Introduction
Digitaria spp. (crabgrasses) are summer annual grass weeds that commonly infest both turfgrass and landscape environments, as well as all crops (Lepschi and Macfarlane Reference Lepschi and Macfarlane1997). Southern crabgrass [Digitaria ciliaris (Retz.) Koeler] is a C4 herbaceous, monocotyledonous annual in the Poaceae family (Bantilan et al. Reference Bantilan, Palada and Harwood1974; Murphy et al. Reference Murphy, Colvin, Dickens, Everest, Hall and McCarty2014; Shetty et al. Reference Shetty, Sivakumar and Ram1982; Watson and Dallwitz Reference Watson and Dallwitz1992). Native to Asia, D. ciliaris is now found in tropical and subtropical areas and distributed throughout the midwestern and southern United States (Gleason and Cronquist Reference Gleason and Cronquist1991). Due to its prodigious seed production and vigorous growth rate, this grass competes with desirable crops. Furthermore, its bunch-type growth and light-green color reduce turfgrass uniformity, and it also has allelopathic properties that can act against crops, other weeds, nitrifying bacteria, and Rhizobium (Ito et al. Reference Ito, Kobayashi and Ueki1987; Ito and Ichikawa Reference Ito and Ichikawa1994).
In modern agriculture, the control of grass weed species, especially the Poaceae family, largely depends on applications of selective herbicides (Délye et al. Reference Délye, Pernin and Michel2011). Acetyl-coenzyme A carboxylase (ACCs or ACCase; EC 6.4.1.2) inhibitors (often referred to as graminicides) are a unique herbicide mechanism of action that is primarily active on grasses. These herbicides are predominantly used as a selective graminicide for POST grass weed control in broadleaf crops; however, certain selectivity exists, allowing for some grass control in turfgrass (Délye Reference Délye2005; Kaundun Reference Kaundun2010; Powles and Yu Reference Powles and Yu2010). Based on chemical structure, ACCase inhibitors (WSSA Group 1 herbicide) are broadly classified into three distinct chemical families, namely, aryloxyphenoxypropionates (APPs or FOPs), cyclohexanediones (CHDs or DIMs), and phenyl-pyrazoline (DEN) (Hochberg et al. Reference Hochberg, Sibony and Rubin2009; Hofer et al. Reference Hofer, Muehlebach, Hole and Zoschke2006; Liu et al. Reference Liu, Harrison, Chalupska, Gornicki, O’Donnell, Adkins, Haselkorn and Williams2007; Powles and Yu Reference Powles and Yu2010; Tang et al. Reference Tang, Zhou, Chen and Zhou2014). The herbicidal action of these herbicides mainly depends on the selective binding of the carboxylase transferase (CT) domain of plastidic ACCase isoforms (Nikolskaya et al. Reference Nikolskaya, Zagnitko, Tevzadze, Haselkorn and Gornicki1999).
Pinoxaden is a selective grass-active compound discovered by Syngenta Crop Protection (Basel, Switzerland), a relatively new chemical, and the only herbicide in the DEN family (Hofer et al. Reference Hofer, Muehlebach, Hole and Zoschke2006; Petit et al. Reference Petit, Bay, Pernin and Délye2010; Senseman Reference Senseman2007; Tang et al. Reference Tang, Zhou, Chen and Zhou2014). In 2006, pinoxaden was globally introduced as AXIAL® for the control of annual grass weeds in cereal crops. At recommended rates of 30 to 60 g ha−1, AXIAL® is active against a wide range of important grass weed species such as blackgrass (Alopecurus myosuroides Huds.), silky windgrass [Apera spica-venti (L.) Beauv.], Avena spp., Lolium spp., Phalaris spp., and Setaria spp. Due to its effective POST activity against a broad spectrum of grass weeds, pinoxaden was originally labeled for annual grass weed control in cereal crops, including wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) (Hofer et al. Reference Hofer, Muehlebach, Hole and Zoschke2006; Kuk et al. Reference Kuk, Burgos and Scott2008; Locke et al. Reference Locke, Reddy and Zablotowicz2002; Porter et al. Reference Porter, Kopec and Hofer2005). Like DIMs and FOPs, pinoxaden interacts with the CT domain of homomeric ACCase enzyme in grass chloroplasts (Hofer et al. Reference Hofer, Muehlebach, Hole and Zoschke2006; Muehlebach et al. Reference Muehlebach, Boeger, Cederbaum, Cornes, Friedmann, Glock, Niderman, Stoller and Wagner2009; Yu et al. Reference Yu, Kim and Tong2010).
Resistance to ACCase-inhibiting herbicides involves a non–target site resistance (NTSR) mechanism, changing the metabolic activity of the protein, and/or a target site–based resistance (TSR) mechanism, altering the target-site protein structure at the herbicide’s binding site, thus rendering it less sensitive to herbicidal activity (Délye Reference Délye2005; Devine Reference Devine1997). Resistance to the grass-specific ACCase-inhibiting herbicides depends on an NTSR mechanism, which is initiated without structural alteration of a target-site protein (Délye et al. Reference Délye, Pernin and Michel2011; Yuan et al. Reference Yuan, Tranel and Stewart2006). NTSR can be, but is not limited to, increased protein expression, increased protein abundance, posttranslational modification of the existing protein, increased herbicide metabolism, reduction of herbicide diffusion rate into the plant, repolarization of plasma membrane electrogenic potential (Em), or reduced rate of herbicide translocation (Bradley et al. Reference Bradley, Wu, Hatzios and Hagood2001; Délye et al. Reference Délye, Jasieniuk and LeCorre2013). Researchers have reported that NTSR, including enhanced degradation, has been a possible mechanism of resistance to ACCase-inhibiting herbicides in the resistant biotypes of A. myosuroides, Lolium spp., wild oat (Avena fatua L.), large crabgrass [Digitaria sanguinalis (L.) Scop.], as compared with susceptible biotypes (Cocker et al. Reference Cocker, Coleman, Blair, Clarke and Moss2000; Cummins et al. Reference Cummins, Moss, Cole and Edwards1997; DePrado et al. Reference DePrado, Osuna, Heredia and DePrado2005; Hidayat and Preston Reference Hidayat and Preston1997; Letouzé and Gasquez Reference Letouzé and Gasquez2003; Mendez and DePrado Reference Mendez and DePrado1996; Petit et al. Reference Petit, Bay, Pernin and Délye2010; Preston and Powles Reference Preston and Powles1998).
TSR is most commonly a single point mutation in the plastidic ACCase gene that produces an amino acid alteration and reduces sensitivity of the ACCase enzyme to these herbicide groups (Preston and Mallory-Smith Reference Preston, Mallory-Smith, Powles and Shaner2001; Yuan et al. Reference Yuan, Tranel and Stewart2006). To date, eight conserved amino acid substitutions at seven positions in the CT domain of the ACCase gene have been documented for the Group 1 herbicide resistance in a variety of grass weed species (Beckie and Tardif Reference Beckie and Tardif2012). The different known amino acid alterations, Gln-1756-Glu, Ile-1781-Leu, Thr-1805-Ser, Lys-1930-Arg, Trp-1999-Cys, Trp-2027-Cys, Ile-2041-Asn/Val, Asp-2078-Gly, Cys-2088-Arg, and Gly-2096-Ala, can confer different patterns of resistance among ACCase inhibitors (Beckie and Tardif Reference Beckie and Tardif2012; Collavo et al. Reference Collavo, Panozzo, Lucchesi, Scarabel and Sattin2011; Délye Reference Délye2005; Délye et al. Reference Délye, Calmès and Matéjicek2002a, Reference Délye, Matéjicek and Gasquez2002b, Reference Délye, Wang and Darmency2000c, Reference Délye, Pernin and Michel2011; Gherekhloo et al. Reference Gherekhloo, Osuna and DePrado2012; Hochberg et al. Reference Hochberg, Sibony and Rubin2009; Kaundun Reference Kaundun2010; Kaundun et al. Reference Kaundun, Bailly, Dale, Hutchings and McIndoe2013; Liu et al. Reference Liu, Harrison, Chalupska, Gornicki, O’Donnell, Adkins, Haselkorn and Williams2007; Petit et al. Reference Petit, Bay, Pernin and Délye2010; White et al. Reference White, Moss and Karp2005; Yu et al. Reference Yu, Collavo, Zheng, Owen, Sattin and Powles2007, Reference Yu, Ahmad-Hamdani, Han, Christoffers and Powles2013; Zhang and Powles Reference Zhang and Powles2006a, Reference Zhang and Powles2006b). While the amino acid alterations Ile-1781-Leu/Val, Asn-2078-Gly, and Cys-2088-Arg confer resistance to all ACCase inhibitors, the amino acid alterations Trp-1999-Cys/Leu, Trp-2027-Cys, Ile-2041-Asn/Val, or Gly- 2096-Ala endow resistance to one or more FOPs but not to DIM or DEN herbicides families (Collavo et al. Reference Collavo, Panozzo, Lucchesi, Scarabel and Sattin2011; Powles and Yu Reference Powles and Yu2010).
Two biotypes of D. ciliaris with suspected resistance to DIM and FOP herbicide groups were identified in centipedegrass [Eremochloa ophiuroides (Munro) Hack.] sod production fields in Georgia, USA (Yu et al. Reference Yu, McCullough and Czarnota2017). Sethoxydim applied at 315 and 945 g ha−1 did not control these D. ciliaris biotypes >20% in field experiments. Yu et al. (Reference Yu, McCullough and Czarnota2017) also reported that the shoot biomass production for D. ciliaris treated with sethoxydim, based on dose–response data, was >64 times higher in both resistant populations than in the susceptible population. Resistance to pinoxaden has already been identified in Italian ryegrass [Lolium perenne L. ssp. multiflorum (Lam.) Husnot], A. myosuroides, A. fatua, and Japanese foxtail (Alopecurus japonicus Steudel) (Kaundun Reference Kaundun2010; Kuk et al. Reference Kuk, Burgos and Scott2008; Martins et al. Reference Martins, Sánchez-Olguín, Perez-Jones, Hulting and Mallory-Smith2014; Mohamed et al. Reference Mohamed, Li, You and Li2012; Petit et al. Reference Petit, Bay, Pernin and Délye2010; Yu et al. Reference Yu, Kim and Tong2010). Pinoxaden is now available for grass control in the U.S. turfgrass market, particularly for bermudagrass [Cynodon dactylon (L.) Pers.], zoysiagrass (Zoysia japonica Steud.), and St. Augustinegrass [Stenotaphrum secundatum (Walter) Kuntze.]. We hypothesized that the two populations of D. ciliaris previously confirmed sethoxydim resistant would be cross-resistant to pinoxaden. Further, based on the pattern of resistance to sethoxydim and other ACCase inhibitors, we hypothesized that these populations would possess a common mutation previously associated with ACCase herbicide resistance. The objectives of our research, therefore, were to evaluate the response of the two primarily sethoxydim-resistant biotypes of D. ciliaris to pinoxaden and determine whether a target-site mutation commonly associated with ACCase resistance is present.
Materials and Methods
Plant Materials and Growth Conditions
This research used two biotypes, R1 and R2, of D. ciliaris with previously confirmed resistance to sethoxydim and FOP herbicides (Yu et al. Reference Yu, McCullough and Czarnota2017) collected from two undisclosed fields of ‘TifBlair’ centipedegrass in Georgia. The fields were around 160 km apart in central Georgia; sethoxydim and FOP herbicides had been used annually for over two decades in these areas, and control failure was evident in these fields. The plants from both biotypes were uninjured by sethoxydim and FOP herbicides at a standard use rate ranging from 315 to 945 g ha−1 applied approximately 3 wk before collection. A separate susceptible (S) biotype of D. ciliaris was collected in Auburn, AL, from an area with no known history of exposure to ACCase inhibitors. Per an agreement with the landowners of the fields where the resistant types were identified, it was agreed that the location of the resistant types would not be reported in any form in the future. The collected plants were propagated separately in a greenhouse environment to increase seed lots for experiments. Seeds from mature plants were collected randomly by hand, then air-dried and stored in paper bags at 4 C until planted.
The research was conducted at the Auburn University Weed Science greenhouse (32.35°N, 85.29°W) in Auburn, AL. Three biotypes were seeded in separate plastic flats containing commercial potting soil and peat moss (2:1 v/v). The plastic flats were placed in a greenhouse set for 32/25 C (day/night) with no supplementary lighting. Plastic flats were overhead-irrigated three times daily (around 0.2 cm per cycle) to prevent moisture deficiencies. Four weeks later, single seedlings at the 3- to 4-leaf stage were transplanted individually into plastic pots (10 cm by 10 cm by 8.5 cm) filled with the surface horizon of a Marvyn sandy loam (fine-loamy, kaolinitic, thermic Typic Kanhapludults) with a pH of 6.5 and 1.1% organic matter. The potted seedlings were irrigated daily and fertilized weekly to promote growth. Plants reached a 1- to 2-tiller growth stage before treatments. Some plants were also allowed to produce flower and seed. All seeds from dehiscent inflorescences were cleaned, air-dried, and stored at 4 C until used in subsequent experiments.
Pinoxaden Response Evaluation
The responses of the three D. ciliaris biotypes were evaluated from a rate titration of pinoxaden (Axial®, Syngenta, Greensboro, NC) herbicide. Axial also contains the safener cloquintocet- mexyl at 25 g L−1 (2.45% w/w). Treatments were applied with a CO2-pressurized sprayer calibrated to deliver 280 L ha−1 from a handheld four-nozzle boom at 32 psi (TeeJet® TP8003VS nozzles with 25 cm spacing; Spraying Systems Company, Wheaton, IL). All treatments included a nonionic surfactant (Induce®, Helena Chemical, Collierville, TN) at 0.25% v/v. Pinoxaden at 0.1, 0.2, 0.4, 0.7, 1.5, 2.9, 5.9, 11.8, and 23.5 kg ha−1 was applied to both resistant and susceptible plants. Nontreated checks of the three biotypes were included as control treatments and sprayed with water. Plants were returned to the greenhouse after herbicide application, and irrigation was withheld for 24 h. Control data were collected at 14, 28, and 42 d after treatment (DAT) on a 0% to 100% scale in which 0% corresponded to no control and 100% corresponded to complete plant death or desiccation. Shoot tiller length was collected by measuring the length of the longest tiller from the base to the termination of the tiller at 42 DAT. The foliar weight of each plant was measured to determine the total aboveground biomass. The shoots, therefore, were clipped at the soil surface and were weighed using an analytical balance.
The greenhouse experimental design was conducted twice in time as a completely randomized factorial design. All treatments were replicated on three plants per biotype. Data analyses were performed using PROC GLM in SAS (v. 9.4, SAS Institute, Cary, NC). Differences between the data of the two experimental runs were not detected in the ANOVA at the 0.05 probability level, so the data were pooled over runs for subsequent analysis. Pinoxaden rates were log transformed to produce equal spacing among treatments before regression analysis. The nontreated control (0 kg ha−1) was transformed to −1.36 to maintain equal spacing among log treatment rates. Tiller length (cm) and aboveground biomass weight (g) were converted to percent relative to the nontreated plants, respectively. The nontreated mean of each biotype was used for conversion calculations to determine relative measures of each treatment, and a model was selected that characterized the relationship of the response curves with pinoxaden herbicide rate after plotting treatment means. All measurements relative to nontreated were used for the regression model. Percent control data were fit to an exponential growth model (Equation 1) with two parameters, and percent data of the tiller length and the aboveground biomass were fit to a sigmoidal equation with three parameters (Equation 2) in SigmaPlot v. 13 (Systat Software, London, UK):
In Equation 1, y is the control (%) of D. ciliaris biotype, x is the log-transformed pinoxaden herbicide rate (kg ha−1), and b is the y-intercept. In Equation 2, y is the length/weight (%) of D. ciliaris biotype, x is the log-transformed pinoxaden herbicide rate (kg ha−1), x 0 is the asymptote, and b is the y-intercept. The 95% confidence intervals (α = 0.05) for the estimates were calculated for regression model parameters. Regression equations were used to calculate inhibition values at 50% and 90% (referred to as I50 and I90 values) compared with those of the nontreated for each biotype, and pinoxaden I50 and I90 R/S values were determined by comparing each resistant biotype with the susceptible biotype.
Target Site–based Resistance
Experiments were conducted to explore the potential target-site mechanism commonly associated with ACCase-inhibiting herbicide sensitivity. RNA for the three D. ciliaris biotypes (R1, R2, and S) was isolated from leaf samples (approximately 0.1 g) using the TRIzol method (Trizol, Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The quality and quantity of total RNA were determined with gel electrophoresis, a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and a Qubit 2.0 fluorometer (Invitrogen, Life Technologies). Then, the high-quality RNA was converted to complementary DNA (cDNA) through a reverse transcriptase-polymerase chain reaction (RT-PCR) conversion using Proto Script II First Strand cDNA Synthesis Kit (New England Biolabs, Ipswich, MA). PCR techniques were employed with some variations previously described by Zhang and Powles (Reference Zhang and Powles2006a). Sections of the ACCase gene were amplified with forward and reverse primers produced using the NCBI design tool and were sequenced to evaluate potential known single-nucleotide polymorphisms (SNPs) for conferring herbicide resistance. The primers (listed in Table 1) were designed to amplify highly conserved regions of D. ciliaris, covering the known resistance-conferring mutation sites using ACCase gene sequences of barnyardgrass [Echinochloa crus-galli (L.) P. Beauv.] (NCBI accession number: KU198448).
Table 1. List of five primer pairs used in this study to detect single-nucleotide polymorphisms in Digitaria ciliaris.

PCR was conducted using plant cDNA amplification in a 25 μl volume. The total volume of each PCR reaction mixture contained 1X standard reaction buffer, 200 µM dNTP, 0.5 µM forward and reverse primers, 250 ng of cDNA, and 0.125 U Taq DNA polymerase (New England Biolabs, Ipswich, MA). The cycling program consisted of an activation step of 95 C for 30 s followed by 35 cycles of 20 s at 95 C; 1 min annealing at 58 to 62 C, depending on the primers used; and 1 min at 68 C, followed by a final extension step of 5 min at 68 C. The PCR product was visualized on a 1.3% agarose gel in Tris-acetate-EDTA buffer and 1% ethidium bromide solution. The specific band size of PCR product from the gel was extracted using the E.Z.N.A.® Gel DNA Extraction Kit. Each extracted PCR product was sequenced in forward and reverse directions at Eurofins Genomic (Louisville, KY). The sequence data of resistant and susceptible D. ciliaris biotypes were compared to determine if there was a nucleotide substitution. Nucleotide sequences predicted from the ACCase gene sequences of D. ciliaris were subjected to the nucleotide (blastn) and amino acid (blastx) searches using NCBI BLAST (National Center for Biotechnology Information Basic Local Alignment Search Tool) to identify nucleotide and transcribed amino acid positions, respectively. The nucleotide sequences for each D. ciliaris biotype then were further aligned and compared using CLC Genomics Workbench v. 6.5.2 (CLC Bio, Aarhus, Denmark).
PCR products generated with Taq polymerase were ligated into the pGEMT®Easy Vector (Promega, Fitchburg, WI) and transformed in Escherichia coli (JM109 High-Efficiency competent cells, Promega). According to the manufacturer’s instructions, a 100 µl aliquot of each bacterial suspension was plated on media containing LB broth, ampicillin, X-gal, and Isopropyl β-d-1-thiogalactopyranoside (IPTG). The white colonies with putative inserts were selected from the transformed plates and incubated at 37 C overnight. The plasmids were purified using QIAprep Spin Miniprep Kit (Qiagen N.V., Venlo, Netherlands). The plasmids containing ACCase putative mutated cDNA fragments were sequenced, analyzed, and compared with cDNA fragments of the susceptible biotype as discussed earlier. As the standard PCR-based resistance mechanism determination in the R2 biotype was unable to detect the amino acid substitution in the Ile-1781 codon, we carried out further studies to investigate the amino acid substitution using transcriptome analysis.
Transcriptome Profiling
Digitaria ciliaris is a suspected polyploid species with potentially multiple copies of plastidic ACCase in subgenomes. To fully account for all expressed plastidic ACCase, mRNA was sequenced using next-generation sequencing (NGS). The two resistant biotypes, R1 and R2, were sequenced in the Genomic Service Laboratory using Illumina HiSeq 2500 platform (Illumina, San Diego, CA) at the HudsonAlpha Institute for Biotechnology (Cummings Research Park, Huntsville, AL). Large quantities of data files were received, and the raw read qualities were checked using FastQC v. 0.11.1 (Andrews Reference Andrews2010). The reads were then processed using Trimmomatic v. 0.33 to remove adaptor contamination, unqualified reads, and sequences (Bolger et al. Reference Bolger, Lohse and Usadel2014). Again, the trimmed reads were qualified for high-throughput sequence data with FastQC. The sequence data were normalized with Trinity’s in silico read normalization (Grabherr et al. Reference Grabherr, Haas, Yassour, Levin, Thompson, Amit, Adiconis, Fan, Raychowdhury, Zeng, Chen, Mauceli, Hacohen, Gnirke and Rhind2011), with maximum coverage of 30 and k-mer of 25. Each biotype was assembled using three de novo transcriptome assemblers:Trinity 2014-04-13p1, Velvet 1.2.08_maxkmer101, and CLC Genomic workbench (Grabherr et al. Reference Grabherr, Haas, Yassour, Levin, Thompson, Amit, Adiconis, Fan, Raychowdhury, Zeng, Chen, Mauceli, Hacohen, Gnirke and Rhind2011; Zerbino and Birney Reference Zerbino and Birney2008). Trinity k-mer size was 25, Velvet k-mer size was 2 to 81 with a step size of 10, and CLC k-mer size was 14 to 64 with a step size of 5. N50s and contig length distributions of the assemblies were calculated for estimating the quality of the assembly with the script Count_fasta.pl. Consensus regions (contigs) bigger than 200 bp were considered from all assemblies.
All the assemblies were pooled into one merged assembly for each species individually. The merged assembly was processed using the EvidentialGene tr2aacds pipeline. Using the transcript fasta file from any of the transcript assemblers, this pipeline can produce the coding DNA sequence (CDS) and amino acid sequences from each input contig. It then can use fastanrdb for reducing duplicate sequences and cd-hit and cd-hit-est and blastn to search the similar CDS among sequences. The output transcripts were three classes: Main (the best transcripts with the unique CDS, which is close to a biologically real set), Alternate (possible isoforms), and Drop (the transcripts did not pass the internal filter). The Main and Alternate sets were submitted to the NCBI Transcriptome Shotgun Assembly (TSA) database. Sequences flagged by TSA as duplicates or moderate to strong matches with Univac vectors were masked or removed as per TSA requirements. For R1 and R2 assemblies, 629 of 485,564 sequences, and 575 of 367,801 were modified, respectively, to meet submission requirements for submission to TSA.
For extracting contigs, each assembly was searched for homologous ACCase gene sequences from A. myosuroides (AJ310767), green foxtail [Setaria viridis (L.) P. Beauv.] (AM408428), and E. crus-galli (KU198448) using the BLAST tool at NCBI (http://www.ncbi.nlm.nih.gov) and SwissProt using ncbi-blast-2.2.29+ with an E-value threshold of 1e−5. The blast outputs were processed with Trinity downstream analyses. Main and alternate sequence sets were annotated with the NCBI nonredundant (Nr) protein database using ncbi-blast-2.2.29+ at E-value threshold of 1e−5, with 20 maximum hits found for each query. The Nr blast results were processed using Blast2 Gene Ontology v. 3.0 (Götz et al. Reference Götz, García-Gómez, Terol, Williams, Nagaraj, Nueda, Dopazo, Talón, Robles and Conesa2008) to analyze the gene functions and for comparison with reference ACCase genes from the three species downloaded from the NCBI. Open reading frames (ORFs) were projected using CLC Genomics Workbench and confirmed by comparison with mRNA of E. crus-galli (KU198448). Digitaria ciliaris ACCase sequences were aligned to E. crus-galli (KU198448) genomic DNA. All reads were mapped to the putative assembled plastidic ACCase contiguous sequence to identify single-nucleotide variations associated with herbicide resistance possibly not previously identified using standard PCR sequencing. Read mapping and single-nucleotide variation detection or other related mutations were conducted using the “map reads to reference” and “probabilistic variant detection” tools in CLC Genomics Workbench v. 6.5.2 (Li Reference Li2013). The mapping parameters were selected to “Mismatch cost = 3, Insertion cost = 3, Deletion cost = 3, Length fraction = 0.95, Similarity fraction = 0.95.” The parameters of variants calling were set to “Minimum coverage = 30, Variant probability = 90.”
Results and Discussion
Pinoxaden Response Evaluation
Pinoxaden herbicide treatment by experimental run interactions was not significant (P > 0.05) for control, tiller length, and aboveground biomass; therefore, data were pooled over experimental run and reported as combined means. However, the pinoxaden herbicide treatment by resistant and susceptible biotypes of D. ciliaris interactions was highly significant, with P < 0.05 observed for control, tiller length, and aboveground biomass. Pinoxaden controlled and reduced the measured variables of R1 and R2 less than those for S across all rates (Figures 1 and 2). At 14 DAT, no control was observed for the R1 and R2 biotypes, whereas, 8% control was observed in the S biotype at a rate of 0.1 kg ha−1. At 0.2, 0.4, and 0.7 kg ha−1, pinoxaden controlled R1 and R2 biotypes less than 5% but controlled the S biotype 32%, 37%, and 40%, respectively. Pinoxaden at 1.5, 2.9, 5.9, 11.8, and 23.5 kg ha−1 controlled the R1 biotype 3%, 20%, 23%, 50%, and 60%, respectively, and the R2 biotype 12%, 20%, 43%, 80%, and 88%, respectively, while the S biotype was controlled 67%, 75%, 97%, 100%, and 100%, respectively. The S biotype was completely controlled at 11.8 and 23.5 kg ha−1, resulting in no aboveground biomass at these rates, while no rate completely controlled the R1 and R2 biotypes at 14 DAT.

Figure 1. Percent visible control response relative to nontreated of resistant and susceptible Digitaria ciliaris biotypes with increasing rates of pinoxaden at 14, 28, and 42 d after treatment (DAT). The response was modeled based on the log rate of pinoxaden to create equal spacing between rates using exponential growth regression. Results were pooled over experimental runs. Vertical bars represent the standard error (P = 0.05) of the mean. Means (n = 6) are represented by different symbols for each biotype, and regression equation models are represented by different line types for each biotype. Digitaria ciliaris biotypes: Resistant biotypes, R1 and R2, Georgia, and susceptible biotype, S, Alabama.

Figure 2. Percent tiller length (A) and percent aboveground biomass (B) response in resistant and susceptible Digitaria ciliaris biotypes at 42 d after treatment with increasing rates of pinoxaden. The tiller length (cm) and aboveground biomass weight (g) were expressed as a percentage (%) of the nontreated control at y-axis labels. The response was modeled based on the log rate of pinoxaden to create equal spacing between rates using sigmoidal regression. Results were pooled over experimental runs. Vertical bars represent the standard error (P = 0.05) of the mean. Means (n = 6) are represented by different symbols for each biotype, and regression equation models are represented by different line types for each biotype. D. ciliaris biotypes: Resistant biotypes, R1 and R2, Georgia, and susceptible biotype, S, Alabama.
A similar trend in the biotype response at 28 and 42 DAT was observed for R1, R2, and S. For example, pinoxaden at 1.5, 2.9, and 5.9 kg ha−1 at 28 DAT controlled the R1 biotype 12%, 32%, and 38%, respectively, the R2 biotype 22%, 33%, and 63%, respectively, compared with 55%, 77%, and 97% for the S biotype, respectively. Pinoxaden at 2.9 to 23.5 kg ha−1 controlled the S biotype 100% at 42 DAT, but R1 and R2 control was only ≤40%. Percent control relative to the nontreated response to pinoxaden rate was modeled for all three biotypes through exponential growth functions. The 50% inhibition (I50) and 90% percent inhibition (I90) values were calculated with the exponential growth function equation (Figure 1) presented in Table 2. The level of resistance was determined by I50 and I90 values of each resistant biotype versus the S biotype presented as an R/S ratio. The S biotype had lower I50 and I90 values than R1 and R2 biotypes. Obtaining 50% inhibition of S biotype by 14 DAT, 28 DAT, and 42 DAT required 1.1, 0.5, and 0.2 kg ha−1, respectively, which were much lower rates than needed for the R1 biotype at 16.2, 7.4, and 8.5 kg ha−1, respectively, and the R2 biotype at 8.3, 4.9, and 6.9 kg ha−1, respectively. The resistant biotypes were significantly less sensitive to pinoxaden than the S biotype. The level of resistance as expressed by R/S ratios at 14 DAT, 28 DAT, and 42 DAT was 14.6, 14.3, and 56.9 for the R1 biotype and 7.4, 9.5, and 46.2 for the R2 biotype, respectively. Similarly, the amount of pinoxaden required was also lower for 90% inhibition at 14 DAT, 28 DAT, and 42 DAT for the S biotype at 9.2, 6.8, and 4.9 kg ha−1, respectively compared with the R1 biotype at 37.7, 18.2, and 19.7 kg ha−1, respectively, and the R2 biotype at 20.8, 15.7, and 16.9 kg ha−1, respectively. The resistant biotypes had high resistance levels to pinoxaden compared with the S biotype. The ratio of R/S at 14 DAT, 28 DAT, and 42 DAT was 4.1, 2.7, and 4.0 for the R1 biotype and 2.2, 2.3, and 3.4 for the R2 biotype, respectively.
Table 2. Comparison of response of resistant and susceptible Digitaria ciliaris biotypes to increasing pinoxaden rate relative to the nontreated control was measured through the model with equations of exponential growth for percent control.a

a The required rate of pinoxaden was also calculated by 50% (I50) and 90% (I90) based on control at 14, 28, and 42 d after treatment (DAT). Parameter estimates, 95% confidence intervals (CI), as well as values of I50 and I90 are presented as means of model comparison.
b Digitaria ciliaris biotypes: R1 and R2, resistant biotypes; S, susceptible biotype.
c In the exponential growth equation, x represents pinoxaden rate, y represents the response variable of control at 14, 28, and 42 d after treatment.
d Inhibition rate: I50 and I90 values were calculated using exponential growth equation; R/S ratios, resistant/susceptible ratios.
Trends similar to those observed for comparisons of control data of R1, R2, and S response were observed in aboveground biomass and tiller length. Pinoxaden at a rate of 1.5 kg ha−1 reduced aboveground biomass of R1 and R2 biotypes 8% and 37%, respectively, compared with 71% for the S biotype. The S biotype produced no tillers at rates ≥2.9 kg ha−1 of pinoxaden, which resulted in no aboveground biomass produced; however, the R1 and R2 biotypes did produce aboveground biomass at 2.9 and 5.9 kg ha−1. Percent maximum tiller length and percent aboveground biomass relative to the nontreated response to the increasing rate of pinoxaden were modeled for all three biotypes using sigmoidal functions. I50 and I90 values were calculated through the sigmoidal equation (Figure 2) presented in Table 3. The S biotype contained less pinoxaden for I50 and I90 reduction than R1 and R2 biotypes both in tiller length and aboveground biomass. In the case of tiller length, pinoxaden I50 for S biotype was 0.7 kg ha−1 compared with 7.2 and 6.9 kg ha−1 for the R1 and R2 biotypes, respectively. The R1 and R2 biotypes were 10.4 and 9.9 times more resistant, respectively, than the S biotype based on R/S ratios. Pinoxaden tiller length I90 for the S biotype was 2.1 kg ha−1 compared with 13.2 and 8.6 kg ha−1 for the R1 and R2 biotypes, respectively. The R1 and R2 biotypes were 6.2 and 4.1 times more resistant, respectively, than the S biotype based on R/S ratio. In the case of aboveground biomass, the pinoxaden I50 value for the S biotype was 1.6 kg ha−1 compared with 7.7 and 7.2 kg ha−1 for the R1 and R2 biotypes, respectively. The R1 and R2 biotypes were 4.8 and 4.5 times more resistant, respectively, than the S biotype based on R/S ratios. Pinoxaden aboveground biomass I90 for the S biotype was 2.3 kg ha−1 compared with 10.2 and 7.9 kg ha−1 for the R1 and R2 biotypes, respectively. The R1 and R2 biotypes were 4.5 and 3.5 times more resistant, respectively, than the S biotype based on R/S ratios.
Table 3. Comparison of response of resistant and susceptible Digitaria ciliaris biotypes to increasing pinoxaden rate relative to the nontreated control measured through the model with equations of sigmoidal for percent length of the tiller and aboveground biomass.a
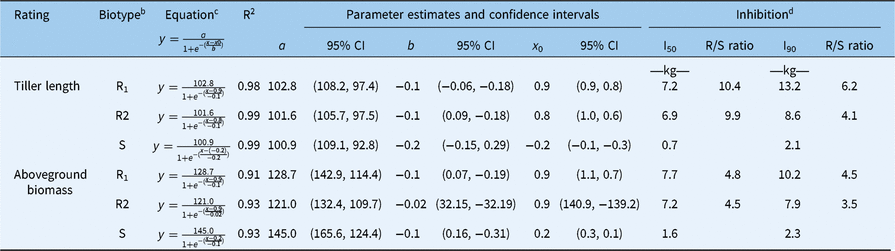
a The required rate of pinoxaden to reduce the measured variables of D. ciliaris biotypes was also calculated by 50% (I50) and 90% (I90). Parameter estimates and 95% confidence intervals (CI) are presented as means of model comparison.
b Digitaria ciliaris biotypes: R1 and R2, resistant biotypes; S, susceptible biotype.
c In the sigmoidal equation, x represents pinoxaden rate, y represents the response variable of tiller length and aboveground biomass relative to the nontreated check.
d Inhibition rate: I50 and I90 values were calculated using sigmoidal equation; R/S ratios, resistant/susceptible ratios.
Target-based Resistance
PCR-based sequencing was conducted in the ACCase CT domain–coding region containing known amino acid substitutions conferring resistance to ACCase-inhibiting herbicides (Délye et al. Reference Délye, Pernin and Michel2011; Liu et al. Reference Liu, Harrison, Chalupska, Gornicki, O’Donnell, Adkins, Haselkorn and Williams2007; Powles and Yu Reference Powles and Yu2010). Sequencing of plastidic ACCase revealed a double peak in the sequencing chromatogram in a single nucleotide for the Ile-1781 codon from cDNA sequencing in the R1 biotype (Figure 3), but not in the R2 or S biotypes (Supplementary Data 1). Cloning of the PCR product was conducted to confirm the resistant and susceptible alleles in the R1 biotype. A 914 bp cDNA fragment containing the Ile-1781 codon was cloned as described earlier. Twelve ACCase putative mutated cDNA fragments were sequenced, analyzed, and compared with the S biotype along with several homologs of ACCase genes from A. myosuroides (AJ310767), E. crus-galli (KU198448), and S. viridis (AM408428). The cloned cDNA fragment from the sequencing resulted in two separate nucleotide sequences surrounding the Ile-1781 codon. Out of the 12 ACCase putative mutated cDNA fragments, eight cloned cDNA fragments contained codon CTA for Leu-1781 amino acid, and four cloned cDNA fragments contained ATA for Ile-1781. Other polymorphisms between cloned amplicons are presented in Supplementary Data 2 and 3. Sequences from ACCase-resistant D. ciliaris were submitted to the NCBI database (accession numbers: MK558087 and MK558088). The presence of a double peak on the sequencing chromatogram can indicate heterozygosity between two homologs of a diploid organism or an allelic difference between homologs of a polyploid organism. Based on sequencing of PCR products generated by various primers, we found only the Ile-1781-Leu substitution in the R1 biotype, representing potential herbicide resistance mutation in D. ciliaris. However, no double peaks or amino acid substitutions were found in the R2 biotype. Considering both R1 and R2 response to ACCase-inhibiting herbicides was similar, we theorized that standard PCR was not able to amplify all homoeologs in the R2 biotype. So, NGS using the Illumina platform was performed to sequence all expressed plastidic ACCase in the R2 biotype.

Figure 3. Chromatogram of nucleotide sequence surrounding ATA codon encoding Ile-1781. The double peak indicates an amino acid substitution of CTA coding for Leu-1781 in the R1 biotype (A). Nucleotide sequences from cloned cDNA fragments of transgenic and nontransgenic alleles surrounding the Ile-1781 codon. An Ile-1781-Leu substitution confers resistance to ACCase-inhibiting herbicides, DIMs, FOPs, and pinoxaden in Digitaria ciliaris. Cloned cDNA fragments show the ATA codon for Ile in nontransgenic allele (wild type) and the CTA codon for Leu in transgenic allele (mutant) at position 1781 (B)
Transcriptome Profiling
Two de novo assembly transcriptomes were assembled separately for both the R1 and R2 biotypes. The number of reads obtained for the R1 and R2 biotypes were 484,935 and 367,226, respectively. ACCase contigs were extracted via BLAST analysis with plastidic ACCase genes from three species, NCBI: A. myosuroides (AJ310767), E. crus-galli (KU198448), and S. viridis (AM408428). The ACCase transcript of D. ciliaris coding for the complete protein was recovered, showing 91.5% similarities and 83.0% identity with A. myosuroides. Sequence analysis produced an ORF containing coding for a 157-amino-acid-long peptide for the ACCase CT-domain with 93.9% homology to E. crus-galli. Mapping of sequence reads from extracted contigs revealed heterozygosity in the Ile-1781 codon conferring a nonsynonymous substitution of Leu at this position (Figure 4) along with 25 synonymous substitutions (T to C at 1456, A to G at 1516, C to T at 1543, G to A at 1555, T to C at 1705, C to T at 1849, A to G at 1852, C to A at 1900, T to G at 1903, T to A at 1969, C to T at 1981, C to T at 2023, A to T at 2293, A to T at 2311, T to A 2323, C to T at 2374, A to G at 2431, T to C at 2474, C to T at 2549, C to T at 2593, A to G at 2617, T to C at 2650, T to C at 2719, C to G at 2932, and C to G at 2971) in the carboxyl transferase domain of the plastid ACCase for both the R1 and R2 biotypes (Supplementary Data 4).

Figure 4. Illumina sequencing with single-nucleotide variation of adenine (A) to cytosine (C) at the 1781 region in both resistant biotypes, R1 and R2. The ACCase gene of Digitaria ciliaris in both resistant biotypes, R1 and R2, contained the CTA codon for the amino acid Leu-1781 instead of the ATA codon for Ile-1781. Comparison of Illumina sequencing of D. ciliaris transcriptome producing 93.9% homology to the reference gene of Echinochloa crus-galli (KU198448). Illumina sequence analysis at the carboxyl transferase domain revealed the amino acid substitution Ile-1781-Leu conferring resistance to DIMs, FOPs, and pinoxaden in D. ciliaris.
Identification of an Ile-1781-Leu substitution in R2 supported our initial hypothesis of target-based resistance mechanism determination and our suspicions of a lack of adequate amplification to all homologues of plastidic ACCase mRNA with standard PCR. Sequencing reads of R1 and R2 biotypes have been submitted to the NCBI Sequence Read Archive (SRA) database (accession numbers PRJNA524359 and PRJNA524643, respectively). The sequence assemblies to the NCBI TSA project have been deposited at DDBJ/EMBL/GenBank under the accession GHOH00000000. The version described in this paper is the first version, GHOH01000000. The amino acid substitution, Ile-1781-Leu, therefore, in the resistant biotypes could be a possible causal resistance mechanism to ACCase-inhibiting herbicides in D. ciliaris. SNPs in ACCase mapping of both resistant biotypes were also found in a 1:2 ratio, with approximately 33% of mapped reads containing SNPs. A 1:2 ratio likely indicates that D. ciliaris is a hexaploid as well, but further research is needed to confirm this hypothesis.
Research Implications
The insensitivity of the ACCase target site has been reported as the most common mechanism of resistance to ACCase-inhibiting herbicides (Kuk et al. Reference Kuk, Burgos and Scott2008). Using a combination of standard PCR-based sequencing, vector sequencing, and Illumina sequencing, we identified a common amino acid Ile-1781-Leu substitution in the plastid ACCase gene of D. ciliaris resistant biotypes. The double peak in the PCR chromatogram (expected as A or C) in the center of the nucleotide at position 1781 in the R1 biotype suggests the resistant biotypes can express at least two different plastidic ACCase genes. Our finding of the Ile-1781-Leu substitution causing ACCase-resistance to DIMs and FOPs is consistent with the vast majority of literature on the subject in A. myosuroides (Brown et al. Reference Brown, Moss, Wilson and Field2002; Délye et al. Reference Délye, Calmès and Matéjicek2002a, Reference Délye, Matéjicek and Gasquez2002b; Petit et al. Reference Petit, Bay, Pernin and Délye2010), rigid ryegrass (Lolium rigidum Gaudin) (Zagnitko et al. Reference Zagnitko, Jelenska, Tevzadze, Haselkorn and Gornicki2001), L. perenne ssp. multiflorum (White et al. Reference White, Moss and Karp2005; Yu et al. Reference Yu, Collavo, Zheng, Owen, Sattin and Powles2007; Zhang and Powles Reference Zhang and Powles2006a, Reference Zhang and Powles2006b), S. viridis (Délye et al. Reference Délye, Wang and Darmency2002c), A. fatua (Christoffers et al. Reference Christoffers, Berg and Messermith2002), sterile oat (Avena sterilis L.) (Liu et al. Reference Liu, Harrison, Chalupska, Gornicki, O’Donnell, Adkins, Haselkorn and Williams2007; Torres-García et al. Reference Torres-García, Tafoya-Razo, Velázquez-Márquez and Tiessen2018), and corn (Zea mays L.) (Genbank accession numbers: AF359517 and AF359518; Zagnitko et al. Reference Zagnitko, Jelenska, Tevzadze, Haselkorn and Gornicki2001). A target-site mutation does not preclude other possible resistance mechanisms contributing to overall resistance response; for example, Maneechote et al. (Reference Maneechote, Preston and Powles1999) found that the FOP-resistant A. sterilis biotype had target-site ACCase mutations and enhanced metabolism, but Cyt-P450 inhibitors reduced its tolerance to diclofop. Yu et al. (Reference Yu, Ahmad-Hamdani, Han, Christoffers and Powles2013) tested hexaploid A. fatua and concluded that the lower level of target-site ACCase resistance in polyploid than diploid weed species, especially the Poaceae family, was due to a herbicide dilution effect. Torres-García et al. (Reference Torres-García, Tafoya-Razo, Velázquez-Márquez and Tiessen2018) found that the ACCase gene alteration Ile-1781-Leu produced enhanced metabolism in A. fatua tested for resistance. All the experiments implied ACCase gene mutation metabolic adaptation, as well as detoxification mechanisms, can contribute to resistance in the resistant biotype.
Based on our findings, we reached three major conclusions: first, the D. ciliaris biotypes (Yu et al. Reference Yu, McCullough and Czarnota2017) previously reported as resistant to sethoxydim and mildly resistant to some FOP herbicides are also resistant to pinoxaden relative to the S biotype. The D. ciliaris biotypes responded similarly to pinoaxden and other ACCase-inhibiting herbicides. Prior selection pressure with DIM and FOP herbicides could result in the evolution of D. ciliaris cross-resistance to pinoxaden herbicide in United States. Second, the amino acid substitution Ile-1781-Leu in the ACCase gene is the likely causal mechanism of resistance in D. ciliaris. Mutation at Ile-1781 is a common substitution that yields resistance to ACCase-inhibiting herbicides in weed species tested. The authors acknowledge that other NTSR mechanisms may be simultaneously occurring and contributing to resistance. Third, while not an initial part of our original research goals, reliance on PCR to amplify all expressed copies of plastidic ACCase in a weed species could lead to erroneous conclusions, whereas NGS transcriptome profiling was able to identify a polymorphism previously missed by the standard PCR. Such a conclusion is especially important with respect to ACCase resistance, where both TSR and NTSR mechanisms can occur simultaneously and a lack of adequate PCR-based amplification could cause one to conclude TSR is absent in a population. This is the first reported case of cross-resistance to pinoxaden from previously identified ACCase-resistant biotypes and of target-based resistance to ACCase-inhibiting herbicides in D. ciliaris species from managed turfgrass.
Acknowledgments
This publication was supported by the Alabama Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. No conflicts of interest have been declared.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/wsc.2019.56









