Introduction
Developmental arrest often occurs at different stages of mammalian preimplantation embryo development in vitro. For example, mouse fertilized eggs arrested at the 2-cell stage cannot divide after the first division (Lawitts & Biggers, Reference Lawitts and Biggers1991). The early stages of embryonic development are closely related to the maternal instructions loaded into the oocyte in the form of mRNAs and proteins (Farley & Ryder, Reference Farley and Ryder2008; Abrams & Mullins, Reference Abrams and Mullins2009). These maternal molecules ultimately induce zygotic genome activation (ZGA) in the oocyte via a series of regulatory steps. Zygotic gene products eventually replace the maternally encoded products after ZGA. Mouse embryonic ZGA begins at the 2-cell stage, and peaks at the 2-cell to 4-cell embryo stages (Hamatani et al., Reference Hamatani, Carter, Sharov and Ko2004). Therefore, most researchers hypothesize that early in vitro embryonic developmental arrest is related to the process of ZGA (Bettegowda et al., Reference Bettegowda, Lee and Smith2008).
After fertilization, a large proportion of maternally derived mRNAs are eliminated via two distinct programmes (Bashirullah et al., Reference Bashirullah, Halsell, Cooperstock, Kloc, Karaiskakis, Fisher, Fu, Hamilton, Etkin and Lipshitz1999; Tadros & Lipshitz, Reference Tadros and Lipshitz2009). Firstly, maternally encoded products initiate the destruction of maternal mRNAs, then microRNAs (miRNAs) act on target maternal mRNA to destroy them. miRNAs are small 19–25 nucleotide (nt) single-stranded non-coding RNAs that can post-transcriptionally regulate gene expression (Bushati & Cohen, Reference Bushati and Cohen2007). Mature miRNAs act on their target mRNAs and lead to deadenylation, degradation and inhibition of mRNA expression via formation of the RNA-induced silencing complex (Wu et al., Reference Wu, Fan and Belasco2006). miRNAs represent one of the most-studied mechanisms of post-transcriptional control, and are thus hypothesized to be actively involved in preimplantation embryo developmental arrest.
It has been demonstrated that miRNAs can modulate gene expression in the early stages of development by inhibition of mRNA transcription (Lee et al., Reference Lee, Feinbaum and Ambros1993). Expression of zygotic miRNAs can significantly increase the efficiency of removal of maternal mRNAs, especially in zebrafish, Xenopus and Drosophila embryos (Giraldez et al., Reference Giraldez, Mishima, Rihel, Grocock, Van Dongen, Inoue, Enright and Schier2006; Bushati et al., Reference Bushati, Stark, Brennecke and Cohen2008; Lund et al., Reference Lund, Liu, Hartley, Sheets and Dahlberg2009). miR-430 and miR-427 are expressed abundantly in zebrafish and Xenopus during ZGA (Giraldez et al., Reference Giraldez, Cinalli, Glasner, Enright, Thomson, Baskerville, Hammond, Bartel and Schier2005; Watanabe et al., Reference Watanabe, Imai and Minami2008; Rosa et al., Reference Rosa, Spagnoli and Brivanlou2009). Knockout of miR-430 reduces the rate of degradation of several hundred maternal mRNAs (Giraldez et al., Reference Giraldez, Mishima, Rihel, Grocock, Van Dongen, Inoue, Enright and Schier2006). In addition, the generation of a miRNA cluster knockout in Drosophila revealed that zygotic expression of the miR-309 cluster directs the degradation of a subset of maternal mRNAs during ZGA (De Renzis et al., Reference De Renzis, Elemento, Tavazoie and Wieschaus2007). Other reports have demonstrated that the miRNAs lin-4 and let-7 can degrade and inhibit the expression of early transcripts Caenorhabditis elegans, allowing the subsequent embryonic developmental stages to progress smoothly (Reinhart et al., Reference Reinhart, Slack, Basson, Pasquinelli, Bettinger, Rougvie, Horvitz and Ruvkun2000; Abbott et al., Reference Abbott, Alvarez-Saavedra, Miska, Lau, Bartel, Horvitz and Ambros2005). Some mRNAs targeted by miRNAs are also transcribed during ZGA and, in this case, miRNAs can reduce the activation of these zygotic mRNAs and also regulate their steady-state expression levels. This situation indicates that miRNAs can control the degradation rate of target mRNAs to maintain homeostasis (Giraldez et al., Reference Giraldez, Mishima, Rihel, Grocock, Van Dongen, Inoue, Enright and Schier2006; Shkumatava et al., Reference Shkumatava, Stark, Sive and Bartel2009).
We speculate that miRNAs can regulate ZGA by regulating their target mRNAs, and that miRNA expression levels are closely related to embryonic developmental arrest. It has been confirmed that miRNAs are expressed in mature mouse eggs and early embryos (Bartel, Reference Bartel2004; Alizadeh et al., Reference Alizadeh, Kageyama and Aoki2005), however there are no reports of miRNA-mediated regulation in mouse embryos. Expression of miRNAs is downregulated by approximately 60% between the 1- and 2-cell stages in mouse embryos, and miRNA expression increases approximately 2.2-fold between the 2-cell stage and 4-cell stage, a finding that suggested that de novo synthesis of miRNAs occurs after ZGA (Tang et al., Reference Tang, Kaneda, O'Carroll, Hajkova, Barton, Sun, Lee, Tarakhovsky, Lao and Surani2007). However, the precise qualitative and quantitative changes in miRNA expression between the mouse 2-cell and 4-cell embryo stages are still unknown. In this study, we used a high-throughput microarray assay to identify the differentially expressed miRNAs in 2-cell and 4-cell ICR mouse embryos, and validated the reliability of the results by quantitative RT-PCR.
Materials and methods
Animals
ICR mice were fed ad libitum with a standard diet and maintained in a controlled 20–22°C environment at 50–70% humidity under a 12 h/12 h light/dark cycle, in accordance with the Animal Research Committee Guidelines of Nanjing Medical University.
Embryo collection
First, 6- to 8-week-old female ICR mice were superovulated by intraperitoneal injection of 10 IU pregnant mare serum gonadotrophin (PMSG, Sansheng Pharmaceuticals, Ningbo, China), followed by 10 IU human chorionic gonadotrophin (hCG, Sansheng Pharmaceuticals, Ningbo, China) 48 h later. The 2-cell embryos were collected from superovulated ICR female mice mated with ICR males and were flushed from the reproductive tract at 43–45 h post-hCG in G-MOPSTM (VitroLife, Gothenburg, Sweden). All good quality embryos were selected for further investigation. Half of the total number of 2-cell embryos were collected into a microcentrifuge tube that contained 10 μl RNALater (Qiagen, Germany) and stored at –80°C until use. The remaining embryos were transferred into G-2 culture media (VitroLife) that contained 10% synthetic serum supplement and were incubated at 37°C in a humidified 6% CO2/air atmosphere until the 4-cell embryo stage, the embryos were then collected in RNALater (Ling et al., Reference Lian, Liu, Liu, Jin, Sun, Li, Xia and Gao2009; Zhang et al., Reference Zeng, Baldwin and Schultz2009).
miRNA microarray
RNA extraction, amplification and labelling total RNA was extracted from the embryos using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the miRNeasy mini kit (Qiagen) according to the manufacturer's instructions. RNA quality and quantity were assessed using a NanoDrop spectrophotometer (ND-1000, NanoDrop Technologies, USA) and the RNA integrity was determined by gel electrophoresis. The microarray assay was created by a commercial service provider (Kangchen Bio-tech, Shanghai, China). RNA (20 ng) was amplified using the NCode miRNA Amplification System (Invitrogen) according to the manufacturer's instructions, and the amplified sense RNA was labelled using the miRCURY™ Hy3™/Hy5™ Power Labelling Kit (Exiqon, Vedbaek, Denmark) according to the manufacturer's guidelines.
miRNA microarray hybridization
The Hy3TM-labelled samples were hybridized onto the miRCURY™ LNA Array (v.14.0; Exiqon). Briefly, 25 μl Hy3TM-labelled RNA was mixed with 25 μl hybridization buffer, denatured for 2 min at 95°C, incubated on ice for 2 min and then hybridized to the microarray for 16–20 h at 56°C in a 12-Bay Hybridization System (Nimblegen Systems, Inc., Madison, WI, USA). After hybridization, the slides were washed several times using the Wash Buffer Kit (Exiqon), dried by centrifugation for 5 min at 400 rpm and scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA).
Microarray data analysis
The scanned images were imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and the miRNAs whose intensities were >50 in every sample were selected to calculate the normalization factor. The expression data were normalized using median normalization, and differentially expressed miRNAs were identified using fold change filtering. Hierarchical clustering was performed using MEV software (ver 4.6, TIGR).
miRNA real-time PCR
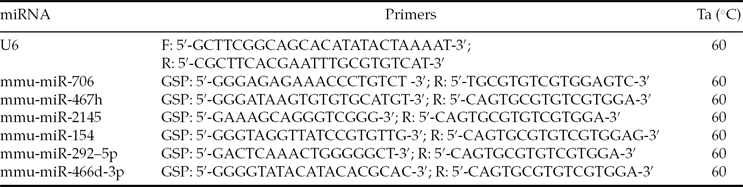
Total RNA was extracted from the embryos using TRIzol (Invitrogen) and the miRNeasy mini kit (Qiagen). Single-stranded cDNA was synthesized by reverse transcription in reactions that contained 50 ng total RNA, 0.3 μl mmu-miRNA reverse primer (Table 1), 2 μl 10× buffer, 2 μl dNTPs (2.5 mmol/l), 0.3 μl M-MLV reverse transcriptase, 0.3 μl RNasin and RNase-free H2O to a total volume of 20 μl. The reaction conditions were 16°C for 30 min, 42°C for 40 min and 85°C for 5 min.
Table 1 Reverse-transcription PCR primer sequences

The 25 μl real-time PCR reaction mixtures contained 1 μl cDNA, 2.5 μl 10× buffer, 2.5 μl dNTPs (2.5 mmol/l), 1 IU Taq polymerase, 1 μl SYBR-Green and 2 μl each of the reverse and forward primers. The reactions were amplified using the Rotor-Gene 3000 Real-Time PCR machine at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, 72°C for 20 s and 78°C for 20 s. The primer sequences are listed in Table 2.
Table 2 Real-time RT-PCR primer sequences

Target prediction analysis
TargetScan 6.0 (http://www.targetscan.org/) was used to perform bioinformatic-based target prediction analysis.
Results
Embryo collection
Approximately 150 ICR female mice were used for embryo collection. The mating success rate was about 70%; and 600 and 300 2-cell and 4-cell embryos were collected, respectively.
miRNA microarray
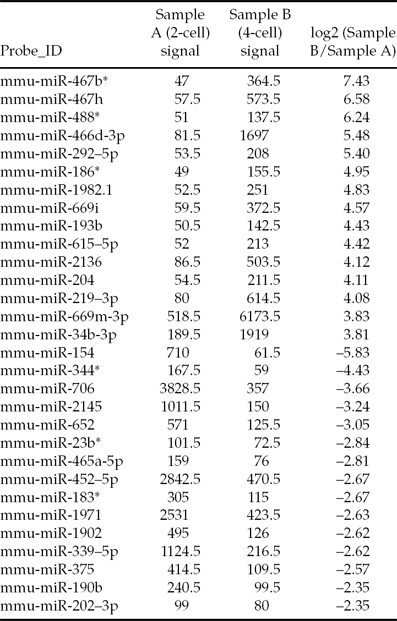
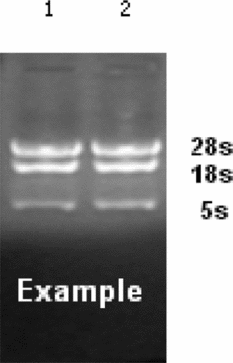
Due to the small number of samples, which lead to the isolation of a small quantity of RNA, we confirmed the quality of the total RNA before the microarray experiment. The total RNA was good quality, with little degradation observed (Fig. 1). The miRNA array contained over 1891 capture probes, and miRNAs with signal intensities over >50 in each sample were chosen for calculation of the normalization factor. Patterns of differentially expressed miRNA were observed (Fig. 2). Analysis of the miRNA microarray data revealed that a total of 192 miRNAs were significantly differentially expressed (>2-fold) in 4-cell embryos compared with 2-cell embryos; 122 of which were upregulated and 70 of which were downregulated. Table 3 presents some of the differentially expressed miRNAs in 2-cell and 4-cell embryos.
Table 3 Partial list of the miRNAs differentially expressed in 2-cell and 4-cell mouse embryos

The signal values are the foreground intensity for each miRNA.

Figure 1 Representative analysis of total RNA integrity using 1.5% formaldehyde denaturing agarose gel electrophoresis.

Figure 2 Representative fluorescence images of the miRNA microarray chips. Left panel: 2-cell embryos; right panel:4-cell embryos.
miRNA real-time PCR
In order to verify the microarray results, we selected six miRNAs that were highly differentially expressed for real-time PCR analysis including mmu-miR-467h, mmu-miR-466d-3p and mmu-miR-292–5p, which were upregulated in 4-cell embryos, and mmu-miR-154, mmu-miR-2145 and mmu-miR-706, which were downregulated in 4-cell embryos. U6 was used as an internal control to calculate the relative miRNA expression levels. The real-time qRT-PCR expression data for each of the six miRNAs selected were comparable with the microarray data (Fig. 3).

Figure 3 Validation of the microarray miRNA expression results by real-time qRT-PCR. Data are normalized to U6 as an endogenous control and expressed as the fold change in 4-cell mouse embryos relative to 2-cell mouse embryos.
Target prediction analysis
We selected the most differentially expressed miRNA, mmu-mir-154, which was downregulated in 4-cell embryos, and predicted its potential target genes. Cdca4 and Tcf12 were identified as putative target genes of mmu-mir-154.
Discussion
Many reports have been published on the relationship between miRNAs and embryonic development in recent years. These studies suggest that miRNAs are expressed in both mouse embryos and embryonic stem cells, and suggest that miRNAs play an important role in mesoderm formation and organ differentiation (Houbaviy et al., Reference Houbaviy, Murray and Sharp2003; Takada et al., Reference Takada, Berezikov, Yamashita, Lagos-Quintana, Kloosterman, Enomoto, Hatanaka, Fujiwara, Watanabe, Soda, Choi, Plasterk, Cuppen and Mano2006; Hong Chen et al., 2010). However, little information is known about the role of miRNAs in mouse embryonic developmental arrest. In this study, we collected 2-cell and 4-cell mouse embryos and extracted total RNA from these embryos. Using a miRNA microarray 192 miRNAs were found to be differentially expressed in 2-cell and 4-cell embryos, 122 of which were upregulated and 70 of which were downregulated in 4-cell embryos. The microarray results were confirmed using real-time qRT-PCR for six miRNAs (mmu-miR-467h, mmu-miR-466d-3p, mmu-miR-292–5p, mmu-miR-154, mmu-miR-2145, and mmu-miR-706). We also identified Cdca4 and Tcf12 as putative miR-154 target genes using target prediction analysis. Cdca4 and Tcf12 are a cell cycle protein and transcription factor, respectively, that are both involved in the regulation of embryonic development (Zeng et al., Reference Zeng, Baldwin and Schultz2004; Cui et al., Reference Cui, Li, Jeong, Jun and Kim2006). Therefore, it is possible that miR-154 plays an important role in embryonic developmental arrest by regulating the cell cycle protein Cdca4 and transcription factor Tcf12.
In this study, expression of mir-466d-3p was upregulated in 4-cell embryos, compared with 2-cell embryos. Previous studies have shown that expression of miR-466d-3p decreased in the liver of C57BL/6 female mice after parasite infection, however the specific function and regulation mechanisms of mir-466d-3p remain unclear (Delić et al., Reference Delić, Dkhil, Al-Quraishy and Wunderlich2011). Redell et al. (Reference Redell, Liu and Dash2009) reported that expression of miR-292–5p increased in the mouse hippocampus 24 h after brain injury. Bioinformatic analysis suggests that miR-292–5p may regulate target genes with functions in a number of biological processes, including signal transduction, transcriptional regulation, cell proliferation and differentiation (Redell et al., Reference Redell, Liu and Dash2009). Early embryonic development is dependent on a number of biological processes, including cell cycle, apoptosis and transcriptional regulation (Zeng et al., Reference Zeng, Baldwin and Schultz2004; Cui et al., Reference Cui, Li, Jeong, Jun and Kim2006). In this study, expression of miR-292–5p increased between the 2-cell stage and the 4-cell stage, a finding that suggested that miR-292–5p may modulate embryonic development by regulating signal transduction, transcription, cell proliferation and differentiation.
Apoptosis plays an important role in embryonic development. Mmu-miR-154 and mmu-miR-706 were both downregulated at the 4-cell stage, and previous reports have confirmed that mmu-miR-706 can inhibit apoptosis by reducing expression of the apoptotic factors caspase-3 and caspase-9 (Lian et al., Reference Lian, Liu, Liu, Jin, Sun, Li, Xia and Gao2010). Wang et al. (Reference Wang, Zhang, Li, Yang, Pei, Xu, Wang, Zhou and Li2009) demonstrated that the treatment of pregnant mice with ethanol significantly increased the fetal malformation rate and significantly decreased expression of miR-154 in the fetal mouse brain. These results suggest that miR-154 is expressed in mouse embryonic tissues, and may be associated with embryonic development. Accordingly, Su et al. (Reference Su, Wang, Qian and Deng2010) analyzed miR-154 using bioinformatic techniques, and found that the expression patterns of miR-154 target genes and embryonic development-related transcription factor target genes were similar, and that both groups of genes could be classified into the same subsets by cluster analysis. Based on these results, we predict that miR-706 and miR-154 are associated with embryonic development and may be closely related to early embryonic developmental arrest. The function of miR-706 and miR-154 and the mechanisms by which they modulate embryonic developmental arrest require further study.
The results of this study have theoretical significance for understanding the molecular mechanisms of early embryonic developmental arrest and the biological function of miRNAs, and have potential for application in the field of reproductive medicine.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81100420, 81270701), the Natural Science Foundation of Jiangsu Province (BK2012520), the Foundation of Nanjing Medical University (10NJMUZ035) and the Nanjing Medical Science and Technique Development Foundation.