financial support
The funding for this research was provided by the Science and Engineering Research Board (SERB), New Delhi under the Young Scientist Scheme (YSS/2014/00321).
Conflict of interest
The authors have no conflict to declare.
Ethical standards
The animal experimental work was carried out as per the regulations provided by the Animal Ethical Committee of the Banaras Hindu University, Varanasi. This study was approved by Institutional Animal Ethical Committee of the University (vide letter no. Dean/2015/CAEC/1517 dated 21/12/2015).