Introduction
Rice (Oryza sativa L.) is one of the most important grain crops in the world, and it is the staple food for more than 3 billion people (Kuenzer and Knauer Reference Kuenzer and Knauer2013). In China, weed infestation results in 10 billion kg of annual rice yield reduction, with an average loss rate of about 15%, and weeds are becoming the major causes of yield losses (Dong et al. Reference Dong, Gao, Fang and Chen2018). Barnyardgrass [Echinochloa crus-galli (L.) P. Beauv.], an annual grass species, is one of the most problematic weeds that infest paddy fields, and it is considered a serious threat to rice production (Zhang et al. Reference Zhang, Gu, Zhao, Yang, Peng, Li and Bai2017).
Acetolactate synthase (ALS) and acetyl-CoA carboxylase (ACCase) inhibitors, represented in this study by penoxsulam and cyhalofop-butyl, respectively, are two groups of herbicides commonly used in rice fields (Dong et al. Reference Dong, Gao, Fang and Chen2018). ALS inhibitors target the key enzyme in the biosynthesis of branched-chain amino acids and can be classified into sulfonylureas, triazolopyrimidines, imidazolinones, sulfonyl-aminocarbonyl-triazolinones, and pyrimidinylthiobenzoates (Tranel and Wright Reference Tranel and Wright2002). ACCase inhibitors prevent fatty-acid synthesis and fall into three chemical families, namely aryloxyphenoxypropionates, cyclohexanediones, and phenylpyraxoline (Sasaki et al. Reference Sasaki, Konishi and Nagano1995). Both ALS and ACCase inhibitors have a high resistance risk in E. crus-galli (Bagavathiannan et al. Reference Bagavathiannan, Norsworthy, Smith and Neve2014). Recurrent use of these herbicides has promoted resistance to ALS and ACCase inhibitors in 165 and 49 weed species, respectively (Heap Reference Heap2021).
Both target site–based resistance (TSR) and non–target site based resistance (NTSR) are involved in resistance to ALS and ACCase inhibitors (Perotti et al. Reference Perotti, Larran, Palmieri, Martinatto and Permingeat2020; Powles and Yu Reference Powles and Yu2010). Molecular mechanisms of TSR to ALS and ACCase herbicides in weeds have been well researched. Twenty-eight amino acid substitutions at the Ala-122, Pro-197, Ala-205, Asp-376, Arg-377, Trp-574, Ser-653, and Gly-654 sites in ALS have been reported to confer resistance to ALS inhibitors (Heap Reference Heap2021). Thirteen amino acid substitutions at seven sites (Ile-1781, Trp-1999, Trp-2027, Ile-2041, Asp-2078, Cys-2088, Gly-2096) in the carboxyltransferase domain of ACCase have been identified to be associated with resistance to ACCase inhibitors (Kaundun Reference Kaundun2014). In the case of NTSR, enhanced herbicide metabolism is frequently reported (Ghanizadeh and Harrington Reference Ghanizadeh and Harrington2017); however, the molecular mechanisms remain largely unknown (Délye Reference Délye2013; Yu and Powles Reference Yu and Powles2014).
Echinochloa spp. are the most troublesome weeds in rice fields and can compete very effectively with rice seedlings, causing huge crop yield losses (Dong et al. Reference Dong, Gao, Fang and Chen2018). However, only a limited number of herbicides are registered and can be used in rice fields. Among them, the auxin herbicide quinclorac was used for nearly 30 yr in China, resulting in high resistance frequency (Gao et al. Reference Gao, Pan, Sun, Li and Dong2019). In addition, the ALS inhibitor bispyribac-sodium and ACCase inhibitor metamifop provided poor crop safety for some rice varieties, making them unpopular among rice growers in China. For these reasons, the foliar-applied herbicides penoxsulam and cyhalofop-butyl have been used widely for the control of Echinochloa spp. in rice. This overreliance on one or two herbicides is likely to result in the development of weed resistance. Penoxsulam-resistant Echinochloa spp. were reported in various rice-growing regions of China, including Jiangsu, Anhui, Hubei, Hunan, Jiangxi, and Heilongjiang provinces (Chen et al. Reference Chen, Wang, Yao, Zhu and Dong2016; Fang et al. Reference Fang, Liu, Zhang, Li and Dong2019a; Liu et al. Reference Liu, Fang, He, Li and Dong2019; Yang et al. Reference Yang, Yu and Li2013). Xia et al. (Reference Xia, Tang, He, Kang, Ma and Li2016) also reported E. crus-galli populations from Hubei Province that had developed resistance to the ACCase inhibitor metamifop.
Ningxia Province, located in the northwest of China, is one of the major rice-producing areas. Weed management in Ningxia rice fields mainly depends on chemical control. However, penoxsulam and/or cyhalofop-butyl cannot effectively control the occurrence of E. crus-galli in some regions of Ningxia Province due to overreliance on a limited number of herbicides. Here, we aimed to (1) investigate the levels of resistance to penoxsulam and cyhalofop-butyl in 12 E. crus-galli populations collected from Ningxia Province, (2) characterize the resistance mechanisms to these two herbicides, and (3) determine the cross-resistance patterns of resistant populations to other ALS and ACCase inhibitors.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of 12 E. crus-galli populations were collected from Ningxia Province in 2017. A total of 11 putative herbicide-resistant populations (NX1 to NX11) were collected from rice fields where penoxsulam and cyhalofop-butyl failed to control the weeds. The known susceptible population (NX12) was collected from an area with no previous herbicide treatment (Figure 1). Seeds of each population were collected randomly from 15 to 20 mature plants and bulked.

Figure 1. Locations of 12 Echinochloa crus-galli populations in this study.
Seeds were stratified in darkness for 3 d at 4 C before being transferred to a growth chamber (30/25 C, 14-h light/10-h dark). After germination, seedlings were transplanted to 7-cm-diameter plastic pots (12 seedlings per pot) containing a 1:1:1 (w/w/w) mixture of sand, vermiculite, and organic matter and grown in growth chamber under conditions of 30/25 C temperature, a 14-h light/10-h dark cycle, light intensity of 12,000 lx, and 75% relative humidity.
Whole-Plant Dose Response to Penoxsulam and Cyhalofop-Butyl
Before herbicide treatment, seedlings were thinned to 8 to 10 plants per pot. When the plants had grown to the 3-leaf stage, herbicides were applied using a laboratory sprayer (machine model: 3WPSH-500D, Nanjing Research Institute for Agricultural Mechanization, Nanjing, Ministry of Agriculture of China) equipped with a cone nozzle delivering 280 L ha−1 at 0.3 MPa. The penoxsulam doses were 0, 1/16×, 1/4×, 1×, 2×, 4×, and 8× the recommended field dose (30 g ai ha−1) for putative resistant populations and 0, 1/64×, 1/16×, 1/4, 1×, and 2× the recommended rate for susceptible population. The cyhalofop-butyl doses were 0, 1/8×, 1/4×, 1/2×, 1×, 2×, 4×, and 8× the recommended field dose (120 g ai ha−1) for putative resistant populations and 0, 1/16×, 1/8×, 1/4×, 1/2×, 1×, and 2× the recommended rate for susceptible population. The fresh weight of aboveground plants was recorded at 2 wk after herbicide treatment. Each herbicide dose had three replications, and each experiment was conducted twice.
Partial ALS and ACCase Gene Cloning and Sequencing
Primers used for the amplification of E. crus-galli ALS genes were designed based on the ALS (accession number LC006058, LC006059, and LC006061) sequences retrieved from the National Center for Biotechnology Information (NCBI) GenBank database (Table 1).
Table 1. Primers used for amplification of Echinochloa crus-galli ALS and ACCase genes.

a T m, melting temperature.
b EcACCase, amplify the carboxyltransferase domains of the orthologous ACCase genes EcACC1, EcACC2, EcACC4, and EcACC6 of E. crus-galli.
c EcACC3, amplify EcACC3; EcACC5, amplify EcACC5.
For ACCase genes, the primer set designed based on ACCase (accession numbers LC006070 to LC006075) sequences of E. crus-galli were used to amplify the carboxyltransferase domains of the orthologous genes EcACC1, EcACC2, EcACC4, and EcACC6 in this study (Table 1). The other orthologous genes, EcACC3 and EcACC5, were specifically amplified by primers described in Iwakami et al. (Reference Iwakami, Hashimoto, Matsushima, Watanabe, Hamamura and Uchino2015).
Survivors from dose–response treatments were selected for gene identification. Approximately 100 mg of shoot tissue from each individual plant was collected for further analyses. Genomic DNA was extracted from each sample using the Hi-DNAsecure Plant Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. The polymerase chain reaction (PCR) mixture contained 2 μl of template DNA, 1 μl (10 μM) of each primer, 25 μl of 2×Taq PCR Master Mix (Tiangen Biotech), and ddH2O to a final volume of 50 μl. The PCR program was set according to the following profile: 3 min at 95 C for DNA denaturation; followed by 35 cycles of 30-s denaturation at 95 C, 30-s annealing at each melting temperature (T M), and 90-s elongation at 72 C; then a final 5 min at 72 C for further DNA elongation. The PCR products were purified using TIANgel Midi Purification Kit (Tiangen Biotech) and cloned into a pLB vector (Tiangen Biotech). The plasmids containing the fragment insertions were transformed into TOP10 chemically competent cells, and sequenced by Sangon Biotechnology (Shanghai, China) with pLB vector sequencing primers (forward: 5′-CGACTCACTATAGGGAGAGCGGC-3′; reverse: 5′-AAGAACATCGATTTTCCATGGCAG-3′). At least 10 transformed clones of each plant were sequenced to obtain partial ALS and ACCase gene sequences that were aligned and compared in DNAMAN v. 5.22 software (Lynnon Biosofe, Quebec, Canada).
Sensitivity to Other ALS- and ACCase-inhibiting Herbicides
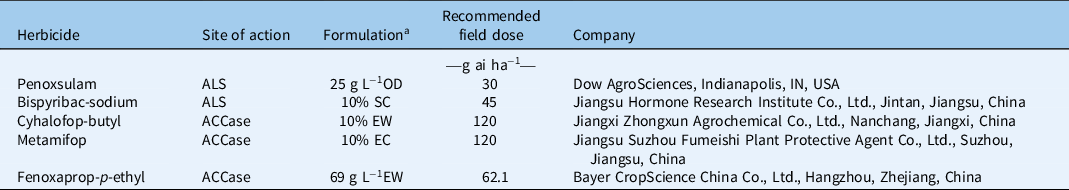
Three ALS- and ACCase-inhibiting herbicides commonly used in rice fields were used to evaluate their effectiveness on all E. crus-galli populations. Bispyribac-sodium, metamifop, and fenoxaprop-p-ethyl at doses of 45, 120, and 20.7 g ai ha−1, respectively were used to differentiate the resistant and susceptible populations. The doses were designed based on the primary experiments; the herbicides tested are listed in Table 2. When the plants grew to the 3-leaf stage, the herbicides were applied as described earlier with three replicates. At 2 wk after treatment, aboveground shoots were collected and their fresh weight recorded.
Table 2. ALS- and ACCase-inhibiting herbicides used in this study.
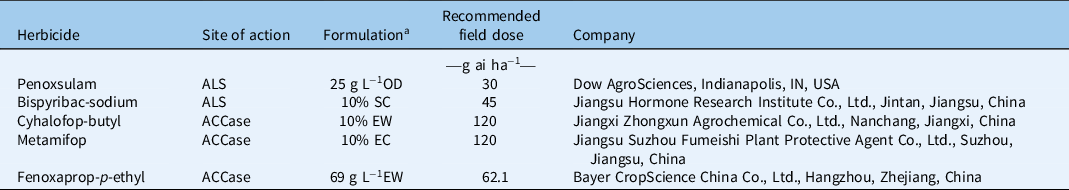
a OD, dispersible oil suspension; SC, suspension concentrate; EW, emulsion in water. EC, emulsifiable concentrate.
Statistical Analysis
The herbicide dose required for 50% growth reduction (GR50) was evaluated using the following logistic-regression model (Equation 1) with SigmaPlot software (v. 12.0, Systat Software, San Jose, CA, USA) (Seefeldt et al. Reference Seefeldt, Jensen and Fuerst1995).
In the equation, y is the fresh weight, expressed as percentage of the nontreated control at dose x of herbicide, b is the slope, C is the lower limit, and D is the upper limit. The resistance index (RI) was calculated as the GR50 of the resistant population divided by that of susceptible population (NX12). The resistance levels of E. crus-galli populations to each herbicide were defined according to a standard (Beckie and Tardif Reference Beckie and Tardif2012) and appropriately modified. The standard was as follows: no resistance (RI < 3), low resistance (3 ≤ RI < 5), moderate resistance (5 ≤ RI < 10), and high resistance (RI ≥ 10). The data for other ALS- and ACCase-inhibiting herbicide sensitivities of the 12 E. crus-galli populations were analyzed with one-way ANOVA (SPSS Statistics v. 20.0, IBM, Armonk, NY, USA).
Results and Discussion
Dose Response to Penoxsulam and Cyhalofop-Butyl
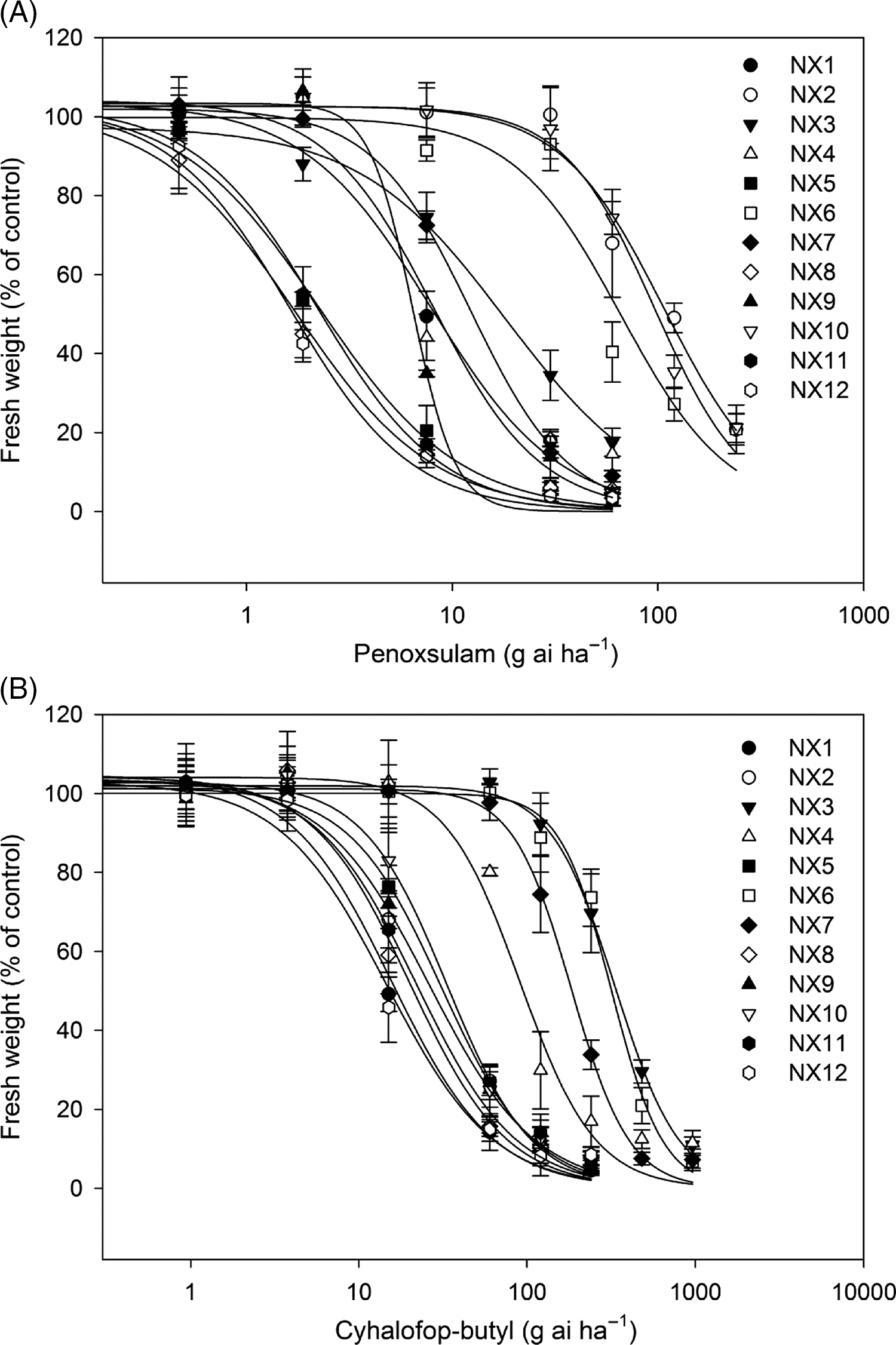
In this study, we surveyed and characterized penoxsulam and cyhalofop-butyl sensitivity of 12 E. crus-galli populations collected from rice fields in Ningxia Province, China. The results showed that eight E. crus-galli populations (NX1, NX2, NX3, NX4, NX6, NX7, NX9, and NX10) displayed resistance to the ALS inhibitor penoxsulam (Table 3; Figure 2A). The susceptible populations (NX5, NX8, NX11, and NX12) were completely killed by the recommended rate of penoxsulam, and the GR50 values were 2.25, 1.71, 2.23, and 1.63 g ai ha−1, respectively. The resistant populations (NX2, NX6, and NX10) were not injured by the recommended rate and exhibited more than 90% growth relative to untreated controls at that dose. The GR50 values of NX2 and NX10 were similar to each other: 107.32 g ai ha−1 and 96.67 g ai ha−1, respectively; and the resistance indexes were 65.84 and 59.30, respectively, compared with the susceptible population (NX12). The GR50 of NX6 was 65.84 g ai ha−1, with 40.39-fold higher resistance than the susceptible population. In addition, there were five populations (NX1, NX3, NX4, NX7, and NX9) that partially survived under the recommended rate application and exhibited approximately 20% growth relative to untreated controls. The GR50 values of these populations were calculated to be 8.04, 18.21, 7.87, 12.25, and 7.40, respectively, showing at least moderate-level resistance to penoxsulam.
Table 3. GR50 and resistance index (RI) values to penoxsulam and cyhalofop-butyl in Echinochloa crus-galli populations.
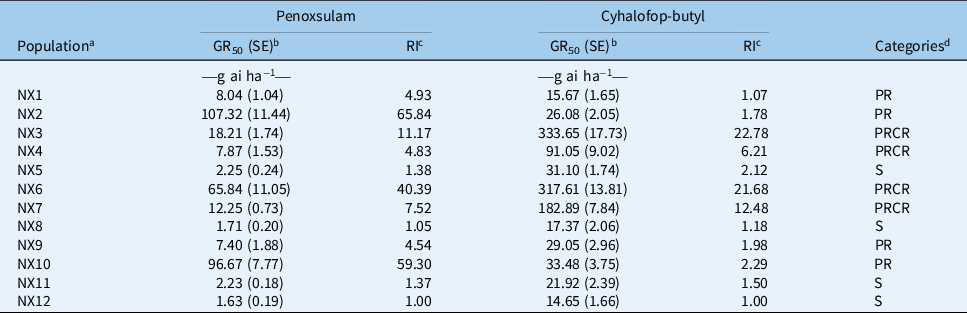
a NX12 was the known susceptible population.
b GR50, herbicide dose causing 50% growth reduction; SE, standard error.
c RI, resistance index, which calculated with GR50 value of resistant population divided by that of susceptible population.
d PR, E. crus-galli population resistant to penoxsulam; CR, population resistant to cyhalofop-butyl.
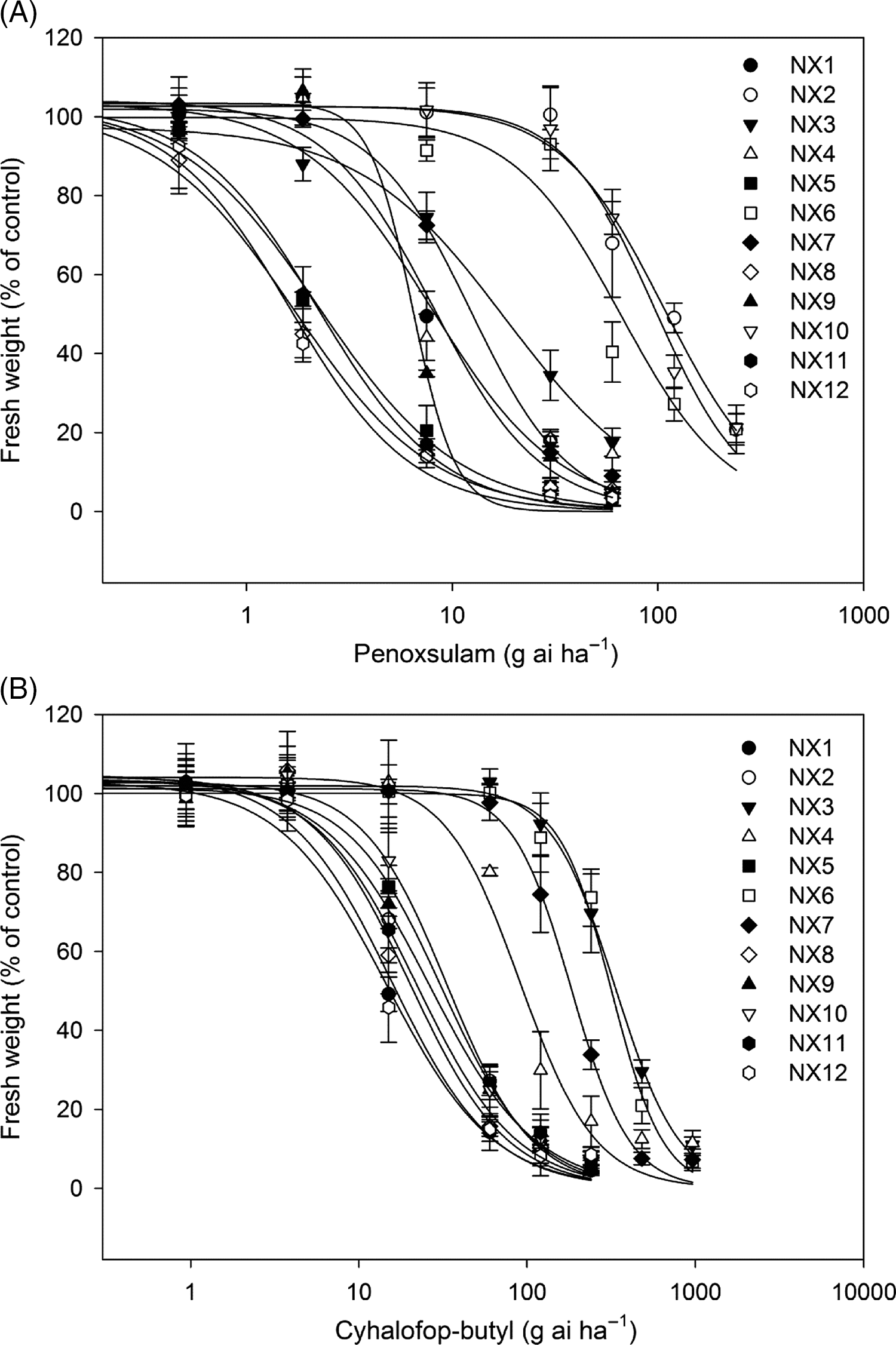
Figure 2. Dose-response curves of 12 Echinochloa crus-galli populations to penoxsulam (A) and cyhalofop-butyl (B). Shoot fresh weight is shown as percentages of the fresh weight of untreated controls. Bars indicate standard error (n = 3).
The results of the cyhalofop-butyl sensitivity assay showed that three (NX3, NX6, and NX7) out of 12 E. crus-galli populations had evolved high level resistance to cyhalofop-butyl, with 22.78, 21.68, and 12.48-fold resistance compared with the susceptible population (NX12) (Table 3; Figure 2B). The NX4 population demonstrated a moderate level of resistance, with an RI value of 6.21. The cyhalofop-butyl–susceptible populations (NX1, NX2, NX5, NX8, NX9, NX10, NX11, and NX12) were completely killed by the recommended application rate of cyhalofop-butyl, and the GR50 ranged from 14.65 to 33.48 g ai ha−1.
Interestingly, the four cyhalofop-butyl–resistant E. crus-galli populations (NX3, NX4, NX6, and NX7) also showed resistance to penoxsulam. The NX6 population exhibited especially high-level resistance to both penoxsulam and cyhalofop-butyl, with 40.39- and 21.68-fold resistance, respectively, relative to susceptible population (Table 3; Figure 2). The other three populations (NX3, NX4, and NX7) had evolved low- to moderate-level resistance to penoxsulam, and the RI values were 11.17, 4.83, and 7.52, respectively. The results indicate that at least two different types of herbicide resistance exist among E. crus-galli plants in Ningxia Province, and the resistance patterns are evolving from single herbicide resistance to multiple resistance. These findings revealed the widespread occurrence of ALS and/or ACCase inhibitor–resistant E. crus-galli populations in China, a situation that requires attention, especially the occurrence of MHR populations.
Cloning and Sequence Analysis of ALS and ACCase Genes
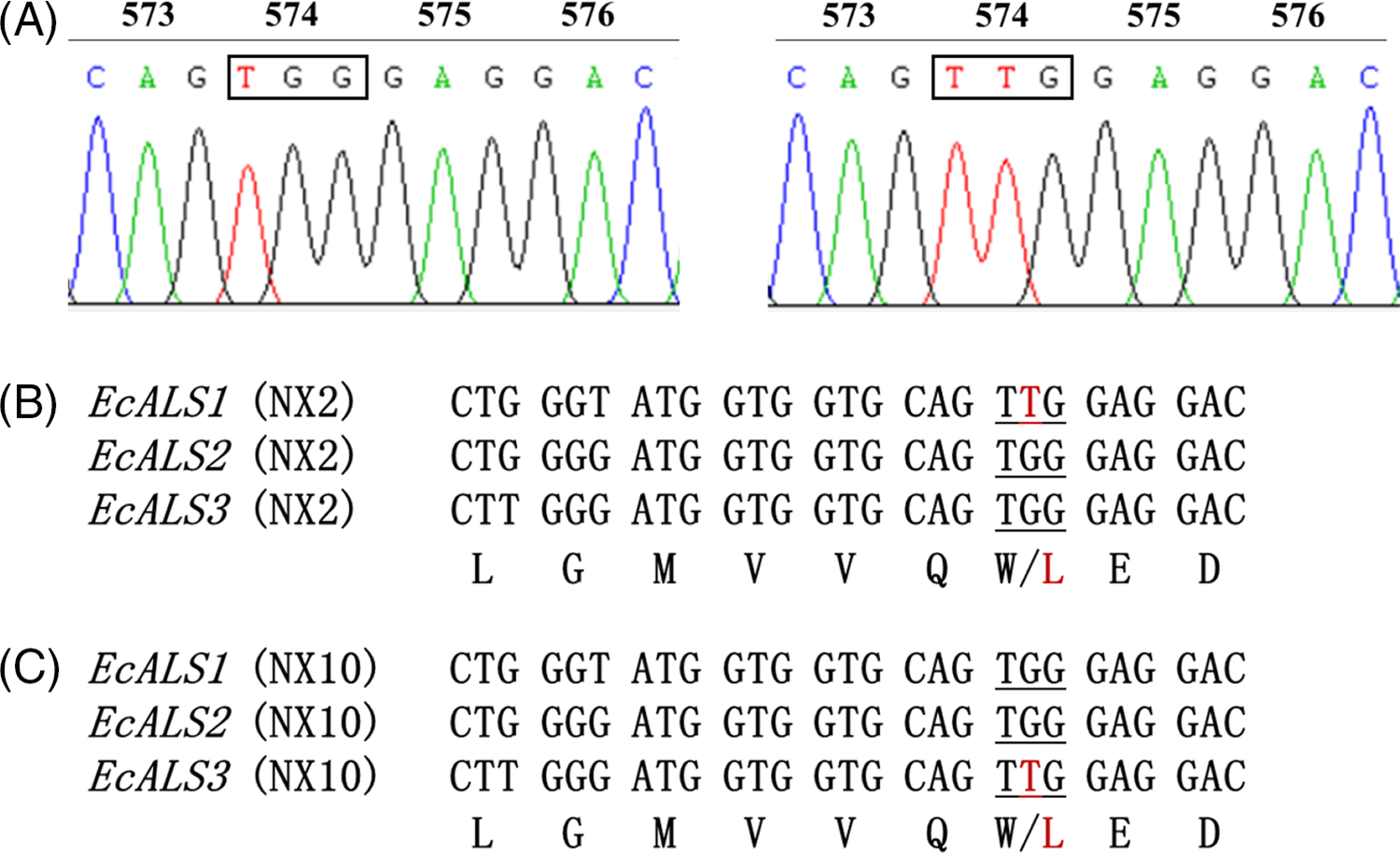
Three partial gene sequences of ALS that covered more than 85% of full-length sequences were isolated from each E. crus-galli population. The isolated region contains all known eight amino acid positions whose substitutions confer ALS-inhibitor resistance (Powles and Yu Reference Powles and Yu2010) and exhibited highest homology with the ALS genes (accession number LC006058, LC006059, and LC006061) of E. crus-galli registered in the NCBI database. A Trp-574-Leu substitution caused by a nucleotide mutation (TGG to TTG) was identified in resistant E. crus-galli populations NX2 and NX10 (Table 4; Figure 3A). Moreover, the Trp-574-Leu substitution in the NX2 population occurred in EcALS1 (Figure 3B), and in the NX10 population it occurred in EcALS3 (Figure 3C). In addition, no known resistance-conferring mutations were found in the highly penoxsulam-resistant population NX6 and other resistant populations (Table 4). In these populations, the NTSR mechanism may be dominant, and further research is needed. The Trp-574-Leu mutation was mainly responsible for penoxsulam resistance in E. crus-galli, and the same substitution has been documented in other ALS inhibitor–resistant weeds, including catchweed bedstraw (Galium aparine L.) (Deng et al. Reference Deng, Di, Cai, Chen and Yuan2019b), common gromwell [Lithospermum arvensis (L.) I.M. Johnst.] common gromwell [Lithospermum arvensis (L.) I.M. Johnst.] (Wang et al. Reference Wang, Ge, Zhang, You, Wang, Liu and Wang2019), johnsongrass [Sorghum halepense (L.) Pers.] (Hernandez et al. Reference Hernandez, Leon, Fischer, Gebauer, Galdames and Figueroa2015), and annual bluegrass (Poa annua L.) (McElroy et al. Reference McElroy, Flessner, Wang, Dane, Walker and Wehtje2013). Multiple ALS gene copies are a common phenomenon in weeds, especially in polyploid weeds. For example, there are two copies of ALS in flixweed (Descurainia sophia L.) (Deng et al. Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017) and shepherd’s purse [Capsella bursa-pastoris (L.) Medik.] (Zhang et al. Reference Zhang, Gu, Zhao, Yang, Peng, Li and Bai2017). The variation in ALS copy number was also observed in shortawn foxtail (Alopecurus aequalis Sobol.) (Iwakami et al. Reference Iwakami, Shimono, Manabe, Endo, Shibaike, Uchino and Tominaga2017) and Japanese foxtail (Alopecurus japonicus Steudel) (Feng et al. Reference Feng, Gao, Zhang, Dong and Li2017). Furthermore, it was demonstrated that any of the ALS gene copies could host the ALS-inhibitor resistance mutation (Deng et al. Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017; Zhang et al. Reference Zhang, Gu, Zhao, Yang, Peng, Li and Bai2017), which may also be the reason why these weeds are prone to develop resistance to ALS inhibitors. The Ala-122-Gly/Asn, Ala-205-Val, Trp-574-Leu, and Ser-653-Asn mutations in ALS have been reported to confer resistance to ALS-inhibiting herbicides in E. crus-galli (Fang et al. Reference Fang, Liu, Zhang, Li and Dong2019a; Matzenbacher et al. Reference Matzenbacher, Bortoly, Kalsing and Merotto2015; Panozzo et al. Reference Panozzo, Scarabel, Tranel and Sattin2013, Reference Panozzo, Scarabel, Rosan and Sattin2017). In addition, the NTSR mechanism was reported in various ALS inhibitor–resistant weeds, such as E. crus-galli, D. sophia, and giant chickweed [Myosoton aquaticum (L.) Moench] (Bai et al. Reference Bai, Zhang, Li, Wang, Wang, Wang, Liu and Bai2019; Fang et al. Reference Fang, Zhang, Liu, Yan, Li and Dong2019b; Yang et al. Reference Yang, Deng, Li, Yu, Bai and Zheng2016). Two homologous cytochrome P450 genes, CYP81A12 and CYP81A21, have been identified as conferring resistance to the ALS inhibitors bensulfuron-methyl and penoxsulam in rice barnyardgrass [Echinochloa phyllopogon (Stapf) Koso-Pol.] (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama and Inamura2014). Therefore, the possible NTSR mechanism involved in the eight penoxsulam-resistant E. crus-galli populations, especially in NX6, could not be ruled out and requires further study.
Table 4. Target-site mutations in ALS genes of penoxsulam-resistant Echinochloa crus-galli populations.
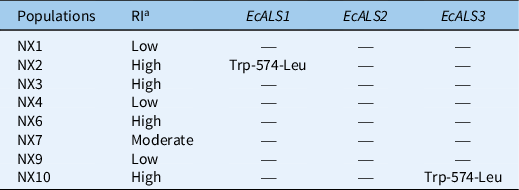
a RI, resistance index: low, 3–5; moderate, 5–10; high, >10.

Figure 3. DNA sequencing results showing TGG for Trp of susceptible Echinochloa crus-galli plants (A, left), TTG for Leu of resistant plants (A, right). Sequence alignment identified a Trp-574-Leu substitution in EcALS1 of the NX2 population (B) and in ECALS3 of the NX10 population (C). The underlined bases indicate the amino acid mutation in codon position 574.
Six partial sequences of ACCase genes were amplified and sequenced from the genomic DNA of each E. crus-galli population (Figure 4). All six ACCase genes showed highest sequence similarity to those registered in the NCBI database (accession numbers LC006070 to LC006075). However, none of the ACCase genes that we sequenced in the four cyhalofop-butyl–resistant populations of E. crus-galli encode proteins with any of the amino acid substitutions that have previously been reported to cause resistance to ACCase inhibitors. The sequencing results suggest that these populations have mechanisms of NTSR to the ACCase inhibitor cyhalofop-butyl. Our results were consistent with a previous report. Two Echinochloa crus-galli var. formosensis lines in Japan exhibited multiple resistance to cyhalofop-butyl and penoxsulam, and no amino acid substitutions conferring herbicide resistance were found in the ACCase and ALS sequences, suggesting an NTSR mechanism of herbicide resistance (Iwakami et al. Reference Iwakami, Hashimoto, Matsushima, Watanabe, Hamamura and Uchino2015). It is noteworthy that two populations (NX3 and NX6) showed more than 20-fold resistance to cyhalofop-butyl. Generally, NTSR, particularly metabolic resistance, confers low to moderate levels of herbicide resistance (<10-fold) (Tranel and Wright Reference Tranel and Wright2002). Now, with the discovery of more NTSR cases in various weeds, it has been realized that NTSR could also endow high-level resistance compared with TSR. For example, an American sloughgrass [Beckmannia syzigachne (Steud.) Fernald] population with metabolic resistance was 63.5-fold resistance to fenoxaprop-pethyl (Bai et al. Reference Bai, Zhao, Zhou, Wang, Li, Luo and Li2020). An NTSR mechanism in Chinese sprangletop [Leptochloa chinensis (L.) Nees] showed 66.7-fold resistance to cyhalofop-butyl (Deng et al. Reference Deng, Cai, Zhang, Chen, Chen, Di and Yuan2019a). In addition, it is worth noting that EcACC4 has a base deletion resulting in an aberrant protein with a frameshift at Thr-1954, which is consistent with a previous report (Iwakami et al. Reference Iwakami, Hashimoto, Matsushima, Watanabe, Hamamura and Uchino2015).

Figure 4. Partial sequences of six ACCase genes from Echinochloa crus-galli. The boxed regions represent seven amino acid positions whose substitutions are known to confer ACCase-inhibitor resistance.
The copy numbers of target genes involved in TSR are negatively correlated with resistance evolution (Iwakami et al. Reference Iwakami, Hashimoto, Matsushima, Watanabe, Hamamura and Uchino2015). Due to the “dilution effect,” a single gene copy with a resistance-conferring mutation is diluted by more amounts of susceptible copies and will not necessarily confer strong resistance in plants with multiple copies of the target gene (Yu et al. Reference Yu, Ahmad-Hamdani, Han, Christoffers and Powles2013). For example, the hexaploid wild oat (Avena fatua L.) carrying a homozygous resistance mutation in one of the three ACCase gene copies shows relatively low-level (2- to 5-fold) resistance to ACCase-inhibiting herbicides compared with the resistance observed in a single-copy species (Yu et al. Reference Yu, Ahmad-Hamdani, Han, Christoffers and Powles2013); one polyploidy E. crus-galli population with a single Asp-2078-Glu mutant allele among the four ACCase gene copies only exhibited 1.7-, 5.7-, and 7.5-fold resistance to the ACCase inhibitors metamifop, cyhalofop-butyl, and fenoxaprop-p-ethyl, respectively (Fang et al. Reference Fang, He, Liu, Li and Dong2020). In such a case, additional mutations in other copies of the target gene and/or additive NTSR mechanisms may be required for the plant to develop resistance to herbicides (Iwakami et al. Reference Iwakami, Hashimoto, Matsushima, Watanabe, Hamamura and Uchino2015). Therefore, the hexaploid E. crus-galli may be more likely to evolve NTSR than TSR to ACCase inhibitors, because six ACCase gene copies are present. This hypothesis may explain why the NX3, NX6, and NX7 populations without any ACCase gene mutations displayed high-level (>10-fold) resistance to cyhalofop-butyl.
Sensitivity to Other ALS and ACCase Inhibitors
Bispyribac-sodium, metamifop, and fenoxaprop-p-ethyl at doses of 1×, 1×, and 1/3× the field recommended application rate, respectively, were used to examine the responses of E. crus-galli to other ALS- and ACCase-inhibiting herbicides. All susceptible populations (NX5, NX8, NX11, and NX12) were well suppressed by the application rates for three herbicides, with fresh weight less than 15% of that of untreated controls (Figure 5).
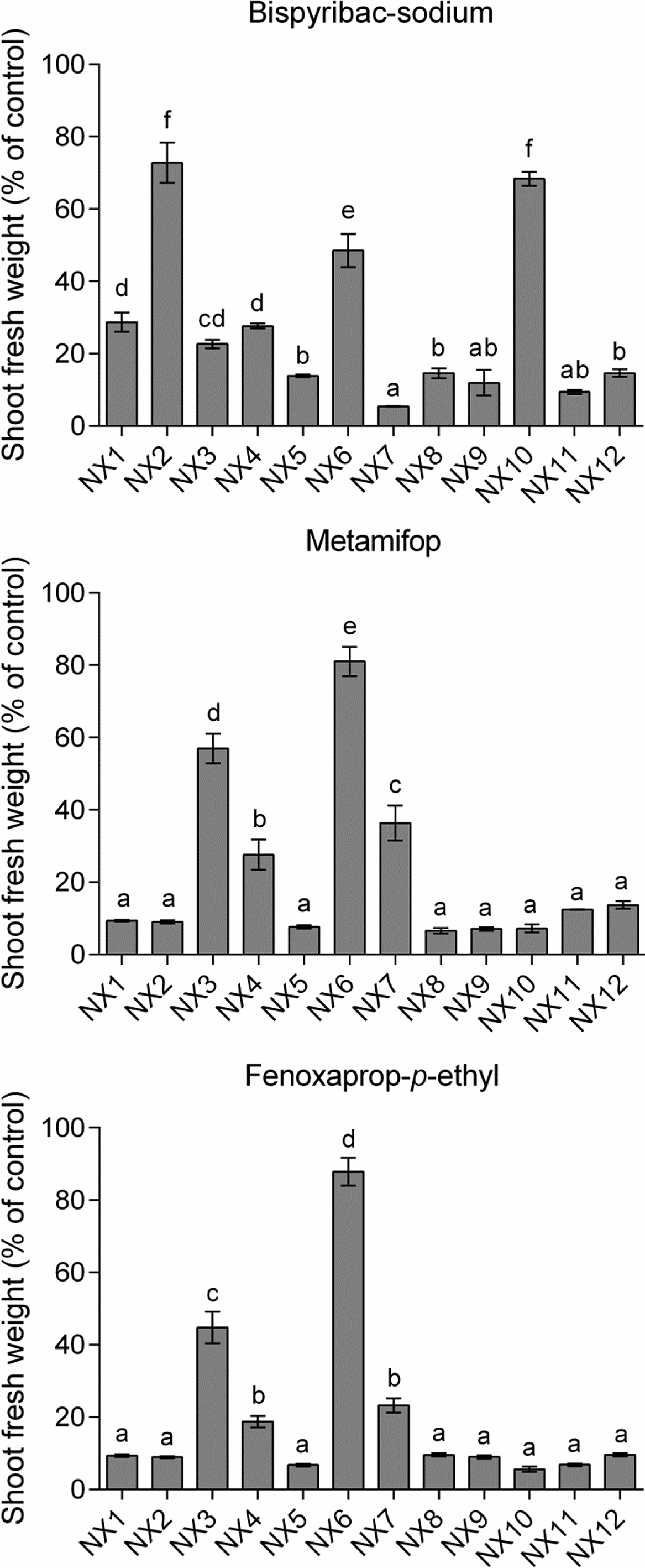
Figure 5. Responses of 12 Echinochloa crus-galli populations to the ALS inhibitor bispyribac-sodium and ACCase inhibitors metamifop and fenoxaprop-p-ethyl. The herbicides were applied at 45, 120, and 20.7 g ai ha−1, respectively. Shoot fresh weight is shown as percentage of untreated control. Bars indicate standard error (n = 3). One-way ANOVA was conducted for each treatment. Bars shown with the same letter are not significantly different (p = 0.05).
The single-dose assays indicated that six (NX1, NX2, NX3, NX4, NX6, and NX10) out of eight penoxsulam-resistant populations showed resistance to the ALS inhibitor bispyribac-sodium, with 28.7%, 72.8%, 22.7%, 27.6%, 48.5%, and 68.4% shoot fresh weight of untreated control, respectively (Figure 5). Among them, the NX2 and NX10 populations with the Trp-574-Leu substitutions were the most resistant.
The four cyhalofop-butyl–resistant populations (NX3, NX4, NX6, and NX7) showed cross-resistance to the ACCase inhibitors metamifop and fenoxaprop-p-ethyl. Of these, NX6 showed the highest level of resistance to metamifop and fenoxaprop-p-ethyl, with more than 80% shoot weight compared with controls in both applications. NX3 exhibited 56.9% and 44.8% weight of controls for metamifop and fenoxaprop-p-ethyl treatments, respectively. In addition, NX4 and NX7 survived treatment with both herbicides, although growth was somewhat inhibited (fresh weight less than 40% of controls in both cases).
In conclusion, 4 of 12 E. crus-galli populations evolved multiple herbicide resistance to both penoxsulam and cyhalofop-butyl. Moreover, three alternative herbicides, bispyribac-sodium, metamifop, and fenoxaprop-p-ethyl, cannot effectively suppress the growth of MHR populations. Therefore, herbicides with other sites of action, rather than ALS and ACCase inhibitors, and efficient PRE herbicides should be considered as alternatives to control the E. crus-galli populations. In addition, in view of the rapid spread of MHR populations in E. crus-galli and the restricted development of new herbicides, diversified and sustainable weed management strategies, such as biological control, precision pesticide application, and automatic weeding technology should be encouraged to counteract resistance in the field.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (2018M642193); the National Natural Science Foundation of China (31901904, 31772183, and 31672042); and the Priority Academic Program of Jiangsu Province (PAPD-2018-87). No conflicts of interest have been declared.