Introduction
A common mode of tolerance to herbicides in agronomic crops is by metabolism brought about by enzyme systems such as cytochrome P450s (CYPs), glutathione S-transferases (GSTs), and glucosyl transferases (GTs). These enzymes, as well as cofactors such as reduced glutathione (GSH), are activated by certain chemicals called safeners (Riechers et al. Reference Riechers, Kreuz and Zhang2010). Safeners are applied in combination with herbicides to provide tolerance in grass crops such as wheat (Triticum aestivum L.), rice (Oryza sativa L.), corn (Zea mays L.), and grain sorghum [(Sorghum bicolor (L.) Moench.] against certain thiocarbamate, chloroacetamide, sulfonylurea (SU), and aryoxyphenoxypropionate (AOPP) herbicides applied PRE or POST. Metabolism of herbicides usually occurs in three phases: a conversion of the herbicide molecule into a more hydrophilic metabolite (Phase 1); followed by conjugation to biomolecules such as glutathione/sugar (Phase 2); and further conjugation/breakup/oxidation reactions with subsequent transport to vacuoles or cell walls, where additional breakdown or sequestration occurs (Phase 3).
The next and most important phase after the confirmation of herbicide resistance in a weed population is the deciphering of the underlying resistance mechanism(s), which can greatly determine the effectiveness of resistance management strategies. One of the common mechanisms of resistance is metabolic deactivation, whereby the herbicide active ingredient is transformed to nonphytotoxic metabolites (Yu and Powles Reference Yu and Powles2014).
An immediate and urgent challenge for weed scientists is to understand and characterize the basis of metabolic resistance to sustain the limited herbicide portfolio and develop integrated weed management strategies. Metabolic resistance research in weeds has mostly been limited to grass species such as rigid ryegrass (Lolium rigidum Gaudin), blackgrass (Alopecurus myusuroides Huds.), and Echinochloa spp. However, dicot species such as waterhemp [Amaranthus tuberculatus (Moq.) J. D. Sauer] and Palmer amaranth (Amaranthus palmeri S. Watson) have evolved resistance to herbicides with different mechanisms of action by enhanced metabolic degradation. Thus, both grass and dicot weed species that develop metabolic herbicide resistance can pose a severe management challenge.
The main objective of this symposium was to gain an understanding of current research on metabolic resistance in weeds by revisiting the history of related research, including crop tolerance; reporting recent advances; and identifying future research opportunities. This report is not an exhaustive all-encompassing review of herbicide metabolism in crops and weeds; it is a compilation of papers presented at a symposium during the 2018 Weed Science Society of America annual meeting.
Complex Signaling, Defense, and Detoxification Pathways in Safener-treated Grain Sorghum Shoots
Induction of herbicide-detoxification enzymes catalyzing Phase I to III metabolic reactions by safeners is well documented (Cummins et al. Reference Cummins, Dixon, Freitag-Pohl, Skipsey and Edwards2011; Theodoulou et al. Reference Theodoulou, Clark, He, Pallett, Cole and Hallahan2003; Zhang et al. Reference Zhang, Xu, Lambert and Riechers2007) and has been reviewed extensively in recent years (Kraehmer et al. Reference Kraehmer, Laber, Rosinger and Schulz2014; Riechers and Green Reference Riechers and Green2017; Riechers et al. Reference Riechers, Kreuz and Zhang2010). However, identification of signaling genes and pathways leading to safener-induced herbicide metabolism has remained mostly elusive. Recent research findings have indicated oxidized lipids, or oxylipins, play an important role in plant defense responses to abiotic and biotic stresses (Hou et al. Reference Hou, Ufer and Bartels2016; Mueller and Berger Reference Mueller and Berger2009) and may also play a key role in safener-mediating signaling (Brazier-Hicks et al. 2018; Matsumoto et al. Reference Matsumoto, Riechers, Lygin, Baluska and Sivaguru2015; Riechers et al. Reference Riechers, Kreuz and Zhang2010; Skipsey et al. Reference Skipsey, Knight, Brazier-Hicks, Dixon, Steel and Edwards2011). In addition to oxylipins, plant hormones such as salicylic acid and jasmonic acid (JA) regulate many plant responses to pathogen attack or herbivore injury (Gao et al. Reference Gao, Zhu, Pradeep Kachroo and Kachroo2015; Koo Reference Koo2018; Larrieu and Vernoux Reference Larrieu and Vernoux2016) and may also function in safener-regulated responses (Behringer et al. Reference Behringer, Bartsch and Schaller2011).
Although a precise signaling cascade has yet to be established for safener-regulated induction of herbicide detoxification in cereal crops, several new hypotheses and research areas have recently emerged involving oxylipins and other signaling molecules that will be the subject of future studies. In addition to unraveling the complex signaling pathways that lead to the induction of enzymes involved in herbicide detoxification, recent research has also shown that tissue- and cell-specific expression of these enzymes may also play an important role in safener mechanisms of action in cereal crops (reviewed by Riechers et al. Reference Riechers, Kreuz and Zhang2010) and may potentially explain why dicot plants do not respond to safener treatments with increased crop tolerance despite increased gene and protein expression (DeRidder and Goldsbrough Reference DeRidder and Goldsbrough2006). These two topics will be the focus of the following sections.
Oxylipin Involvement in Safener-mediated Signaling
A key finding from research in the mid-2000s was that several classes of oxylipins (Mosblech et al. Reference Mosblech, Feussner and Heilmann2009; Mueller Reference Mueller2004) are detected in plants following exposure to stresses, and subsequent work demonstrated that oxylipins induce the expression of plant defense and detoxification genes that mimic safener-induced genes and proteins (Loeffler et al. Reference Loeffler, Berger, Guy, Durand, Bringmann, Dreyer, von Rad, Durner and Mueller2005; Mueller et al. Reference Mueller, Hilbert, Dueckershoff, Roitsch, Krischke, Mueller and Berger2008; Riechers et al. Reference Riechers, Kreuz and Zhang2010; Zhang et al. Reference Zhang, Xu, Lambert and Riechers2007). Two major categories of oxylipins have been detected in plants (Cuyamendous et al. Reference Cuyamendous, Leung, Durand, Lee, Oger and Galano2015; Durand et al. Reference Durand, Bultel-Poncé, Guy, El Fangour, Rossi and Galano2011; Mosblech et al. Reference Mosblech, Feussner and Heilmann2009; Mueller and Berger Reference Mueller and Berger2009): (1) phytoprostanes and phytofurans, which are categorized based on their nonenzymatic formation via interaction of reactive oxygen species with α-linolenic acid (ALA); and (2) enzymatic conversion of ALA to 12-oxo-phytodienoic acid (OPDA) and subsequent ß-oxidation to yield JA (Figure 1). Interestingly, the enzyme catalyzing conversion of OPDA to 3-oxo-2-(2-pentenyl)-cyclopentaneoctanoic acid (OPC-8:0, the precursor of JA), OPDA reductase (OPR), has been frequently identified in transcript- or protein-profiling studies of plant responses to stress (Okazaki and Saito Reference Okazaki and Saito2014; Taki et al. Reference Taki, Sasaki-Sekimoto, Obayashi, Kikuta, Kobayashi, Ainai, Yagi, Sakurai, Suzuki, Masuda, Takamiya, Shibata, Kobayashi and Ohta2005; Yan et al. Reference Yan, Christensen, Isakeit, Engelberth, Meeley, Hayward, Emery and Kolomiets2012), including safener-treated plants and tissues (Riechers et al. Reference Riechers, Kreuz and Zhang2010; Rishi et al. Reference Rishi, Muni, Kapur, Nelson and Goyal2004; Zhang et al. Reference Zhang, Xu, Lambert and Riechers2007).

Figure 1 Representative structures of oxidized lipids (oxylipins) formed in plants. Two classes of oxylipins are generated from α-linolenic acid as substrate; either nonenzymatically formed (A, generalized phytofuran; or B, phytoprostane) via interaction with reactive oxygen species or enzymatically synthesized (C, jasmonic acid). For more details on structures and biosynthetic pathways see Cuyamendous et al. (Reference Cuyamendous, Leung, Durand, Lee, Oger and Galano2015), Durand et al. (Reference Durand, Bultel-Poncé, Guy, El Fangour, Rossi and Galano2011), and Mosblech et al. (Reference Mosblech, Feussner and Heilmann2009).
Recent research has investigated possible links between oxylipin-mediated defense signaling and safener mechanism of action. The tau-class AtGSTU19 enzyme catalyzed the conjugation of GSH to OPDA (Dixon and Edwards Reference Dixon and Edwards2010), leading to a reduction in GSH reactivity. As mentioned earlier, OPR enzymes reduce the double bond in the cyclopentenone ring of OPDA, resulting in an analogous reduction in reactivity (i.e., electrophilicity) but also leading toward biosynthesis of JA (Mueller and Berger Reference Mueller and Berger2009). Root cultures from Arabidopsis mutants defective in fatty-acid desaturation (fad3-2/fad7-2/fad8), which are impaired in forming the oxylipin precursor ALA, demonstrated a decreased ability to respond to safener treatment when AtGSTU24 expression was measured and compared with expression in wild-type Arabidopsis (Skipsey et al. Reference Skipsey, Knight, Brazier-Hicks, Dixon, Steel and Edwards2011). Because these fad mutants accumulate linoleic acid (18:2) instead of ALA (18:3), they are unable to synthesize OPDA or phytoprostanes from ALA substrate released via lipase activities (Christeller and Galis Reference Christeller and Galis2014). The decreased ability of these mutant lines to respond to safener treatment via induction of GST expression is consistent with a link between safener-regulated responses and endogenous oxylipin signaling.
Based on the literature regarding oxylipin-regulated gene expression (Mueller and Berger Reference Mueller and Berger2009) and recent results with fad mutants in Arabidopsis (Skipsey et al. Reference Skipsey, Knight, Brazier-Hicks, Dixon, Steel and Edwards2011), it was postulated that certain oxylipins may not only rapidly induce genes involved in herbicide detoxification pathways but may also confer safener activity in cereals (Brazier-Hicks et al. 2018; Riechers et al. Reference Riechers, Kreuz and Zhang2010). To directly test this hypothesis, a series of compounds modeled on oxylipin structures were chemically synthesized and tested for biological activity as herbicide safeners in rice (Brazier-Hicks et al. 2018), in comparison with the commercial rice safener fenclorim. Three of the 21 compounds tested rapidly induced GST expression in Arabidopsis, but only showed minor whole-plant safening activity against pretilachlor herbicide in rice seedlings. In addition to possible species-specific differences in responses to these potential crop-safening compounds (Brazier-Hicks et al. 2018), metabolic pathways and turnover rates of oxylipins (Dueckershoff et al. Reference Dueckershoff, Mueller, Mueller and Reinders2008) may differ significantly from those of commercial safeners in tissues of cereal crop seedlings (Miller et al. Reference Miller, Irzyk, Fuerst, McFarland, Barringer, Cruz, Eberle and Föry1996; Riechers et al. Reference Riechers, Kreuz and Zhang2010), therefore requiring further investigation.
Organ-, Tissue-, and Cell-Specific Expression of Safener-induced Detoxification Enzymes
As described previously, although the precise signaling pathway(s) that regulate gene expression within herbicide detoxification pathways have not been elucidated, previous research demonstrated that tau-class GST proteins and GST enzyme activities involved in herbicide detoxification are highly expressed in the outermost cells of wheat seedling coleoptiles after safener treatment (Riechers et al. Reference Riechers, Zhang, Xu and Vaughn2003). Interestingly, similar results were found in safener-treated sorghum coleoptiles using the same tau-class wheat GST antiserum (Figure 2). Additional research examining stress-responsive gene expression in Arabidopsis cell cultures (Mueller et al. Reference Mueller, Hilbert, Dueckershoff, Roitsch, Krischke, Mueller and Berger2008) and protein abundance in leaves (Dueckershoff et al. Reference Dueckershoff, Mueller, Mueller and Reinders2008) showed that oxylipins (such as phytoprostanes or OPDA) trigger detoxification and defense responses in a manner similar to safener treatments. Current experiments were designed to test the hypothesis that safeners and phytoprostanes induce GST activity and the expression of genes related to plant defense and detoxification in sorghum shoot coleoptiles in an analogous manner (Riechers et al. Reference Riechers, Ma, Baek, Goodrich, Lygin and Brown2018). A cryostat-microtome sectioning method was developed to extract high-quality RNA from the outermost cells of frozen coleoptiles (excluding leaf tissues) for transcript profiling to enrich for safener- and phytoprostane-responsive mRNAs at different time points after treatment (Riechers et al. Reference Riechers, Ma, Baek, Goodrich, Lygin and Brown2018). Current localization experiments are using an antiserum raised against a specific phi-class sorghum GST isozyme (SbGSTF1) to further investigate tissue-specific expression of different GST subclasses (Labrou et al. Reference Labrou, Papageorgiou, Pavli and Flemetakis2015) in safener-treated grain sorghum seedling tissues (as shown in Figure 2).

Figure 2 Tissue distribution of glutathione S-transferase (GST) proteins in a cross section of etiolated grain sorghum seedlings, probed with an antiserum raised against the tau-class TtGSTU1 protein from wheat (Riechers et al. Reference Riechers, Zhang, Xu and Vaughn2003). (A) Unsafened (DMSO only) seedling, no primary antiserum (negative control); (B) unsafened (DMSO only) seedling, probed with a 1:500 dilution of primary antiserum raised against TtGSTU1; (C) seedling treated with 10 µM fluxofenim safener for 12 h, probed with a 1:500 dilution of primary antiserum raised against TtGSTU1. Red arrows in C mark the massive accumulations of immunoreactive GST proteins in the outermost coleoptile and epidermal cells. Abbreviations: CL, coleoptile; LP, inner leaf primordia.
Initial RNA-seq results have identified >10-fold increases in transcripts of several detoxification genes, including multiple GSTs, CYPs, and GTs, in safener-treated seedlings compared with untreated controls (unpublished data). Moreover, transcripts encoding proteins related to plant development and defense were highly upregulated by safener, such as enzymes involved in lipid signaling (including OPRs), hormone-related processes (i.e., synthesis of benzoic acid and salicylic acid), or auxin metabolism and homeostasis. Transcripts encoding biosynthetic enzymes possibly involved in chemical defense mechanisms in roots (Cook et al. Reference Cook, Rimando, Clemente, Schröder, Dayan, Nanayakkara, Pan, Noonan, Fishbein, Abe, Duke, Scott and Baerson2010) and shoots (Busk and Möller Reference Busk and Möller2002; Halkier and Möller Reference Halkier and Möller1989) of sorghum seedlings were also strongly induced by safener treatment in coleoptile tissues (unpublished data). These results indicate that safeners may be utilizing signaling pathways and enzymatic mechanisms related to generating allelochemicals (Baerson et al. Reference Baerson, Sanchez-Moreiras, Pedrol-Bonjoch, Schulz, Kagan, Agarwal, Reigosa and Duke2005) or other defense chemicals against abiotic or biotic stresses, as well as upregulating enzymes with the putative function of preventing autotoxicity from these chemicals in sorghum seedlings (Bjarnholt et al. Reference Bjarnholt, Neilson, Crocoll, Jørgensen, Motawia, Olsen, Dixon, Edwards and Møller2018).
Future Research Directions
Ongoing analyses using bioinformatics and comparative gene expression approaches are aimed at further mining these RNA-seq data to provide additional insight into how transcriptional responses are reprogrammed in sorghum coleoptiles following safener treatment (unpublished data). An emerging hypothesis is that safeners regulate a specific, coordinated, and rapid defense and detoxification response in cereal crop seedlings, which includes both up- and downregulation of gene expression. This research helps to elucidate the yet-to-be discovered mechanisms that trigger specific detoxification responses related to safener-regulated protection of cereal crops and, as mentioned previously, may also provide insights into the perplexing question of why safeners do not protect dicot crops from herbicide injury (DeRidder and Goldsbrough Reference DeRidder and Goldsbrough2006; Riechers and Green Reference Riechers and Green2017).
In summary, herbicide safeners are unique organic molecules used for crop protection. Safeners increase the success of commercializing new herbicides by providing a chemical tool to enhance crop tolerance and/or crop–weed selectivity for active ingredients that otherwise might be removed from primary screens due to inadequate crop safety (Riechers and Green Reference Riechers and Green2017), therefore providing an alternative to creating genetically modified crops (Goodrich et al. Reference Goodrich, Butts-Wilmsmeyer, Bollero and Riechers2018; Kraehmer et al. Reference Kraehmer, Laber, Rosinger and Schulz2014). Furthermore, safeners may expand the utility of existing herbicides that do not exhibit adequate crop tolerance without a safener as well as expand our basic knowledge of plant responses to abiotic stresses.
Contributions of Metabolism to Clomazone Activity and Selectivity
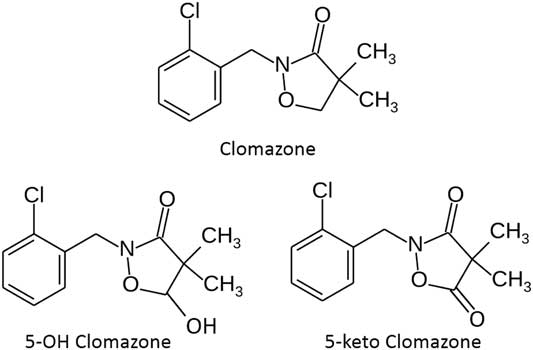
Clomazone (Figure 3), a 3-isoxazolidinone, was initially introduced by FMC Corporation in the 1980s for weed management in soybean [Glycine max (L.) Merr.] (Chang et al. Reference Chang, Knoz, Aly, Sticker, Wilson, Krog and Dickinson1987). Since that time, use of clomazone (also known in the literature as FMC 57020 and dimethazone) expanded to several additional crops (Anonymous 2018). Clomazone injury manifests itself as bleaching of new leaves (Duke and Kenyon Reference Duke and Kenyon1986). However, attempts to tie the clomazone mechanism of action to inhibition of phytoene desaturase or steps in the cytoplasmic isoprenoid biosynthesis pathway were unsuccessful (Croteau Reference Croteau1992; Lutzov et al. Reference Lutzov, Beyer and Kleining1990; Weimer et al. Reference Weimer, Balke and Buhler1992).

Figure 3 Structures of clomazone, 5-OH clomazone, and 5-keto clomazone.
Seeking to expand the uses of clomazone shortly after its commercialization, FMC explored the use of safeners. Naphthalic anhydride seed treatment afforded some protection from clomazone injury to corn, but this system was never commercially developed. However, the organophosphate insecticides phorate and disulfoton could protect cotton (Gossypium hirsutum L.) from clomazone injury (Culpepper et al. Reference Culpepper, York, Marth and Corbin2001). This is still a commercial practice. The clomazone label (Anonymous 2018) contains specific language regarding use of the insecticides to protect cotton from clomazone damage: “Do not apply Command 3ME Herbicide to cotton unless disulfoton or phorate organophosphate insecticide is applied in-furrow with the seed at planting time” and “Failure to apply either disulfoton or phorate insecticides with Command in accordance with in-furrow label use directions can result in crop phytotoxicity (bleaching) and/or stand reduction.”
Phorate Effects on Clomazone Injury and Metabolism
A series of experiments were initiated, in cooperation with FMC, to understand the mechanism of organophosphate safening of cotton from clomazone, and the results were originally published in two articles (Ferhatoglu et al. [Reference Ferhatoglu, Avdiushko and Barrett2005] and Ferhatoglu and Barrett [Reference Ferhatoglu and Barrett2006]). Briefly, the experimental system employed was to place 7-d-old cotton seedlings into hydroponic solution with or without clomazone and with or without phorate. The chlorophyll and carotenoid content of the leaves emerging after the beginning of the treatment was measured 6 d after the start of the experiment. Complete experimental details are in Ferhatoglu et al. (Reference Ferhatoglu, Avdiushko and Barrett2005).
Clomazone (100 nM) reduced the levels of both chlorophyll and carotenoids in the new cotton leaves approximately 80% (Figure 4). Phorate (50 μM) partially reversed this reduction, while 0.5 and 5 μM phorate were ineffective.

Figure 4 Effect of phorate on chlorophyll and carotenoid levels in new leaves of cotton seedlings treated with 100 nM clomazone for 6 d.
Phorate and other organophosphate insecticides are known inhibitors of CYPs (Baerg et al. Reference Baerg, Barrett and Polge1996; Diehl et al. Reference Diehl, Stoller and Barrett1995; Kreuz and Fonne-Pfister Reference Kreuz and Fonne-Pfister1992; Mougin et al Reference Mougin, Polge, Scalla and Cabanne1991). They can act as herbicide synergists by blocking the CYP-mediated detoxification of an active herbicide molecule (Ahrens Reference Ahrens1990; Chample and Shaner Reference Chample and Shaner1982).
To test whether phorate affected clomazone metabolism in cotton plants, the roots of cotton seedlings were incubated for 8 h in [14C]clomazone with or without 50 μM phorate; this was followed by a 16-h chase period. The phorate reduced clomazone metabolism in the shoots, but not the roots (Table 1). The phorate treatment had no effect on the unextracted radioactivity. Phorate also reduced clomazone metabolism in excised cotton shoots fed [14C]clomazone with or without phorate through the cut stem (Ferhatoglu et al. Reference Ferhatoglu, Avdiushko and Barrett2005).
Table 1 Effect of 50 μM phorate on [14C]clomazone metabolism in shoots and roots of 7-d-old cotton seedlings. a

a Seedling roots in hydroponic solution were exposed to [14C]clomazone with and without phorate for 8 h followed by a 16-h chase period before extraction.
b Mean ± SD.
c A significant difference at P ≤ 0.05 compared with the control (no phorate) within the same tissue and within the same column.
Clomazone Metabolism in Microsomes
Isolated microsomes are an experimental system that can be used for in vitro studies of pesticide, including herbicide, metabolism by plant CYPs. While cotton microsomes with the capacity to metabolize herbicides had been isolated (Frear Reference Frear1968; Frear et al. Reference Frear, Swanson and Tanaka1969), microsomes isolated from etiolated corn shoots were used. Phorate does reduce the effects of clomazone on chlorophyll and carotenoid levels in new leaves of corn seedlings (Ferhatoglu et al. Reference Ferhatoglu, Avdiushko and Barrett2005).
Clomazone metabolism was present in microsomes prepared from etiolated 3-d-old corn seedlings (Ferhatoglu et al. Reference Ferhatoglu, Avdiushko and Barrett2005). Three clomazone metabolites eluting at 12.6, 15.4, and 23 min were produced in the microsomes (Table 2). Naphthalic induced activity for the metabolites eluting at 15.4 and 23 min but not at 12.6 min (Ferhatoglu et al. Reference Ferhatoglu, Avdiushko and Barrett2005). The metabolite eluting at 12.6 min was not NADPH dependent, so it is not a product of CYP activity. Production of the metabolite eluting at 23 min was totally inhibited by phorate, while the production of the metabolite at 15.4 was unaffected. This showed that there were two NADPH-dependent clomazone metabolism activities present in the corn microsomes, presumably CYP mediated, and that one was sensitive to phorate inhibition while the other was not. The clomazone metabolite standards 2-chlorobenzyl alcohol and 5-OH clomazone, supplied by FMC, eluted at 15.4 and 23 min, respectively. Therefore, the phorate sensitive activity is presumed to be the production of 5-OH clomazone from clomazone.
Table 2 Induction of [14C]clomazone metabolism in corn microsomes by seed treatment with naphthalic anhydride (0.5% w/w), seedling treatment with ethanol (10% v/v), or a combination of the two.

a Mean ± SD. Means within a column followed by different letters are significantly different at P ≤ 0.05.
The 5-OH clomazone can also cause bleaching in cotton seedlings, reducing both chlorophyll and carotenoid levels in the plants (unpublished data). The 5-OH clomazone was approximately 10% as toxic as clomazone, which is consistent with data presented by Chang et al. (Reference Chang, Knoz, Aly, Sticker, Wilson, Krog and Dickinson1987). However, phorate was ineffective as a safener for 5-OH clomazone.
Bioactivation of Clomazone
From this information, a working hypothesis was formed that phorate inhibited the CYP responsible for the conversion of clomazone to 5-OH clomazone (Figure 3), but phorate was ineffective in preventing the formation of the actual toxicant, 5-keto clomazone (Figure 3). This hypothesis was based on the metabolic pathway for clomazone in soybean (El-Naggar et al. Reference El-Naggar, Creekmore, Schocken, Rosen and Robinson1992), which has multiple pathways for clomazone degradation, including the formation of 5-keto clomazone. In addition, 5-keto clomazone is phytotoxic (Chang et al. Reference Chang, Knoz, Aly, Sticker, Wilson, Krog and Dickinson1987). Finally, with the discovery of the plastidic isoprenoid pathway (Lichtenthaler Reference Lichtenthaler1999; Lichtenthaler et al. Reference Lichtenthaler, Schwender, Disch and Rohmer1997), it was possible to show that 5-keto-clomazone, but not clomazone or 5-OH clomazone, inhibits plant 1-deoxy-D-xylulose-5-phosphate synthase (DXP synthase; Ferhatoglu and Barrett Reference Ferhatoglu and Barrett2006), the first step in this pathway.
Clomazone Selectivity Is Complicated
In summary, for clomazone to be active, it must be bioactivated to its 5-keto clomazone metabolite to be phytotoxic at its site of action, DXP synthase, the first step in the chloroplastic isoprenoid biosynthesis pathway. The first step in the conversion of clomazone to 5-keto clomazone is the CYP-catalyzed formation of 5-OH clomazone. Organophosphate insecticides such as phorate inhibit this process and can safen a crop like cotton from clomazone. While the formation of 5-keto clomazone from 5-OH clomazone is also likely CYP catalyzed, additional studies would be required to establish this. It would also be interesting to test whether CYP inhibitors other than organophosphates could prevent this conversion. It was apparent from this research that phorate, and presumably other organophosphate insecticides, did not prevent the conversion of 5-OH clomazone to 5-keto clomazone, as the insecticide was ineffective as a safener for 5-OH clomazone.
This all means that clomazone selectivity is complicated. The rates of conversion of clomazone conversion to 5-OH clomazone, 5-OH clomazone to 5-keto clomazone, and the conversion of all three of these compounds to their own metabolites will combine to determine how much 5-keto clomazone will be present in a plant and for how long. The clomazone metabolic pathway in tolerant soybean demonstrates this (El-Naggar et al. Reference El-Naggar, Creekmore, Schocken, Rosen and Robinson1992). More recently, Yasuor et al. (Reference Yasuor, Zou, Tolstikov, Tjeerdema and Fischer2010) proposed that changes in the rates of the conversion of 5-OH clomazone to dihydroxy-clomazone and clomazone to hydroxymethylclomazone plus 3′-hydroxyclomazone all contribute to clomazone resistance in rice barnyardgrass [Echinochloa phyllopogon (Stapf.) Koss].
Glyphosate Metabolism in Crops and Weeds
Glyphosate was introduced to the herbicide market in 1974. It has become the most heavily used herbicide in the world, in large part because of the huge success of glyphosate-resistant (GR) crops (Duke Reference Duke2018). Duke (Reference Duke1988) summarized the literature on glyphosate through the mid-1980s, including its metabolic degradation, and concluded that the evidence for plant metabolism of glyphosate was not conclusive because of the low or no levels of degradation reported, sometimes over long periods of time. Evidence of its degradation by microbes was clear, and it was speculated that reported plant metabolism might have actually been microbial metabolism, either in the plant samples or after extraction. Early work was also limited by difficult analytical methods that have improved with time (Koskinen et al. Reference Koskinen, Marek and Hall2016). It later became unequivocal that many plant species can metabolize glyphosate, especially to aminomethylphosponic acid (AMPA) and glyoxylate (Duke Reference Duke2011). Indeed, metabolism of glyphosate to AMPA found in microbe-free cell cultures of soybean, wheat, and corn proved that plant cells can metabolize glyphosate (Komoßa et al. Reference Komoßa, Gennity and Sandermann1992). In this system, soybean more readily metabolized glyphosate than did wheat or corn.
The two most commonly reported routes of glyphosate metabolism are to AMPA and glyoxylate by a glyphosate oxidoreductase (GOX) and to sarcosine via a glyphosate C-P lyase. In most cases of plant or microbe metabolism of glyphosate, the only metabolite reported is AMPA. This may be because sarcosine is rarely looked for; however, when it has been sought, it has almost always not been found (e.g., Ribeiro et al. Reference Ribeiro, Nandula, Dayan, Rimando, Duke, Reddy and Shaw2015). Metabolism of glyphosate to AMPA has been verified in many plant species now, but the capability for metabolism varies dramatically. For example, Reddy et al. (Reference Reddy, Rimando, Duke and Nandula2008) found a broad range of glyphosate to AMPA metabolism in 11 plant species, ranging from AMPA levels being half, one-fourth, or one-sixth the concentration of glyphosate measured in treated tissues 7 d after application in Illinois bundleflower [Desmanthus illinoensis (Michx.) MacMill. ex B. L. Rob. and Fernald], sicklepod [Senna obtusifolia (L.) H. S. Irwin & Barneby], and cowpea [Vigna unguiculata (L.) Walp.], respectively, to no detectable AMPA in Italian ryegrass [Lolium perenne L. ssp. multiflorum (Lam.) Husnot], corn, and hemp sesbania [Sesbania herbacea (Mill.) McVaugh]. In that study, each species was treated with the glyphosate rate that inhibited growth 50%, so that differential phytotoxicity of glyphosate was unlikely to account for any differences in metabolism. Later work found that corn did metabolize glyphosate to AMPA when GR corn was treated with full recommended rates, and the AMPA as a proportion of glyphosate was very low at most times after application (Bernal et al. Reference Bernal, Martin, Soto, Nozal, Marotti, Dinelli and Bernal2012; Reddy et al. Reference Reddy, Cizdziel, Williams, Maul, Rimando and Duke2018). So species that are not reported to metabolize glyphosate may metabolize it very slowly with levels of AMPA too low to detect at specific times after application. GR crops are ideal for studying glyphosate metabolism, because high rates of glyphosate are not phytotoxic, allowing enzymatic degradation to occur unhindered with ample substrate. Glyphosate metabolism in GR soybeans, maize, and canola (Brassica napus L.) has been studied.
Metabolism of glyphosate to AMPA is significant in GR soybean, with levels of ~2 to 3 and 7 to 25 μg of glyphosate and AMPA g−1 of dry harvested seed, respectively, from plants treated with 1.26 kg ae ha−1 of glyphosate at full bloom or 8 wk after planting (Duke et al. Reference Duke, Rimando, Pace, Reddy and Smeda2003). This is not surprising, as glyphosate is phloem mobile, translocating to metabolic sinks such as meristems and developing seeds (Duke Reference Duke1988). Others have found similar levels of glyphosate and AMPA in seed for GR soybean (Bohm et al. Reference Bohm, Rombaldi, Genovese, Castilhos, Alves and Rumjanek2014; Bøhn et al. Reference Bøhn, Cuhra, Traavik, Sanden, Fagan and Primicerio2014). In another study in which GR soybean was treated with 0.87 kg ha−1 glyphosate at both 5 and 7 wk after planting, very high levels of glyphosate (8 to 15 μg g−1 ) but very little AMPA (~0.1 μg g−1) was detected in leaf tissues (Duke et al. Reference Duke, Rimando, Reddy, Cizdziel, Bellaloui, Shaw, Williams and Maul2018). Harvested seed had much lower levels of glyphosate, but relatively more AMPA. Whether the AMPA in the seed was translocated there or was formed by metabolism of glyphosate in the seed is unknown.
GR crops are essentially unharmed by glyphosate at rates recommended for weed management (Nandula et al. Reference Nandula, Reddy, Rimando, Duke and Poston2007). But AMPA is moderately phytotoxic (Hoagland Reference Hoagland1980), and GR crops are not resistant to AMPA (Reddy et al. Reference Reddy, Duke and Rimando2004). Thus, if enough AMPA were formed in a GR crop, it might cause phytotoxicity. Under some environmental conditions, farmers have observed moderate chlorosis after treatment of GR soybean with glyphosate (termed “yellow flash” by farmers). This is a transient effect that does not ultimately influence grain yield. In greenhouse studies, this effect was found with AMPA and glyphosate treatments that resulted in the same in vivo AMPA concentrations, whether from AMPA or glyphosate treatment (Reddy et al. Reference Reddy, Duke and Rimando2004). These results suggest that yellow flash is due to accumulation of sufficient AMPA for noticeable chlorosis.
GR corn metabolizes glyphosate to AMPA, but at a much lower rate than GR soybean does (Bernal et al. Reference Bernal, Martin, Soto, Nozal, Marotti, Dinelli and Bernal2012: Reddy et al. Reference Reddy, Cizdziel, Williams, Maul, Rimando and Duke2018). The first GR canola commercially grown was the only GR crop that contained a transgene for a microbial GOX, in addition to a gene for a microbial GR 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), the target of glyphosate (Green Reference Green2009). Very little glyphosate is converted to AMPA in non-GR canola, whereas virtually all of the glyphosate applied to GR canola is converted to AMPA within 7 d, when glyphosate is supplied in small amounts (Corrêa et al. Reference Corrêa, Dayan, Owen, Rimando and Duke2016) (Figure 5). Only AMPA and no glyphosate was found in untreated leaves of treated plants. Whether the AMPA was translocated to untreated leaves or formed by glyphosate metabolism in those leaves was not determined. The relative contributions to resistance of the GOX and GR EPSPS genes are unknown in GR canola, but current commercial varieties of GR canola contain only a GR EPSPS transgene, as do all other GR crops. Thus, whether the canola GOX gene alone would provide adequate resistance for a commercial GR crop is unknown.

Figure 5 Metabolism of [14C]glyphosate 1 DAT in conventional (A, B) and glyphosate-resistant canola (C, D). The data are expressed as percent distribution of total radioactivity in each leaf on the left side and as actual disintegrations per minute per gram dry weight (dpm g−1 DW) of the leaves on the right. Black bars, glyphosate; gray bars, AMPA; white bars, unknown metabolite(s). Error bars are ±1 SE. With permission from Corrêa et al. (Reference Corrêa, Dayan, Owen, Rimando and Duke2016).
Several glyphosate metabolism transgenes have been proposed for generation of GR crops. These include transgenes for an engineered bacterial oxidase (Nicolia et al. Reference Nicolia, Ferradini, Molla, Biagetti, Pollegioni, Veronesi and Rosellini2014), a modified bacterial glyphosate acetyltransferase (GAT) (Siehl et al. Reference Siehl, Castle, Gorton and Keenan2007), and a decarboxylase-type enzyme that inactivates glyphosate (Hammer et al. Reference Hammer, Hinson, Duck and Koziel2007). None of these transgenes have been used in commercial GR crops, although the GAT genes came close to commercialization (Green Reference Green2009).
Two generalizations can be made from existing glyphosate metabolism literature regarding glyphosate metabolism in non-transgenic plants. Legumes tend to have higher rates of glyphosate metabolism than other species, and Poaceae species usually have very low rates of metabolism. However, no methodical, systematic survey exists that uses a consistent experimental design and reliable assay methods of the capacity for glyphosate metabolism in a wide range of taxonomically diverse plant species.
Glyphosate is nonselective, but some species are more tolerant than others. Because glyphosate is perhaps the slowest-acting commercial herbicide, the plant has more time to metabolize it before severe toxicity occurs. Thus, rapid metabolism could be a tolerance mechanism. However, there is no good evidence that this is the case. The ability to metabolize glyphosate did not correlate well with the glyphosate I50 values for 10 species (Reddy et al. Reference Reddy, Rimando, Duke and Nandula2008), although it was speculated that it might contribute to tolerance in some species. A problem with this study is that glyphosate and AMPA were determined at only one point in time after glyphosate application. The dynamics of metabolism are highly likely to vary between species, so determination of the proportion of glyphosate metabolized over a critical period for herbicide damage would be needed to better evaluate the role of metabolism in producing tolerance.
Morningglory species (Ipomoea spp.) are among the most glyphosate-tolerant weeds. Although there is no proof of evolved resistance (Heap Reference Heap2018), some claims have been made without conclusive evidence (e.g., Debban et al. Reference Debban, Okum, Pieper, Wilson and Baucom2015). Glyphosate tolerance of pitted morningglory (Ipomoea lacunosa L.) biotypes varied by as much as 2.6-fold, but this was not explained by differential metabolism (Ribeiro et al. Reference Ribeiro, Nandula, Dayan, Rimando, Duke, Reddy and Shaw2015). The most and least susceptible biotypes both readily metabolized glyphosate to AMPA at about the same rate, but there was differential translocation that was consistent with the differences in susceptibility.
Clearly, many plant species have a means of metabolizing glyphosate to AMPA. Considering the tremendous selection pressure that has resulted in several mechanisms of evolved resistance to glyphosate (Sammons and Gaines Reference Sammons and Gaines2014), one would expect the enzyme(s) responsible for AMPA production to be selected for, either to produce a mutated enzyme with greater efficiency as a GOX or to generate greater amounts of GOX enzyme. In their review, Sammons and Gaines state that metabolism of glyphosate in plants is rare and are skeptical of reports that metabolism is involved in reported resistance of sourgrass [Digitaria insularis (L.) Mez ex Ekman] and horseweed (Erigeron canadensis L.) to glyphosate, partly by conversion to sarcosine (de Carvalho et al. Reference de Carvalho, Alves, González-Torralva, Cruz-Hipolito, Rojano-Delgado, De Prado, Gil-Humanes, Barro and de Castro2012; Gonzalez-Torralva et al. Reference Gonzalez-Torralva, Rojano-Delgado, de Castro, Muellender and De Prado2012b). Similar results were reported by some of the same authors for glyphosate tolerance of velvet bean [Mucuna pruriens (L.) DC.] (Rojano-Delgado et al. Reference Rojano-Delgado, Cruz-Hipolito, de Prado, de Castro and Franco2012). These papers are unusual, both in reporting sarcosine and glyoxylate as plant metabolites of glyphosate and by invoking metabolism as a resistance mechanism. Both sarcosine and glyoxylate are plant metabolites that can be found in untreated plants, so without 14C-labeling of these compounds from [14C]glyphosate, the results are inconclusive. However, they detected AMPA, a compound absent in plants not treated with glyphosate, to support their claims. Using [14C]glyphosate, others have found no metabolism of glyphosate in either susceptible or resistant E. canadenesis or hairy fleabane (Erigeron bonariensis L.) (Feng et al. Reference Feng, Tran, Chiu, Sammons, Heck and CaJacob2004; Dinelli et al. Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006, Reference Dinelli, Marotti, Bonetti, Catizone, Urbano and Barnes2008). Sammons and Gaines (Reference Sammons and Gaines2014) did not include glyphosate metabolism as a proven mechanism of evolved resistance to glyphosate.
More rapid metabolism of glyphosate in GR weeds has been searched for without result in many papers looking for the mechanism of glyphosate resistance. For example, although there were no statistical differences in AMPA found in susceptible and two GR A. palmeri biotypes, the resistant biotypes tended to have more metabolism than the susceptible biotypes (Nandula et al. Reference Nandula, Reddy, Koger, Poston, Rimando and Duke2012). However, none of the biotypes accumulated much AMPA at 7 d after treatment, with AMPA not reaching 1% of the glyphosate levels found in the tissues, except in the susceptible biotype treated with a very low I50 rate (90 g ha−1).
There are still important, unanswered questions about glyphosate metabolism in plants, including the nature of the enzyme(s) that metabolizes glyphosate to AMPA. Glycine oxidases from microbes can act as a GOX enzyme (e.g., Pollegioni et al. Reference Pollegioni, Schonbrunn and Siehl2011). At least some legumes can oxidize glycine in nodules via a leghaemoglobin-associated reaction (Lehtovaara Reference Lehtovaara1978). But, nonnodulated species and leaf tissues of legumes can convert glyphosate to AMPA and glyoxylate. D-amino acid oxidase from Bradyrhizobium japonicum can also oxidize glyphosate to AMPA and glyoxylate, and the transgene for this enzyme can provide glyphosate resistance to Arabidopsis thaliana L. (Han et al. Reference Han, Zhu, Fu, You, Wang, Li, Zhao, Peng and Yao2015). However, equivalent glycine and D-amino acid oxidases have not been identified in plants. The clear ability of some plant species to oxidize glyphosate to AMPA indicates that they have an enzyme similar to these microbial enzymes. The enzymatic or other potential mechanisms of glyphosate conversion to AMPA are still a mystery.
An alternative explanation to direct metabolism of glyphosate by plants is that some plants might have endophytes that can metabolize glyphosate. Evidence is growing that endophytes can contribute to metabolic degradation of herbicides (Tétard-Jones and Edwards Reference Tétard-Jones and Edwards2016), and some plant-associated microbes can clearly metabolize glyphosate (e.g., Kryuchkova et al. Reference Kryuchkova, Burygin, Gogoleva, Gogolev, Chernyshova, Makarov, Fedorov and Turkovskaya2014). The soybean endophyte Burkholderia gladioli is resistant to glyphosate (Kuklinsky-Sobral et al. Reference Kuklinsky-Sobral, Araújo, Mendes, Pizzirani-Kleiner and Azevedo2005), but whether this is due to metabolism is unknown. There is no available proof of any endophyte-mediated glyphosate metabolism in planta. Endophyte infection increases with glyphosate resistance in L. perenne (Handayani et al. Reference Handayani, Tanno, Yamashita, Tobina, Ichihara, Ishida and Sawada2017), but glyphosate metabolism in the populations was not determined, and metabolism has been found to be very low and uninvolved in Lolium perenne ssp. multiflorum resistance to glyphosate in other places in the world (Barroso et al. Reference Barroso, de S Costa, Neto, dos Santos, Balbuena, Carbonari and Alves PLCA2018; Gonzalez-Torralva et al. Reference Gonzalez-Torralva, Gil-Humanes, Barro, Brants and De Prado2012a). It is possible that there has been horizontal gene transfer of a gene for an enzyme with GOX activity. But, it does not appear that the extreme selection pressure caused by glyphosate in recent decades has caused this to occur, as metabolism is not a firmly established mechanism of weed resistance to glyphosate. This supports the view that horizontal gene transfer from microbes to plants is not a common occurrence.
Bioactivation of Natural Phytotoxins: The Exception or the Rule?
Evolution of Bioactive Natural Products
Within ecosystems, plants cohabit in association with a wide variety of microorganisms, insects and nematodes, and other plants. These constant multitrophic interactions have led to the coevolution of positive interactions such as mutualistic and symbiotic relationships and negative interactions such as competitive and parasitic relationships. Within this context, pathogens have adapted to their plant hosts by deploying virulence proteins that either suppress plant immune responses or increase plant susceptibility. Plant–pathogen interactions also involve a form of chemical warfare derived from novel metabolic pathways (Schueffler and Anke Reference Schueffler and Anke2014) that aims at strategically using one’s opponent’s weakness to one’s benefit (Maor and Shirasu Reference Maor and Shirasu2005; Verhoeven et al. Reference Verhoeven, Biere, Harvey and van der Putten2009). There is great interest in using these natural phytotoxins to develop new herbicide chemical classes or discover novel mechanisms of action (Duke and Dayan Reference Duke and Dayan2015).
These secondary metabolic pathways evolve through gene duplication. This slow evolutionary process most often leads to loss of function of the duplicated genes (pseudogenization), but it occasionally leads to a beneficial gain of a new function (neofunctionalization) (Moore and Purugganan Reference Moore and Purugganan2005). Over time, this has resulted in a vast array of structurally diverse biologically active molecules (Flagel and Wendel Reference Flagel and Wendel2009; Ober Reference Ober2005). This evolutionary process is similar to the high-throughput screens developed by all major agrochemical companies searching for new herbicides. While conventional in vitro screens test a large number of compounds on a single enzyme target very rapidly, natural high-throughput processes allow for the identification and customization of molecules that optimize their in vivo activities. More than 200,000 secondary metabolites have been characterized, and it is anticipated that many more will be discovered (Clevenger et al. Reference Clevenger, Bok, Ye, Miley, Verdan, Velk, Chen, Yang, Robey, Gao, Lamprecht, Thomas, Islam, Palmer, Wu, Keller and Kelleher2017).
Natural phytotoxins are typically small molecules that explore chemical spaces not covered by herbicides obtained through the usual organic synthetic approaches. Many of these chemicals have novel mechanisms of action that target enzymes and/or pathways that also exist in the organisms producing them. Consequently, these molecules are often synthesized and/or stored as inactive protoxins that are bioactivated in the target organisms. Bioactivation may take different forms, such as removing protective groups or adding functional groups. Alternatively, some organisms produce the phytotoxins in their active forms but protect themselves from autotoxicity by sequestering them in subcellular compartments or specialized cells (Figure 6). A few examples of the various mechanisms of bioactivation are examined in the following sections.

Figure 6 Various strategies used by organisms that produce protoxins requiring bioactivation before interacting with their respective target sites.
Bioactivation by Removing Protective Groups
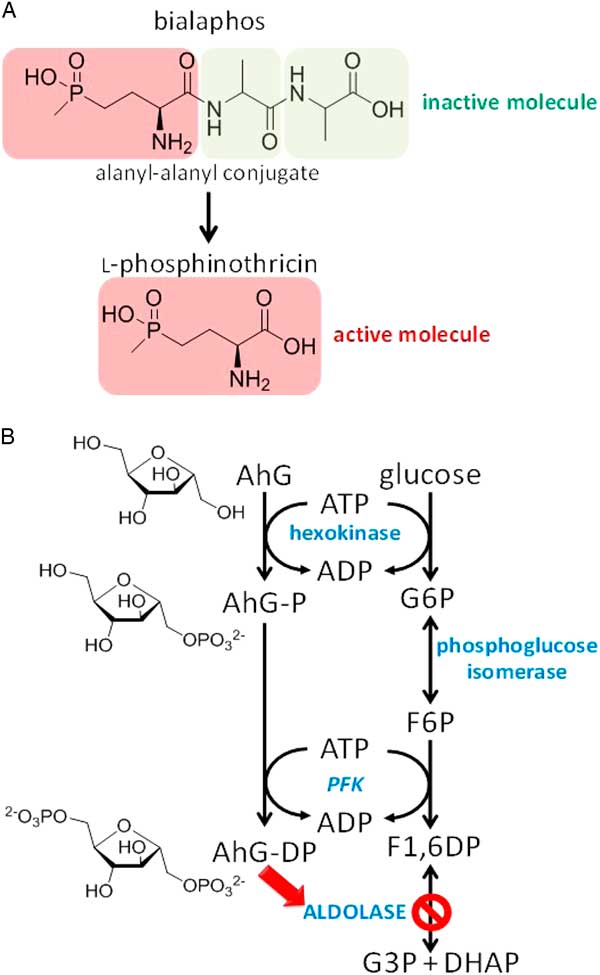
Some organisms produce toxins as protoxins that possesses additional groups to protect themselves from autotoxicity. This is often necessary, because these organisms have enzymes that are sensitive to the toxin they produce. The conversion of the protoxin bialaphos to the herbicidal l-phosphinothricin, the active enantiomer of glufosinate, is perhaps the most well-known and relevant example of bioactivation via cleavage of protective groups (Figure 7A). Bialaphos is a tripeptide (l-alanyl-l-alanyl-phosphinothricin) produced by the soil bacteria Streptomyces hygroscopicus and Streptomyces viridochromogenes. This metabolite is very rapidly bioactivated in planta by removing two alanine residues to release l-phosphinothricin. Phosphinothricin is a potent inhibitor of glutamine synthetase (Wild and Ziegler Reference Wild and Ziegler1989), a key enzyme responsible for glutamine biosynthesis and ammonia detoxification. Organisms producing bialaphos also have specific acetylases that rapidly deactivate l-phosphinothricin as another mechanism of protection against the toxic effect of this bioactive natural product. Another common bioactivation by cleavage involves the removal of glycosides by glucosidases to release herbicidal aglycones, such as is observed with podophyllotoxin produced by the plant belonging to the Podophyllum genus. Podophyllotoxin is an aryltetralin lignin that acts as a natural mitotic inhibitor. While this molecule is used as a precursor for the semisynthesis of several anticancer pharmaceutical drugs because of its antimitotic activity, podophyllotoxin would cause great damage to the plants producing it. Consequently, mayapple (Podophyllum peltatum L.) conjugates podophyllotoxin with a glucose and stores it within the vacuole, preventing it from interacting with the physiologically active cytosol and interfering with numerous microtubule-driven processes. Interestingly, there is a glucosidase present in the cytosol with high specificity for podophyllotoxin-O-glucoside that is mostly inactive at physiological pH of 7 but has its greatest activity at pH 4 (Dayan et al. Reference Dayan, Kuhajek, Canel, Watson and Moraes2003). These biochemical characteristics suggests that this glucosidase would be recruited for the hydrolysis of the glucoside to release the active aglycone upon injury to the leaf, which would cause the pH to drop as the large volume of the vacuole mixes with the cytoplasm.

Figure 7 (A) Example of bioactivation of a protoxin by removal of a protective group. Removal of the alanyl-alanyl conjugate from the inactive protoxin bialaphos produced by Streptomyces hygroscopicus to release the active molecule l-phosphinothricin, a potent inhibitor of plant glutamine synthetases. (B) Example of hijacking of plant biochemical machinery to bioactivate a protoxin by adding an active functionality. Phosphorylation of 2,5-anhydro-d-glucitol produced by Fusarium solani NRRL 18883 via the action of the plant hexokinase and phosphofructokinase, leading to the formation of a fructose-1,6-diphosphate analogue inhibitor of aldolase. Abbreviations: AhG, 2,5-anhydro-d-glucitol; DHAP, dihydroxyacetone phosphate; F-1,6-DP, fructose-1,6-diphosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde 3-phosphate; G6P, glucose-6-phosphate; PFK, phosphofructokinase.
Bioactivation by Adding Functional Groups
Other organisms produce toxins as protoxins that require the addition of functional groups to activate the structures. Some strains of Fusarium solani produce 2,5-anhydro-d-glucitol, a sugar analogue that requires bioactivation to exert activity on its target site (Dayan et al. Reference Dayan, Rimando, Tellez, Scheffler, Roy, Abbas and Duke2002). 2,5-Anhydro-d-glucitol has no known biological activity. However, up to milllimolar concentrations of this molecule is released by F. solani as it invades a plant. Through the process of coevolution this pathogen has exploited the biochemical machinery of its host plant to bioactivate 2,5-anhydro-d-glucitol into a phytotoxin. 2,5-Anhydro-d-glucitol is a sugar analogue that serves as a substrate for two glycolytic kinases endogenous to the host plant (namely hexokinase and phosphofructokinase) (Figure 7B). The biochemical functions of these kinases have been exploited to produce a diphosphate derivative of 2,5-anhydro-d-glucitol. This bioactivated toxin acts as a substrate analogue that inhibits fructose-1,6-diphosphate aldolase, a key step in glycolysis (Dayan et al. Reference Dayan, Rimando, Tellez, Scheffler, Roy, Abbas and Duke2002).
This type of protoxin bioactivation occurs by hijacking native plant kinases to produce active phosphorylated phytotoxins. For example, certain strains of S. hygroscopicus produce hydantocidin. Upon phosphorylation, hydantocidin becomes an analogue of inosine monophosphate, which is a potent inhibitor of adenylosuccinate synthetase, an enzyme involved in purine biosynthesis (Fonne-Pfister et al. Reference Fonne-Pfister, Chemla, Ward, Girardet, Kreuz, Honzatko, Fromm, Schar, Grutter and Cowan-Jacob1996). Similarly, carbocyclic coformycin is a protoxin produced by Sacchavothvix spp., whose mechanism of action requires phosphorylation of its 5′-hydroxy group to produce an irreversible inhibitor of adenosine monophosphate deaminase (Dancer et al. Reference Dancer, Hughes and Lindell1997).
Bioactivation by Altering the Shape of a Phytotoxin
Sometimes, protoxins can be bioactivated as the result of more subtle changes. For example, the gram-negative β-proteobacteria Burkholderia sp. A396 produces large amount of romidepsin, a 16-membered cyclic depsipeptide bridged by a 15-membered macrocyclic linked via a disulfide bridge. Reduction of the disulfide bridge is catalyzed in planta by native enzymes and opens up the 15-membered macrocyclic structure to release a long side chain. This increases the potency of romidepsin on plant histone deacetylases.
Bioactivation by Release from Cellular Sequestration
Not all natural phytotoxins require bioactivation. In these cases, the metabolites are already highly toxic, and organisms have devised schemes to protect themselves from the lethal effect of these compounds by compartmentalizing or exuding them. For example, leptospermone is a metabolite produced by several plant species (e.g., Callistemon spp., Leptospermum spp., and Eucalyptus spp.). This molecule and several other analogues produced by these plants are potent inhibitors of p-hydroxyphenylpyruvate dioxygenase (HPPD) (Dayan et al. Reference Dayan, Duke, Sauldubois, Singh, McCurdy and Cantrell2007). In fact, leptospermone served as a template for the development of HPPD-inhibiting herbicides (Beaudegnies et al. Reference Beaudegnies, Edmunds, Fraser, Hall, Hawkes, Mitchell, Schaetzer, Wendeborn and Wibley2009). This β-triketone natural product is produced exclusively in schizogenous glands (Figure 8A). This allows the production of a potent toxin in a cellular compartment isolated from the rest of the physiologically active portion of the plant that possesses endogenous HPPD enzyme sensitive to its own toxin.

Figure 8 Examples of cellular compartmentalizations. (A) Leptospermone is a potent p-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitor produced by a number of plant species, including Callistemon spp., Leptospermum spp., and Eucalyptus spp. This natural β-triketone is a potent inhibitor of HPPD. Plants produce it in specialized glands isolated from the rest of the cells to avoid autotoxicity problems. (B) Sorgoleone is a potent phytotoxin produced by most members of the Sorghum genus. This lipid benzoquinone is produced exclusively in root hairs and exuded into the rhizosphere.
As another example, sorgoleone is produced in most of the species in the Sorghum genus. This lipid benzoquinone is a potent photosystem II (PSII) inhibitor and competes for the binding site of plastoquinone on the Qb binding site in fashion similar to the herbicide atrazine (Gonzalez et al. Reference Gonzalez, Kazimir, Nimbal, Weston and Cheniae1997). Isolated chloroplasts of sorghum are just as sensitive to sorgoleone as other plant species. However, sorghum is able to produce large amounts of this toxin by compartmentalizing its biosynthesis to specialized root hair cells that rapidly exude it from the root into the rhizosphere (Dayan et al. Reference Dayan, Howell and Weidenhamer2009) (Figure 8B). Because sorgoleone does not translocate from the root to the foliage, sorghum remains uninjured while repressing the growth of small-seeded weeds germinating within its rhizosphere.
Bioactivation of Natural Phytotoxin—The Exception or the Rule?
The short review above clearly shows that many phytotoxins are produced as protoxins or must be sequestered by the producing organism to avoid autotoxicity. This is particularly true if the pathogen producing the toxin possess the enzyme target site affected by the active form of the toxin. These complex systems are the results of coevolution between plant biotic interactions that cannot be achieved by the current approaches used by the agrochemical industry. As to whether bioactivation of natural phytotoxins is the exception or the rule, it is difficult to make a definitive statement. Many toxins are active in themselves and do not require bioactivation. However, evolutionary forces have clearly identified processes that can be hijacked to bioactivate certain molecules. These processes enable organisms to produce protoxins that will only be active in the target organism.
Glutathione Transferases Associated with Non–Target Site Resistance to Herbicides in Alopecurus myosuroides
GSTs (EC 2.5.1.18) are a superfamily of detoxifying enzymes that have evolved into six discreet clades classified as the zeta, theta, phi, tau, lambda, and dehydroascorbate reductase classes in plants (Dixon and Edwards Reference Dixon and Edwards2010). GSTs from the tau (GSTU) and phi (GSTF) classes have long been associated with herbicide tolerance and selectivity in crops through their ability to rapidly conjugate and detoxify a range of chemistries (Cummins et al. Reference Cummins, Dixon, Freitag-Pohl, Skipsey and Edwards2011). Normally, the expression of GSTs is an order of magnitude lower in weeds than in crops, leading to a corresponding reduced ability to detoxify herbicides (Dixon and Edwards Reference Dixon and Edwards2010). Repeated selection with herbicides can lead to enhanced GST expression in weeds, leading in turn to resistance. Consistent with this, transcripts encoding GSTFs and GSTUs have been recently identified as being enhanced in populations exhibiting non–target site herbicide resistance (NTSR) in several weed species; notably wild oat (Avena fatua L.) (Keith et al. Reference Keith, Burns, Bothner, Carey, Mazurie, Hilmer, Biyiklioglu, Budak and Dyer2017), American sloughgrass [Beckmannia syzigachne (Steud.) Fernald] (Pan et al. Reference Pan, Wang, Cai, Gao, Zhao and Dong2016), shortawn foxtail (Alopecurus aequalis Sobol.) (Zhao et al. Reference Zhao, Li, Bai, Guo, Yuan, Wang, Liu and Wang2017), annual ryegrass (Lolium spp.) (Duhoux et al. Reference Duhoux, Carrère, Duhoux and Délye2017a), and A. myusuroides (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). Recent studies have demonstrated that the enhanced expression of GSTs able to conjugate and detoxify atrazine is responsible for evolved resistance to this herbicide in populations of A. palmeri (Nakka et al. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017a) and A. tuberculatus (Evans et al. Reference Evans, O’Brien, Ma, Hager, Riggins, Lambert and Riechers2017). However, in A. myusuroides and A. fatua, the GSTs upregulated in the NTSR populations have little activity toward herbicides, even though their enhanced expression is intimately linked to resistance (Burns et al. Reference Burns, Keith, Rafai, Bothner and Dyer2017; Cummins et al. Reference Cummins, Cole and Edwards1999). Recent studies in A. myosuroides demonstrated that the phi AmGSTF1, which has little conjugating activity toward herbicides, was uniquely associated with all NTSR populations identified to date (Tetard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). Overexpression of AmGSTF1 in Arabidopsis (A. thaliana ecotype Columbia-0), enhanced tolerance to several herbicides, notably those that did not undergo glutathionylation in the course of their detoxification (Cummins et al. Reference Cummins, Wortley, Sabbadin, He, Coxon, Straker, Sellars, Knight, Edwards, Hughes, Kaundun, Hutchings, Steel and Edwards2013; Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). This suggested that GSTs must possess alternative NTSR protective activities that do not directly involve herbicide detoxification. However, it is known that the complex NTSR trait in different weed species is multigenic (Duhoux et al. Reference Duhoux, Pernin, Desserre and Délye2017b; Keith et al. Reference Keith, Burns, Bothner, Carey, Mazurie, Hilmer, Biyiklioglu, Budak and Dyer2017; Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018), suggesting that a single GST cannot confer multiple resistance. In further investigating the roles of these proteins in NTSR, we now report the detailed characterization of the all detectable GSTs (the “GSTome”), associated with herbicide resistance using a combination of transcriptomics, proteomics, and functional expression of the respective recombinant enzymes.
Analysis of AmGSTF1 Variants in Alopecurus myosuroides
Recent proteomic studies confirmed the presence of elevated levels of the phi A. myosuroides AmGSTF1 polypeptides in multiple NTSR populations, as compared with herbicide-sensitive (HS) populations (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). Closer examination of the AmGSTF1 polypeptides revealed they were derived from two of the four known isoforms (Supplementary Figure S1), namely the c and d variants, previously identified in the screening of a cDNA expression library prepared from the NTSR A. myosuroides population, ‘Peldon’ (Cummins et al. Reference Cummins, Cole and Edwards1999). Whereas only the c and d variants were expressed as polypeptides, analysis of the transcriptomic data showed that all four isoforms were present as mRNAs in NTSR A. myosuroides (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). Differences were observed in the relative abundance of the AmGSTF1 transcripts, with the c form generally more abundant in leaves and the d form dominant in the stems. Previous enzymatic and transgenic studies have exclusively concentrated on the characterization of the AmGSTF1c isoform (Cummins et al. Reference Cummins, Wortley, Sabbadin, He, Coxon, Straker, Sellars, Knight, Edwards, Hughes, Kaundun, Hutchings, Steel and Edwards2013). As the proteomic studies now showed that alternative isoforms were also being expressed (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018), it was clearly important to determine whether the variants were all functionally equivalent. AmGSTF1a was selected for comparative enzymatic analysis with AmGSTF1c, as the most variant isoform (Supplementary Figure S1). The GST sequences were then subcloned into the pET-STRP3 vector and expressed as Strep-tag II fusion proteins in Escherichia coli (Dixon et al. Reference Dixon, Hawkins, Hussey and Edwards2009). The affinity-purified enzymes were then assayed for glutathione transferase and glutathione peroxidase (GPOX) activity, with the enzymes showing similar activities and kinetic constants (Supplementary Table S1). It was concluded that while AmGSTF1 is present as multiple isoenzymes in A. myosuroides, the isoenzymes are functionally identical and, as such, all further references to AmGSTF1 refer to the c isoform.
GST Genes Expression in NTSR Alopecurus myosuroides
To identify the full range of GST genes associated with NTSR in A. myosuroides, the respective transcriptome contig sequences of NTSR (‘Peldon’) as compared with the HS (‘Rothamsted’) populations reported previously were examined (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). Using this approach, a total of 53 contigs corresponding to GST genes were identified and analyzed by Blast (BlastX) searches against the respective translated proteome database (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). This approach clustered the contigs into 15 distinct GSTs, which on online protein blast (BlastP) analysis, yielded eight GSTUs, six GSTFs, and one lambda (L) GSTL gene (Supplementary Table S2). Of the phi GSTs, isoforms of the previously undescribed AmGSTF2 and AmGSTF3 genes were identified in addition to AmGSTF1. Of the eight GSTU genes, one sequence corresponded to AmGSTU1 (Cummins et al. Reference Cummins, Bryant and Edwards2009). The remaining new unigenes were named AmGSTU2 to AmGSTU7, of which AmGSTU2 was the most highly represented in the NTSR populations (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018).
AmGSTU2 was selected for further characterization, with multiple sequence variants, termed AmGSTU2a-f, recovered from the Peldon cDNA library (Cummins et al. Reference Cummins, Cole and Edwards1999). Phylogenetic analysis identified orthologues in wheat, maize, and barley (Hordeum vulgare L.). The phylogenetic analysis showed that AmGSTU2a and AmGSTU2b (95% similarity) were most likely derived from a lineage-specific duplication (Figure 9). Both were members of a clade, also containing AmGSTU1, that is unique to monocots (Brazier-Hicks et al. 2018). In contrast, AmGSTU3 to AmGSTU7 were more evolutionarily diverse, sharing protein sequence identities ranging from 36% to 76% and aligning to several distinct tau class clades (Figure 9). Of the 15 upregulated GST genes, AmGSTU2a was the most abundant at both transcript and expressed protein levels (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018; Figure 10); as such, it was selected for further characterization. The coding sequence of AmGSTU2a was isolated by PCR from the Peldon cDNA library and expressed as the respective Strep-tag II fusion protein in E. coli. Enzyme activity assays of the recombinant protein showed that while AmGSTU2a conjugated 1-chloro-2,4-dinitrobenzene and the herbicides fenoxaprop-ethyl and metolachlor, it was inactive as a GPOX (Table 3). Similarly, the related AmGSTU1 from A. myosuroides and an orthologue TaGSTU4 (69% identity) in wheat (Thom et al. Reference Thom, Cummins, Dixon, Edwards, Cole and Lapthorn2002), also showed activity toward multiple herbicides, suggesting this clade of tau GSTs is important in detoxification.

Figure 9 Phylogenetic analysis of tau class of glutathione S-transferase (GSTUs) proteins in grass weeds and crop plants. Amino acid sequences of GSTUs from Alopecurus myosuroides (red), barley (black), wheat (green), and maize (blue) were used for maximum-likelihood alignment for phylogenic analysis. The number on the branch represents the bootstrap support values above 50%. The scale bar indicates the inferred number of substitutions per site. Clades comprising exclusively barley sequences were collapsed into triangles.

Figure 10 Relative abundance of GSTs in blackgrass determined in stem and leaf tissue in NTSR populations isolated from the field (‘Peldon’, ‘Oxford’) and from forced selection with the herbicides pendimethalin, or fenoxaprop. Significant differences in fold-abundance (p<0.05, fold change >1.5) were relative to equivalent herbicide sensitive (HS) plants. The difference in transcript abundances (NTSR vs. HS) are presented in different color codes with red representing enhanced and green representing suppressed transcript abundance. (NA=not analyzed).
Table 3 Activities of purified recombinant glutathione S-transferase (rGST) enzymes from Alopecurus myusuroides in conjugating 1-chloro-2,4-dinitrobenzene (CDNB) and herbicide substrates and as a glutathione peroxidase acting on t-butyl hydroperoxide.

GST Protein Expression in Alopecurus myosuroides
Proteomics was used to monitor changes in the GSTome in NTSR populations either derived from different geographical locations in the United Kingdom (Peldon, ‘Essex’, and ‘Oxford’) or by repeated selection with the herbicides pendimethalin or fenoxaprop as compared with HS plants (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018). While 15 distinct GSTs showed induced transcriptional expression, only four of these genes were accompanied by increased protein expression as determined by differential two-dimensional proteomics of stem and leaf tissue (Figure 10). In the leaves, the c and d isoforms of AmGSTF1 were the most highly abundant GSTs detected by proteomics (Figure 10). In the case of AmGSTU2, the 140-fold enhancement in transcripts in the stems of Peldon versus HS plants was associated with only a 6-fold increase in the respective protein abundance. In the stem tissue, the enhancement of AmGSTF2, AmGSTU2, and the AmGSTF1c/d isoforms in the NTSR Peldon, Oxford, and ‘pendimethalin’ populations, was clearly distinct from that determined in the plants selected on fenoxaprop (Figure 10). This suggested that the GSTome associated with NTSR in A. myosuroides could vary depending on the history of herbicide selection.
As GSTUs and GSTFs are associated with a wide range of plant stress responses (Dixon and Edwards Reference Dixon and Edwards2010), it was then of interest to determine whether the NTSR-associated GSTs were perturbed by biotic and abiotic stress in A. myosuroides. As described previously (Tétard-Jones et al. Reference Tétard-Jones, Sabbadin, Moss, Hull, Neve and Edwards2018), HS plants were exposed to a range of stress treatments, including wounding, heat, and drought, along with exposure to the herbicide safener cloquintocet mexyl. The results demonstrated that the changes in the GSTome associated with NTSR could not be replicated by any of the stresses, with the abundance of AmGSTU2 actually suppressed by these treatments (Figure 10).
Conclusions on the Roles of GSTs in NTSR Alopecurus myosuroides
A characteristic feature of the GSTs induced by NTSR was their relative abundance and multiplicity in isoforms identified in the transcriptome studies, as compared with the proteome experiments. While the major phi AmGSTF1 was encoded by at least four sequences (a to d isoforms) and the tau AmGSTU2 by six open reading frames (a to f isoforms), at the level of protein expression they were represented by only two and one isoforms, respectively (Figure 10). Similarly, the relative enhancement of the transcripts encoding these GSTs was at least an order of magnitude greater than the changes determined in the abundance of the respective proteins (Figure 10). The discrepancy between transcriptome and proteome data for the GSTs demonstrates that the respective genes are subject to major posttranscriptional regulation either at the mRNA level or during translation or protein turnover. Similar discrepancies in transcriptome and proteome expression patterns have been reported for GSTs in other plants responding to stress conditions and may reflect the manner in which these genes are evolving through the process of gene duplication (Dixon and Edwards Reference Dixon and Edwards2010). In this regard, it is interesting that AmGSTU2 is orthologous to TaGSTU4, an enzyme that displays a range of detoxifying activities following minor changes in its coding sequence (Govindarajan et al. Reference Govindarajan, Mannervik, Silverman, Wright, Regitsky, Hegazy, Purcell, Welch, Minshull and Gustafsson2015). We conclude that while AmGSTF1 and its variants have the potential to evolve new signaling-related roles in NTSR, the AmGSTU2 isoforms could rapidly develop new herbicide-detoxifying activities. Further characterization of the functional diversity of these GSTs in the future may help explain the multiple herbicide-resistant phenotypes observed in different NTSR A. myosuroides populations.
Metabolic Herbicide Resistance in Lolium spp.
Resistance to small grain-selective acetolactate synthase (ALS)- and acetyl CoA carboxylase (ACCase)-inhibiting herbicides is common in Lolium spp. (L. rigidum, L. perenne) in the United States, Europe, and especially Australia (Boutsalis et al. Reference Boutsalis, Gill and Preston2012; Broster and Pratley Reference Broster and Pratley2006; Heap Reference Heap2018; Owen et al. Reference Owen, Martinez and Powles2014). Both target-site resistance (TSR) and NTSR mechanisms have evolved, and these often co-occur within individuals and within populations (Han et al. Reference Han, Yu, Owen, Cawthray and Powles2016; Yu and Powles Reference Yu and Powles2014). Metabolism-based resistance in L. rigidum from Australia has evolved to wheat-selective ACCase and ALS herbicides with a mosaic of different cross-resistance and inheritance patterns indicative of multiple resistance genes and pathways (reviewed in Yu and Powles Reference Yu and Powles2014). Resistance to the ACCase inhibitor diclofop has been experimentally evolved from initially susceptible individuals (Manalil et al. Reference Manalil, Busi, Renton and Powles2011; Neve and Powles Reference Neve and Powles2005), and the high level of resistance achieved is due to enhanced diclofop metabolism (Yu et al. Reference Yu, Han, Cawthray, Wang and Powles2013). Inheritance of the resistance is polygenic (Busi et al. Reference Busi, Neve and Powles2013), consistent with the hypothesis that multiple genes may contribute to metabolic resistance. Interestingly, diclofop-susceptible populations were selected for even greater sensitivity (Manalil et al. Reference Manalil, Busi, Renton and Powles2012), and protection against diclofop due to enhanced metabolism could be induced in susceptible individuals by 2,4-D application (Han et al. Reference Han, Yu, Cawthray and Powles2013). Metabolic resistance to pyroxasulfone was experimentally evolved from a population that already had metabolic resistance to ACCase, ALS, and mitosis inhibitors (Busi et al. Reference Busi, Gaines, Walsh and Powles2012), but no pyroxasulfone resistance evolved following repeated selection on a susceptible population. The experimentally evolved pyroxasulfone resistance appeared to be controlled by a single gene (Busi et al. Reference Busi, Gaines, Vila-Aiub and Powles2014), and the population was also cross-resistant to prosulfocarb and triallate, with a subsequent selection using prosulfocarb resulting in higher prosulfocarb resistance (Busi and Powles Reference Busi and Powles2013, Reference Busi and Powles2016). Pyroxasulfone resistance in the experimentally evolved population is due to glutathione conjugation and subsequent metabolic steps on pyroxasulfone, with the first step likely mediated by GSTs (Busi et al. Reference Busi, Porri, Gaines and Powles2018). Some insecticides can reverse metabolic resistance in L. rigidum by inhibiting CYPs, including malathion for chlorsulfuron resistance (Christopher et al. Reference Christopher, Preston and Powles1994) and amitrole for diclofop resistance (Preston and Powles Reference Preston and Powles1998). Phorate reverses chlorsulfuron, pyroxasulfone, and trifluralin resistance, but antagonizes the efficacy of prosulfocarb and triallate that normally are effective on L. rigidum (Busi et al. Reference Busi, Gaines and Powles2017). Enhanced metabolism for herbicide resistance is still not completely understood and can complicate L. rigidum management due to unpredictable cross-resistance patterns (Preston Reference Preston2004).
Metabolic resistance in L. rigidum has been associated with increased expression of genes with roles in metabolism, such as CYPs, GSTs, and GTs (Duhoux et al. Reference Duhoux, Carrère, Gouzy, Bonin and Délye2015; Gaines et al. Reference Gaines, Lorentz, Figge, Herrmann, Maiwald, Ott, Han, Busi, Yu, Powles and Beffa2014). Although not yet reported, metabolic herbicide resistance could also be due to nonsynonymous mutations in metabolism genes, resulting in faster herbicide metabolism due to changes in substrate affinity. An RNA-seq transcriptome analysis was used to identify genes conferring enhanced metabolic resistance in a L. rigidum population experimentally evolved for diclofop resistance (Gaines et al. Reference Gaines, Lorentz, Figge, Herrmann, Maiwald, Ott, Han, Busi, Yu, Powles and Beffa2014). Candidate transcripts identified as overexpressed in resistant plants cosegregated with the resistance phenotype in an F2 population, including CYPs, GSTs, and GTs. A set of four transcripts (two CYPs, one nitrogen monooxygenase, and one GT), initially identified from the experimentally evolved diclofop-resistant population, were upregulated in individuals from nine unrelated L. rigidum populations with metabolic resistance (Gaines et al. Reference Gaines, Lorentz, Figge, Herrmann, Maiwald, Ott, Han, Busi, Yu, Powles and Beffa2014). A cluster analysis demonstrated the potential of these four transcriptional markers in resistance diagnostics when multiple individuals were sampled from different populations, as herbicide-susceptible individuals clustered together and populations containing metabolic resistant individuals could be classified as resistant (Figure 11). Candidate resistance genes from this L. rigidum population are being functionally validated, and additional populations from Australia with different metabolic resistance patterns are being evaluated with transcriptomics for candidate resistance genes. An RNA-seq analysis of ALS-resistant L. rigidum from France identified a set of 19 transcripts associated with metabolic ALS resistance (Duhoux et al. Reference Duhoux, Carrère, Gouzy, Bonin and Délye2015). These 19 transcriptional markers provided diagnostic prediction of herbicide resistance in French Lolium spp. populations (Duhoux et al. Reference Duhoux, Carrère, Duhoux and Délye2017a). The functional role of transcriptional marker genes in Lolium spp. has yet to be evaluated.

Figure 11 Transcriptional markers for metabolic herbicide resistance can be used to diagnose populations as resistant or susceptible (adapted from Gaines et al. Reference Gaines, Lorentz, Figge, Herrmann, Maiwald, Ott, Han, Busi, Yu, Powles and Beffa2014). A cluster analysis of expression levels of four transcriptional markers (increased expression of two cytochrome P450s, one nitrogen monooxygenase, and one glucosyl transferase) differentiates herbicide-susceptible individuals (samples ending in S, highlighted in boxes) from populations containing metabolic resistant individuals, which could be classified as resistant based on their clustering. The results highlight the importance of sampling multiple individuals for transcriptional marker diagnostics.
A major question for metabolic herbicide resistance is that some genes associated with resistance across multiple populations through RNA-seq studies may not have a functional role in resistance, that is, they may not be the genes responsible for direct metabolic modifications to herbicides. These genes could be genetically linked to the major resistance gene(s), they could be co-regulated by a common transcription factor, or they could be co-regulated through chromatin structural changes. These genes may have other functions in resistance besides direct herbicide metabolism, or they could have no functional role in resistance at all. Improved basic knowledge about the molecular genetic basis of metabolic resistance and linked or co-regulated genes is necessary to develop metabolic resistance diagnostics based on molecular markers.
Mechanisms of Multiple Resistance in Echinochloa phyllopogon
Herbicide resistance in E. phyllopogon has been reported in populations in France, Greece, Brazil, South Korea, and the United States (Délye et al. Reference Délye, Causse, Gautier, Poncet and Michel2015; Kaloumenos et al. Reference Kaloumenos, Chatzilazaridou, Mylona, Polidoros and Eleftherohorinos2012; Matzenbacher et al. Reference Matzenbacher, Bortoly, Kalsing and Merotto2014; Song et al. Reference Song, Lim, Yook, Kim and Kim2017). The populations in France, Greece, and Brazil are resistant to ALS inhibitors due mainly to TSR mechanisms, although additional NTSR mechanisms were not excluded. Conversely, populations in South Korea and the United States are known to have NTSR mechanisms and exhibit resistance to several herbicides with different modes of action. Here, the study on the resistance mechanism of E. phyllopogon found in the United States is reviewed.
Multiple Herbicide–Resistant Echinochloa phyllopogon
The Sacramento Valley of California is one of the largest rice production areas in the United States. Since the introduction of molinate and thiobencarb in the 1960s and 1980s, respectively, the two thiocarbamate herbicides were preferentially used for Echinochloa control (Fischer et al. Reference Fischer, Ateh, Bayer and Hill2000a). In 1994, another option from Group 1, fenoxaprop-ethyl, was introduced for POST control of Echinochloa spp. (Williams Reference Williams2000). Soon after, the failure in control of E. phyllopogon by herbicides that previously controlled it was reported by farmers (Fischer et al. Reference Fischer, Ateh, Bayer and Hill2000a). Greenhouse experiments conducted on the plants collected from fields in 1997 revealed that the plants exhibited resistance to fenoxaprop-ethyl, molinate, thiobencarb, and bispyribac-sodium, a herbicide that was not released yet (Fischer et al. Reference Fischer, Ateh, Bayer and Hill2000a; Osuna et al. Reference Osuna, Vidotto, Fischer, Bayer, De Prado and Ferrero2002). Later, the research group revealed that the resistant E. phyllopogon also exhibited resistance to cyhalofop-butyl, penoxsulam, bensulfuron-methyl, quinclorac, and clomazone, most of which had not been in use by 1997.
Mechanism of Resistance to ALS Inhibitors
Metabolism-based Resistance
Multiple herbicide–resistant E. phyllopogon is resistant to three ALS-inhibiting herbicides, bispyribac-sodium, penoxsulam, and bensulfuron-methyl, each belonging to a different chemical family. There are several reports of resistance factors that were estimated from different experiments in different cultivation conditions or with different lines (Fischer et al. Reference Fischer, Ateh, Bayer and Hill2000a, Reference Fischer, Bayer, Carriere, Ateh and Yim2000b; Osuna et al. Reference Osuna, Vidotto, Fischer, Bayer, De Prado and Ferrero2002; Yasuor et al. Reference Yasuor, Osuna, Ortiz, Saldain, Eckert and Fischer2009). When compared under the same experimental conditions, the resistance factor for line 511, which was the line used for molecular analysis, was about 1,100-fold for bensulfuron-methyl, 6.2-fold for penoxsulam (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a), and 1.8-fold for bispyribac-sodium (unpublished data).
No difference was observed in the sensitivity of the target site to these herbicides in vitro (Fischer et al. Reference Fischer, Bayer, Carriere, Ateh and Yim2000b; Osuna et al. Reference Osuna, Vidotto, Fischer, Bayer, De Prado and Ferrero2002; Yasuor et al. Reference Yasuor, Osuna, Ortiz, Saldain, Eckert and Fischer2009). In accordance with the enzyme assay, the nucleotide sequences of the two copies of ALS genes were identical between resistant and sensitive plants (Iwakami et al. Reference Iwakami, Uchino, Watanabe, Yamasue and Inamura2012). These studies confirm that ALS resistance is not caused by TSR. Studies on metabolism were mainly conducted on penoxsulam using [14C]penoxsulam. Resistant plants metabolized penoxsulam into polar metabolites 2.8 times faster than sensitive plants (Yasuor et al. Reference Yasuor, Osuna, Ortiz, Saldain, Eckert and Fischer2009). A CYP inhibitor, malathion, inhibited the metabolism of penoxsulam. Similarly, faster metabolism in resistant plants was confirmed in bensulfuron-methyl. The resistant plants metabolized bensulfuron-methyl into a demethylated form (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a), which is known to lose ALS-inhibiting activity (Takeda et al. Reference Takeda, Erbes, Sweetser, Hay and Yuyama1986). Although metabolism was not tested for bispyribac-sodium, Fischer et al. (Reference Fischer, Bayer, Carriere, Ateh and Yim2000b) revealed application of CYP inhibitors partially reversed bispyribac-sodium resistance.
Isolation of CYP81A CYP Genes from Echinochloa phyllopogon
CYPs comprise a superfamily with hundreds of genes in plant genomes (Nelson et al. Reference Nelson, Schuler, Paquette, Werck-Reichhart and Bak2004). A recent study by Guo et al. (Reference Guo, Qiu, Ye, Jin, Mao, Zhang, Yang, Peng, Wang, Jia, Lin, Li, Fu, Liu, Chen, Shen, Wang, Chu, Wu, Wu, Xia, Zhang, Zhou, Wang, Wu, Song, Wang, Shu, Aoki, Yumoto, Yokota, Miyamoto, Okada, Kim, Cai, Zhang, Lou, Qian, Yamaguchi, Yamane, Kong, Timko, Bai and Fan2017) revealed that a hexaploid close relative of E. phyllpogon, barnyardgrass [Echinochloa crus-galli (L.) P. Beauv.], possesses as many as 917 CYP genes in its genome, suggesting the number is also large in tetraploid E. phyllopogon. The very large number of genes has made the analysis of genes difficult, especially in weed species with no reference genome sequence. Although a CYP gene involved in rice tolerance to some herbicides has been identified (Pan et al. Reference Pan, Zhang, Liu, Zhang, Wu, Zhu and Tu2006), no CYP genes involved in herbicide resistance in weeds were identified before the study conducted in E. phyllopogon. The CYP gene belongs to CYP81A subfamily (CYP81A6) and is involved in tolerance to various ALS-inhibiting herbicides, including bensulfuron-methyl.
Attempts to isolate CYP genes from E. phyllopogon were started by nested degenerate PCR using the very few conserved regions of P450 genes, resulting in the isolation of 39 putative P450 genes, including seven CYP81A members (Iwakami et al. Reference Iwakami, Uchino, Kataoka, Shibaike, Watanabe and Inamura2014b). Later, degenerate PCR was used to exclusively amplify CYP81A genes and succeeded in the isolation of five more CYP81A genes from resistant plants (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a). Among the 12 CYP81A genes, 3 are putative pseudogenes with indels that cause frameshifts. Comparisons of full-length sequences between the resistant and sensitive plants found nonsynonymous substitutions in CYP81A21, CYP81A22, and CYP81A26.
Characterization of CYP81A Genes
Transcript levels of the nine CYP81A genes, except the three putative pseudogenes, were compared between resistant and sensitive plants (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a). Two genes, CYP81A12 and CYP81A21, were constitutively highly expressed in the resistant plants. The two genes share extremely high similarity and were estimated as homologues based on phylogenetic tree analysis. The estimation of homologous relationships was also supported by chromosome linkage analysis of the two genes (Iwakami et al., Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a). The analysis was performed using nucleotide polymorphisms found in both genes between the resistant and sensitive plants. The results indicated there was no linkage between the two genes, which implies that the two genes are on different chromosomes, as expected from the homologous relationship. Overexpression was also observed in CYP81A22, although the overexpression was restricted to the roots of resistant plants.
Transgenic Arabidopsis expressing CYP81A12 or CYP81A21 exhibited significant resistance to bensulfuron-methyl (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a). The resistance factors of the highly expressing lines were more than 1,000-fold, suggesting the CYPs metabolize bensulfuron-methyl very effectively. An in vivo bensulfuron-methyl metabolism experiment using yeast revealed that the two CYPs demethylate bensulfuron-methyl. The results explain the higher amount of demethylated form of bensulfuron-methyl in the resistant E. phyllopogon. Association of bensulfuron-methyl resistance and the higher expressions of the two genes were investigated in the F6 generation of the progenies of the resistant and sensitive E. phyllopogon. The experiment showed that bensulfuron-methyl resistance was associated with higher expression in all the F6 lines tested. In contrast to CYP81A12 and CYP81A21, CYP81A22 did not confer bensulfuron-methyl resistance in Arabidopsis. Higher expression of CYP81A22 observed in the resistant parent did not cosegregate with resistance in the study of the F6 generation. Genotyping of F6 lines also revealed that amino acid substitutions found in CYP81A21, CYP81A22, and CYP81A26 were not associated with resistance. All the results strongly suggest that bensulfuron-methyl resistance in E. phyllopogon is caused by overexpression of CYP81A12 and CYP81A21.
The overexpression mechanism of the two genes was investigated from the aspects of the promoter sequence and copy number (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a). Polymorphisms between the resistant and sensitive lines were observed in the promoter sequences of the two genes, but neither of them cosegregated with resistance in the F6 lines. Also, no difference was detected in the copy number by Southern blot analysis. Considering the two genes are on different chromosomes and the resistance is under the control of a single gene (or locus), it is likely that a single trans-element simultaneously regulates the expression of both genes.
Mechanism of Penoxsulam Resistance
It is reasonable to infer that the same mechanism that controls bensulfuron-methyl resistance also controls the penoxsulam resistance, based on the metabolism and inheritance studies presented. As expected, bensulfuron-methyl and penoxsulam resistance did not segregate in F6 lines (Iwakami et al. Reference Iwakami, Endo, Saika, Okuno, Nakamura, Yokoyama, Watanabe, Toki, Uchino and Inamura2014a). Arabidopsis lines transformed with CYP81A12 or CYP81A21 exhibited significant resistance to penoxsulam. Interestingly, the Arabidopsis that exhibited >1,000-fold resistance to bensulfuron-methyl exhibited ~10-fold resistance to penoxsulam. These results suggest that CYP81A12 and CYP81A21 metabolize bensulfuron-methyl 100-fold more efficiently than penoxsulam. This observation is consistent with the resistance levels to bensulfuron-methyl (1,100-fold) and penoxsulam (6.2-fold) in E. phyllopogon. In summary, the two CYP81A genes play a major role in not only bensulfuron-methyl, but also penoxsulam resistance.
Bispyribac-Sodium Resistance Mechanism Is Not Clear
In contrast to bensulfuron-methyl and penoxsulam, Arabidopsis expressing CYP81A12 or CYP81A21 did not show any significant resistance to bispyribac-sodium (unpublished data). The failure of endowing resistance to Arabidopsis does not necessarily indicate the genes are not involved in bispyribac-sodium resistance. RNAi knockdown rice of CYP81A6, an ALS-inhibitor tolerance gene in rice, has slightly higher sensitivity to bispyribac-sodium, although the difference is not very clear (Saika et al. Reference Saika, Horita, Taguchi-Shiobara, Nonaka, Nishizawa-Yokoi, Iwakami, Hori, Matsumoto, Tanaka, Itoh, Yano, Kaku, Shimizu and Toki2014). Therefore, CYP81A CYPs may have functions of bispyribac-sodium metabolism. More detailed studies will be required to investigate the involvement of the P450 in resistance to bispyribac-sodium in E. phyllopogon.
Mechanism of Resistance to ACCase Inhibitors
Resistant E. phyllopogon plants exhibit resistance to AOPPs, fenoxaprop-ethyl and cyhalofop-butyl, with resistance factors of 10 and 19, respectively (Bakkali et al. Reference Bakkali, Ruiz-Santaella, Osuna, Wagner, Fischer and DePrado2007; Ruiz-Santaella et al. Reference Ruiz-Santaella, De Prado, Wagner, Fischer and Gerhards2006). On the other hand, a cyclohexanedione ACCase inhibitor, profoxydim, can effectively control resistant plants (Ruiz-Santaella et al. Reference Ruiz-Santaella, Fisher and De Prado2003). ACCase sensitivity to fenoxaprop-ethyl did not differ between the resistant and sensitive lines. Nucleotide sequences of the carboxy transferase domain of ACCase, where all the resistance-conferring amino acid substitutions were found in other species, were compared between the resistant and sensitive E. phyllopogon. In accordance with the enzyme sensitivity, no resistance-conferring mutations were observed in the four copies of ACCase genes in E. phyllopogon. These results indicate that the ACCase resistance mechanism is non–target site based.
A metabolism study was performed for fenoxaprop-ethyl and cyhalofop-butyl. Rapid accumulation of GSH-conjugated metabolites in resistant plants strongly suggests that fenoxaprop-ethyl resistance is caused by enhanced activity of GST (Bakkali et al. Reference Bakkali, Ruiz-Santaella, Osuna, Wagner, Fischer and DePrado2007). Resistant plants accumulated more polar metabolites of cyhalofop-butyl than sensitive plants (Ruiz-Santaella et al., Reference Ruiz-Santaella, De Prado, Wagner, Fischer and Gerhards2006). The results suggest that a mechanism in addition to overexpression of CYP81A12 and CYP81A21 endows multiple-herbicide resistance in E. phyllopogon. The research to identify specific GSTs is ongoing.
Mechanism of Resistance to Clomazone
Echinochloa phyllopogon in rice fields had no prior exposure to clomazone. The resistance factor is not very high (2 times), so clomazone resistance eluded classification in the first screening (Fischer et al. Reference Fischer, Ateh, Bayer and Hill2000a; Yasuor et al. Reference Yasuor, Tenbrook, Tjeerdema and Fischer2008). However, after the introduction of clomazone in the Sacramento Valley, control failures with clomazone were observed, leading to the findings of low-level resistance (Yasuor et al. Reference Yasuor, Tenbrook, Tjeerdema and Fischer2008). A detailed study of clomazone resistance revealed that the resistant plants more rapidly metabolize clomazone into oxidative forms than sensitive plants, as described earlier: 5-OH clomazone is converted to dihydroxy-clomazone and clomazone to hydroxymethylclomazone and 3′-hydroxyclomazone (Yasuor et al. Reference Yasuor, Zou, Tolstikov, Tjeerdema and Fischer2010). Therefore, it is possible that CYPs are involved in the reaction.
Mechanism of Resistance to Quinclorac
Enhanced metabolism has not been proposed as a weed resistance mechanism to quinclorac (Yasuor et al. Reference Yasuor, Milan, Eckert and Fischer2012). Therefore, factors related to the mechanism of action of auxin herbicides were addressed.
The resistance factor to quinclorac is different between the methods of quinclorac application: foliar spray application, 6-fold; hydroponic root application, 17-fold (Yasuor et al. Reference Yasuor, Milan, Eckert and Fischer2012). As with other auxin herbicides, quinclorac is known to induce high levels of ethylene production, which is highly related with plant sensitivity to quinclorac (Grossmann and Kwiatkowski Reference Grossmann and Kwiatkowski1993). This correlation is not fully understood, but it is thought to be caused by hydrogen cyanide (HCN) accumulation in plants as a by-product of ethylene production. Yasuor et al. (Reference Yasuor, Milan, Eckert and Fischer2012) compared the ethylene production of resistant and sensitive E. phyllopogon and found resistant plants produced significantly lower ethylene. Another interesting finding from their research was that resistant plants have higher activity of β-cyanoalanine synthase (β-CAS), an enzyme that detoxifies HCN. The authors concluded that quinclorac resistance in the resistant E. phyllopogon is caused by insensitivity along the ethylene production pathway and enhanced β-CAS activity.
Future Work
As reviewed here, extensive work on ALS-inhibitor resistance resulted in the identification of CYP involvement in resistance in E. phyllopogon. These genes may explain resistance to other herbicides such as clomazone, for which enhanced oxidation of clomazone was reported. On the other hand, the involvement of other herbicide-metabolizing genes, namely GSTs, are suggested in the case of fenoxaprop-ethyl resistance. Furthermore, even non–metabolism based resistance is suggested in quinclorac resistance. A hint to elucidate the apparently complicated mechanism(s) of multiple-herbicide resistance may come from analyses of thiocarbamate (molinate and thiobencarb) resistance, which have not yet been investigated. Although findings of resistant populations in the Sacramento Valley occurred right after introduction of fenoxaprop-ethyl, the driving force of resistance evolution may be continuous application of thiocarbamates for more than 20 yr. Therefore, elucidation of the resistance mechanism to thiocarbamates might provide an insight into how the evolution of multiple-herbicide resistance occurred in E. phyllopogon. A preliminary work suggested concerted upregulation of several gene families involved in herbicide metabolism (unpublished data), as has been reported in other metabolism-based resistant weeds (Duhoux et al. Reference Duhoux, Carrère, Gouzy, Bonin and Délye2015, Reference Duhoux, Carrère, Duhoux and Délye2017a; Gaines et al. Reference Gaines, Lorentz, Figge, Herrmann, Maiwald, Ott, Han, Busi, Yu, Powles and Beffa2014; Gardin et al. Reference Gardin, Gouzy, Carrere and Delye2015). Approaches such as genomics and transcriptomics will shed further light on the mystery of rapid evolution of resistance to multiple herbicides in E. phyllopogon.
Multiple Resistance and Metabolic Resistance Mechanisms in Junglerice (Echinochloa colona)
A junglerice [(Echinochloa colona (L.) Link] biotype MS1, collected from a rice field in Sunflower County, MI, was resistant to ALS-inhibiting imazethapyr and cross-resistant to other ALS inhibitors such as imazamox (3.3-fold), penoxsulam (9.4-fold), and bispyribac-sodium (7.2-fold) (Riar et. al. Reference Riar, Norsworthy, Bond, Bararpour, Wilson and Scott2012). In preliminary experiments, the addition of malathion to penoxsulam and imazethapyr reduced shoot dry weight and/or increased mortality compared with the respective herbicides applied alone, indicating possible involvement of herbicide metabolism driven by CYPs as a mechanism of resistance. ALS enzyme assays or ALS gene-sequencing analysis did not indicate a modified target site–based resistance in the MS1 biotype (Riar et. al. Reference Riar, Norsworthy, Srivastava, Nandula, Bond and Scott2013). Lower levels of translocation of [14C]bispyribac and [14C]imazamox in the MS1 biotype compared with a susceptible biotype were recorded, perhaps indicating that metabolism is involved (Riar et. al. Reference Riar, Norsworthy, Srivastava, Nandula, Bond and Scott2013). Additionally, the MS1 biotype was resistant to fenoxaprop-P-ethyl, an ACCase inhibitor (11-fold; but susceptible to sethoxydim and clethodim at field rates) (Wright et. al. Reference Wright, Nandula, Grier, Showmaker, Bond, Peterson, Ray and Shaw2016). Sequencing of ACCase of MS1 did not reveal the presence of any known resistance-conferring point mutations. An enzyme assay confirmed that the ACCase in the MSI biotype was herbicide sensitive. Further investigations with two CYP inhibitors, malathion and piperonyl butoxide, and a GST inhibitor, 4-chloro-7-nitrobenzofurazan (CNBF), did not indicate involvement of any metabolic enzymes inhibited by these compounds (Wright et al. Reference Wright, Nandula, Grier, Showmaker, Bond, Peterson, Ray and Shaw2016).
No reference genome is available for E. colona. RNA-seq analysis of changes in gene expression in the MS1 and a susceptible biotype before and after imazamox treatment was conducted to generate a reference leaf transcriptome (Wright et al. Reference Wright, Sasidharan, Koski, Rodriguez-Carres, Peterson, Nandula, Ray, Bond and Shaw2017). Differentially expressed transcripts between resistant MS1 and susceptible plants included transcription factors, protein-modifying enzymes, and enzymes involved in metabolism and signaling, which are involved in abiotic stress response in other plants. These results suggest that imazamox exposure induced a stress response. A time-course study examining a subset of transcripts showed that expression peaked within 4 to 12 h and then returned to untreated levels within 48 h of exposure. Two additional biotypes showed a similar change in gene expression at 4 h after herbicide exposure compared with the resistant and sensitive biotypes. Thus, within 48 h, E. colona mounted a stress response to imazamox exposure (Wright et al. Reference Wright, Sasidharan, Koski, Rodriguez-Carres, Peterson, Nandula, Ray, Bond and Shaw2017).
The MSI biotype was resistant to propanil and quinclorac as well (Wright et al. Reference Wright, Rodriguez-Carres, Sasidharan, Koski, Peterson, Nandula, Ray, Bond and Shaw2018). Differential gene expression analysis of resistant and sensitive plants revealed that 170 transcripts were upregulated in resistant plants relative to sensitive plants, and 160 transcripts were upregulated in sensitive plants. In addition, 507 transcripts were only expressed in resistant plants, and 562 only in sensitive plants. A subset of these transcripts was investigated further using quantitative PCR (qPCR) to compare gene expression in resistant plants with expression in additional sensitive biotypes. The qPCR analysis identified two transcripts, a kinase and a GST that were significantly upregulated in resistant plants compared with the sensitive plants. A third transcript, encoding an F-box protein, was downregulated in the resistant plants relative to the sensitive plants. As no CYPs were differentially expressed between the resistant and sensitive plants, a single-nucleotide polymorphism analysis was performed, revealing several nonsynonymous point mutations of interest. These candidate genes will require further study to elucidate the resistance mechanisms present in the resistant biotype (Wright et al. Reference Wright, Rodriguez-Carres, Sasidharan, Koski, Peterson, Nandula, Ray, Bond and Shaw2018).
Metabolism-based Multiple Resistance in Amaranthus palmeri
As a result of extensive and intensive selection of PRE and/or POST use of most commonly used herbicides in cropping systems, A. palmeri has evolved resistance to multiple modes of action, for example, microtubule, EPSPS, ALS, PSII, HPPD, and, more recently, protoporphyrinogen oxidase (PPO) inhibitors (Heap Reference Heap2018). Besides herbicide selection, other factors such as biological characteristics of weed species, genetic factors, characteristics of herbicides, and agronomic practices also play an important role in the evolution and spread of herbicide resistance in weed species (Powles and Yu Reference Powles and Yu2010). Amaranthus palmeri characteristics such as high fecundity, germination percentage, wide window of emergence, seed dispersal, and short longevity facilitate the evolution of resistance in response to herbicide selection. Recent advances in agronomic practices have increased adoption of no-till or reduced-tillage practices in crop production to prevent soil erosion and conserve moisture (Kihara et al. Reference Kihara, Bationo, Waswa, Kimetu, Vanlauwe, Okeyo, Mukalama and Martius2012). Consequently, the use of herbicides for weed management became indispensable in crop production in many parts of the world, creating greater herbicide selection.
Evolution of Multiple Herbicide Resistance in Amaranthus palmeri in Kansas
Amaranthus palmeri populations resistant to four mechanism of actions of herbicides—PSII, ALS, EPSPS, and HPPD inhibitors—have been reported in Kansas (Heap Reference Heap2018). Several populations of A. palmeri with resistance to at least two of these herbicide modes of action are common in Kansas. However, a single population of A. palmeri (KSR) in central Kansas (Stafford County) with resistance to PSII and HPPD inhibitors was first confirmed in 2012 (Thompson Reference Thompson, Peterson and Lally2012) in a field where there was no previous history of applications of HPPD inhibitors but a long history of PSII- and ALS-inhibiting herbicides. This population was originally found resistant to Huskie®, a premix of pyrasulfotole (HPPD inhibitor) and bromoxynil (PSII inhibitor) in the field. Later, resistance to atrazine (Nakka et al. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017a; Thompson et al. Reference Thompson, Peterson and Lally2012), chlorsulfuron (Nakka et al. Reference Nakka, Godar, Wani, Thompson, Peterson, Roelofs and Jugulam2017b), and several HPPD inhibitors (e.g., mesotrione, tembotrione, and topramezone) (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c; Thompson et al. Reference Thompson, Peterson and Lally2012) was confirmed and characterized in this population. Investigation of the mechanism of resistance to atrazine, chlorsulfuron, and mesotrione in this population enabled addressing why this population was predisposed to evolve resistance to HPPD inhibitors even though there was no selection of HPPD inhibitors.
Mechanism of Atrazine Resistance in KSR Amaranthus palmeri
Several mutations in the psbA gene, which codes for D1 protein, the target site of PSII inhibitors (triazine, triazinone, uracil, nitrile, etc.), resulted in the evolution of resistance or cross-resistance to different classes of PSII inhibitors (Arntz et al. Reference Arntz, DeLucia and Jordan2000; Gronwald Reference Gronwald1994; Oettmeier Reference Oettmeier1999). Furthermore, such resistance to atrazine has been found to be associated with fitness costs as well (Conard and Radosevich Reference Conard and Radosevich1979). However, enhanced metabolism of atrazine or simazine via GST activity has also been reported in many triazine-resistant weed species (Burnet et al. Reference Burnet, Loveys, Holtum and Powles1993; Cummins et al. Reference Cummins, Cole and Edwards1999; Gray et al. Reference Gray, Balke and Stoltenberg1996; Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013). Similarly, crops such as corn or sorghum are also naturally tolerant to triazines due to rapid metabolism of these herbicides mediated by GST activity.
A high level of resistance to atrazine was confirmed in KSR A. palmeri exhibiting up to 200-fold resistance relative to a known susceptible population (Nakka et al. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017a). No known mutation in the psbA gene conferring the most common substitution (Ser-264-Gly) was found in KSR A. palmeri (Nakka et al. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017a). Also, the resistance trait is not maternally inherited, but rather is transmitted by a nuclear gene in this population (Nakka et al. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017a). On the other hand, the KSR A. palmeri rapidly conjugated [U-14C] atrazine, possibly via GSH mediated by GST activity, within 4 h after treatment (Nakka et al. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017a).
Mechanism of Chlorsulfuron Resistance in KSR Amaranthus palmeri
The ALS enzyme, which is the target site of ALS inhibitors, catalyzes an important step in the biosynthesis of the branched-chain amino acids valine, leucine, and isoleucine in plants and microorganisms (Dailey and Cronan Reference Dailey and Cronan1986; Shaner Reference Shaner1991). SU herbicides such as chlorsulfuron are active on several weed species, including A. palmeri. However, A. palmeri resistance to ALS inhibitors is widespread across many states in the United States, including Kansas (Heap Reference Heap2018). ALS inhibitor–resistant A. palmeri was first documented in Kansas in 1993 (Horak and Peterson Reference Horak and Peterson1995).
A high level of resistance to ALS inhibitors due to several point mutations in the ALS gene has been reported in many weed species (Foes et al. Reference Foes, Liu, Vigue, Stoller, Wax and Tranel1999; Guttieri et al. Reference Guttieri, Eberlein and Thill1995; Patzoldt and Tranel Reference Patzoldt and Tranel2007; Shaner Reference Shaner1991; Tranel et al. Reference Tranel, Wright and Heap2016; Varanasi et al. Reference Varanasi, Godar, Currie, Dille, Thompson, Stahlman and Jugulam2015). Nonetheless, enhanced metabolism of ALS inhibitors contributing to resistance has also been found in a number of weeds, for example, L. rigidum (Christopher et al. Reference Christopher, Powles, Liljegren and Holtum1991; Cotterman and Saari Reference Cotterman and Saari1992), wild mustard (Sinapsis arvensis L.) (Veldhuis et al. Reference Veldhuis, Hall, O’Donovan, Dyer and Hall2000), and A. tuberculatus (Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015). Increased activity of CYPs in the metabolism of this group of herbicides has been reported (Brown Reference Brown1990; Christopher et al. Reference Christopher, Preston and Powles1994).
The KSR A. palmeri exhibited >275 times more resistance to chlorsulfuron relative to a known susceptible population (Nakka et al. Reference Nakka, Godar, Wani, Thompson, Peterson, Roelofs and Jugulam2017b). Such a high level of resistance to ALS inhibitors has also been reported in other Amaranthus species, including redroot pigweed (Amaranthus retroflexus L.) (Scarabel et al. Reference Scarabel, Varotto and Sattin2007), Powell amaranth (Amaranthus powellii S. Watson) (Ferguson et al. Reference Ferguson, Hamill and Tardif2001), prostrate pigweed (Amaranthus blitoides S. Watson) (Sibony et al. Reference Sibony, Michel, Haas, Rubin and Hurle2001; Sibony and Rubin Reference Sibony and Rubin2003), smooth pigweed (Amaranthus hybridus L.) (Whaley et al. Reference Whaley, Wilson and Westwood2006, Reference Whaley, Wilson and Westwood2007), and common waterhemp (Amaranthus rudis J. D. Sauer) (Patzoldt and Tranel Reference Patzoldt and Tranel2007).
Upon sequencing of an ~2-kb length of the ALS gene covering all known mutations at eight codon positions in KSR A. palmeri, only 30% of plants showed the single-nucleotide polymorphism, resulting in an amino acid substitution of proline (CCC) to serine (TCC) at position 197 of the ALS gene. The remaining 70% of KSR plants did not show any mutations (Nakka et al. Reference Nakka, Godar, Wani, Thompson, Peterson, Roelofs and Jugulam2017b). Further, whole-plant response of KSR A. palmeri treated with a combination of chlorsulfuron and malathion (an organophosphate insecticide, known to inhibit the activity of CYPs) showed reduction in biomass accumulation when compared with plants that were treated with either malathion or chlorsulfuron alone or nontreated plants (Nakka et al. Reference Nakka, Godar, Wani, Thompson, Peterson, Roelofs and Jugulam2017b), indicating the synergistic effect of malathion on chlorsulfuron. These data suggest that KSR A. palmeri exhibits two different mechanisms of ALS-inhibitor resistance: (1) detoxification of chlorsulfuron, possibly as a result of CYP activity; and (2) a point mutation (Pro-197-Ser) in the ALS gene. However, metabolism-based resistance appears to exist predominantly in KSR A. palmeri (Nakka et al. Reference Nakka, Godar, Wani, Thompson, Peterson, Roelofs and Jugulam2017b). The KSR A. palmeri population is a prime example of a weed population exhibiting coexistence of both TSR and metabolism-based resistance to ALS inhibitors.
Mechanism of Mesotrione Resistance in KSR Amaranthus palmeri
Herbicides such as mesotrione that inhibit HPPD enzyme are widely used to control a broad spectrum of weeds in agriculture. Mesotrione inhibits carotenoid biosynthesis, resulting in pigment degradation and, eventually, plant death. However, crops such as corn can rapidly metabolize these herbicides via ring hydroxylation mediated by cytochrome P450 monooxygenase(s) combined with reduced uptake (Mitchell et al. Reference Mitchell, Bartlett, Fraser, Hawkes, Holt, Townson and Wichert2001). To date, only two weed species, A. rudis and A. palmeri, have evolved resistance to HPPD inhibitors (Heap Reference Heap2018). HPPD inhibitor–resistant A. rudis was first reported in Illinois in 2009 (Hausman et al. Reference Hausman, Singh, Tranel, Riechers, Kaundun, Polge, Thomas and Hager2011). This biotype of A. rudis was also found to be resistant to atrazine. Detoxification of mesotrione, possibly mediated by CYP and atrazine via GST-mediated conjugation, has been attributed to mesotrione and atrazine resistance, respectively, in this A. rudis population (Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013).
The HPPD-inhibitor resistance in KSR A. palmeri was documented in Kansas in 2012 (Thompson et al. Reference Thompson, Peterson and Lally2012), and later in Nebraska in a cornfield that had a history of continuous use of HPPD inhibitors (Sandell et al. Reference Sandell, Amit and Kruger2012). As mentioned earlier, the field where KSR A. palmeri was found had no previous history of use of HPPD inhibitors. The KSR A. palmeri was up to 18 times more resistant to mesotrione compared with a known sensitive population (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c). There was no difference in uptake or translocation of mesotrione or its metabolites between KSR and a known susceptible A. palmeri biotype (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c). However, similar to mesotrione-resistant A. rudis, KSR A. palmeri also rapidly metabolized [14C]mesotrione. At 4 and 24 h after treatment about 50% and 90% of parent [14C]mesotrione was metabolized, respectively, in KSR plants (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c). Furthermore, the KSR plants were found to detoxify 50% of mesotrione in a shorter time compared with corn or A. rudis (Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013; Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c).
To assess the possibility of coexistence of TSR and NTSR to HPPD inhibitors in KSR A. palmeri, as was observed for ALS inhibitors, the HPPD gene was sequenced and amplified for KSR and a susceptible A. palmeri. However, no mutations or amplification of the HPPD gene that can confer resistance to mesotrione was found (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c). Interestingly, the KSR plants exhibited increased constitutive expression of HPPD transcript and protein (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c). The mesotrione-resistant KSR plants showed at least 8- to 12-fold increase in HPPD mRNA levels (normalized against β-tubulin and carbamoyl-phosphate synthase) relative to susceptible plants. Also, the HPPD protein expression correlated with the transcript expression (Nakka et al. Reference Nakka, Thompson, Peterson and Jugulam2017c). The upregulation of HPPD transcript in KSR plants could have occurred via changes in the cis- or trans-acting elements or alterations in the promoter region of the HPPD gene. Overall, the mesotrione resistance in KSR A. palmeri is conferred predominantly because of rapid detoxification of mesotrione, although increased HPPD gene and protein expression also plays a role in the resistance mechanism.
The Predominance of Metabolism-based Multiple-Herbicide Resistance in KSR
The KSR A. palmeri clearly showed the predominance of metabolism-based resistance to multiple herbicides (Figure 12). The exact chronology of the evolution of resistance to PSII or ALS inhibitors, in this population is unknown. However, in Kansas, resistance to ALS inhibitors in A. palmeri was documented before resistance to PSII inhibitors (Heap Reference Heap2018), but the resistance to HPPD inhibitors evolved more recently in this population (Thompson Reference Thompson, Peterson and Lally2012). It is believed that resistance to ALS inhibitors may have evolved before resistance to PSII inhibitors in the KSR A. palmeri population as well. Regardless, the predominance of metabolism-based resistance to ALS and PSII inhibitors mediated by CYP or GST activity during the phase I and II detoxification process suggests the presence of increased activity of these enzymes, which potentially predisposed this population to detoxify other xenobiotics such as HPPD inhibitors, even though no selection pressure from this herbicide was imposed on this population. The role of specific genes of the CYP enzyme family in detoxification of chlorsulfuron or GSTs in atrazine metabolism is yet to be uncovered. Thus, the prevalence of metabolism-based resistance to ALS and PSII inhibitors may have predisposed this population to evolve resistance to HPPD inhibitors. Current research suggests that the metabolism-based herbicide resistance can be a serious threat to weed management, especially in weeds such as A. palmeri, which is one of the top-ranked economically important weeds across the United States.

Figure 12 Predominance of metabolism-based resistance to photosystem II (PSII), acetolactate synthase (ALS), and p-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitors in a multiple- resistant Amaranthus palmeri evolved in Stafford County, KS.
Biochemical Mechanisms Conferring Multiple Herbicide Resistance in Amaranthus tuberculatus
Amaranthus tuberculatus is a problematic, summer annual broadleaf weed species severely affecting maize, soybean, sorghum, and cotton production in the United States (Hager et al. Reference Hager, Wax, Stoller and Bollero2002; Heap Reference Heap2018; Steckel and Sprague Reference Steckel and Sprague2004). The C4 carbon-fixation pathway and prolonged emergence period are two factors that allow A. tuberculatus to compete with crops in the field, especially under hot and dry weather conditions (Steckel Reference Steckel2007). No new herbicide modes of action have been commercialized in the past three decades (Duke Reference Duke2012). Genes conferring resistance are easily spread throughout natural A. tuberculatus populations by pollen flow due to the obligate-outcrossing nature of A. tuberculatus, which makes effective herbicide options for management of this weed even more limited (Costea et al. Reference Costea, Weaver and Tardif2005; Steckel Reference Steckel2007).
An A. tuberculatus population (named MCR) from central Illinois was the first reported natural weed population to evolve resistance to HPPD-inhibiting herbicides (Hausman et al. Reference Hausman, Singh, Tranel, Riechers, Kaundun, Polge, Thomas and Hager2011), chronologically representing the fifth herbicide site-of-action group for resistance in A. tuberculatus (Heap Reference Heap2018). A different A. tuberculatus population from Nebraska (named NEB) also demonstrated resistance to HPPD-inhibiting herbicides (Kaundun et al. Reference Kaundun, Hutchings, Dale, Howell, Morris, Kramer, Shivrain and Mcindoe2017), and another A. tuberculatus population from Nebraska exhibited resistance to the synthetic auxin herbicide 2,4-D (Figueiredo et al. Reference Figueiredo, Leibhart, Reicher, Tranel, Nissen, Westra, Bernards, Kruger, Gaines and Jugulam2018). In each of these cases, rapid herbicide metabolism contributed to or conferred resistance in the population, as described in the following sections.
In addition to HPPD-inhibitor resistance in MCR, this population is resistant to s-triazines, ALS-inhibiting herbicides, and the foliar-applied PPO-inhibiting herbicide carfentrazone-ethyl (Ma et al. Reference Ma, Skelton and Riechers2015; unpublished data). Neither altered target sites (HPPD for mesotrione or psbA encoding the D1 protein in PSII for s-triazines) nor increased uptake of mesotrione or atrazine was detected in the MCR population. Biochemical studies using excised leaves and whole plants derived from vegetative clones demonstrated that elevated rates of metabolism, through distinct detoxification pathways, contribute to herbicide resistance in MCR (Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013). These metabolic pathways include: (1) oxidative metabolism (presumably via CYPs) for two HPPD-inhibiting herbicides, mesotrione (Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013) and topramezone (Ma et al. Reference Ma, Evans, O’Brien, Obenland, Lygin, Kaundun and Riechers2018), as well as the ALS-inhibiting herbicide primisulfuron-methyl (Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015; Ma et al. Reference Ma, Skelton and Riechers2015); and (2) GSH conjugation catalyzed by GSTs for atrazine (Evans et al. Reference Evans, O’Brien, Ma, Hager, Riggins, Lambert and Riechers2017; Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013; Ma et al. Reference Ma, Evans and Riechers2016). The use of an excised leaf assay and vegetatively cloned plants ensured that the herbicide-metabolism assay was independent of whole-plant translocation patterns and that identical A. tuberculatus genotypes were analyzed within a biochemical time-course analysis (Ma et al. Reference Ma, Skelton and Riechers2015). The CYP inhibitors malathion and tetcyclacis decreased mesotrione metabolism and further reduced the biomass of MCR plants when applied with mesotrione (Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013). Additionally, malathion resulted in a greater injury to MCR from ALS-inhibiting herbicides, including primisulfuron, cloransulam, sulfometuron, pyrithiobac, and imazethapyr (Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015). Treatment with the GST inhibitor CNBF followed by an atrazine PRE or POST application significantly enhanced biomass reduction in another atrazine-resistant A. tuberculatus population from central Illinois (ACR) compared with the atrazine-only treatment (Ma et al. Reference Ma, Evans and Riechers2016). These increased herbicide activities when applied with metabolic inhibitors to resistant A. tuberculatus further support metabolism-based resistance mechanisms to multiple herbicides in A. tuberculatus. Similarly, rapid mesotrione metabolism via 4-hydroxylation of the dione ring was detected in the NEB population compared with a sensitive population (Kaundun et al. Reference Kaundun, Hutchings, Dale, Howell, Morris, Kramer, Shivrain and Mcindoe2017). Rapid metabolism of 2,4-D contributes to resistance in another A. tuberculatus population from Nebraska, which is also potentially mediated by CYP activity (Figueiredo et al. Reference Figueiredo, Leibhart, Reicher, Tranel, Nissen, Westra, Bernards, Kruger, Gaines and Jugulam2018).
Genetic and Molecular Basis of Multiple-Herbicide Resistance in A. tuberculatus
Metabolism-based herbicide resistance can confer unpredictable and complicated cross- or multiple resistance to herbicides with the same or different sites of action, which could be controlled by quantitative trait(s) (Délye Reference Délye2013). A genetic study using F2 segregating lines of multiple herbicide–resistant (MCR) and sensitive A. tuberculatus populations demonstrated that atrazine resistance in the MCR population is conferred by a single, incompletely dominant nuclear gene, whereas the inheritance of mesotrione resistance in the MCR population is more complex and appears to be a multigenic trait (Huffman et al. Reference Huffman, Hausman, Hager, Riechers and Tranel2015). Following up on the atrazine resistance trait in the MCR and ACR populations, traditional protein purification and proteomic methods tested the hypothesis that enhanced metabolic detoxification of atrazine occurs by a distinct GST isozyme (Evans et al. Reference Evans, O’Brien, Ma, Hager, Riggins, Lambert and Riechers2017), which ultimately confers atrazine resistance in the MCR population. Several GST proteins were identified by liquid chromatography–mass spectrometry in affinity-purified fractions from A. tuberculatus using peptide sequence similarity with GSTs from Arabidopsis or other dicots. Elevated, constitutively expressed transcript levels of one phi-class GST (named AtuGSTF2) strongly correlated with atrazine resistance in A. tuberculatus (MCR and ACR populations) and an F2 population that segregates for metabolic atrazine resistance (Huffman et al. Reference Huffman, Hausman, Hager, Riechers and Tranel2015). This correlation indicates that AtuGSTF2 is the predominant GST protein that confers atrazine resistance in A. tuberculatus (Evans et al. Reference Evans, O’Brien, Ma, Hager, Riggins, Lambert and Riechers2017), although additional research involving gene cloning, promoter analysis, and generation of transgenic plants overexpressing AtuGSTF2 is required to further support this hypothesis.
Ongoing Research Investigating Unique Mechanisms Conferring Resistance to Topramezone and Carfentrazone-Ethyl in Amaranthus tuberculatus
The MCR population also demonstrated resistance to topramezone (Hausman et al. Reference Hausman, Singh, Tranel, Riechers, Kaundun, Polge, Thomas and Hager2011), another HPPD inhibitor having a distinct pyrazole structure (Ndikuryayo et al. Reference Ndikuryayo, Moosavi, Yang and Yang2017) compared with the triketone structures of mesotrione and tembotrione (Figure 13). Initial biochemical studies with excised leaves and whole plants indicated that an elevated rate of oxidative metabolism confers topramezone resistance in the MCR population relative to two HPPD-sensitive A. tuberculatus populations (Ma et al. Reference Ma, Evans, O’Brien, Obenland, Lygin, Kaundun and Riechers2018). However, the metabolic route for topramezone identified in MCR is different from the rapid initial N-demethylation reaction that occurs in tolerant maize (Grossmann and Ehrhardt Reference Grossmann and Ehrhardt2007). Recent research determined that two hydroxy-topramezone metabolites are predominantly formed in MCR, which are only present in minor amounts in maize and HPPD-sensitive A. tuberculatus (Lygin et al. Reference Lygin, Kaundun, Morris, McIndoe, Hamilton and Riechers2018). Finally, MCR displayed foliar resistance to carfentrazone-ethyl but sensitivity to diphenylethers, a different class of PPO inhibitors compared with carfentrazone-ethyl (an aryl triazinone). Carfentrazone-ethyl resistance in the MCR population is not due to a glycine codon deletion or arginine substitution in the PPO2 enzyme; it is more likely conferred through an NTSR mechanism such as enhanced oxidative metabolism (unpublished data), although additional research is required to test this hypothesis.

Figure 13 Chemical structures of (A) tembotrione, (B) mesotrione, and (C) topramezone. Mesotrione and tembotrione are examples of triketone p-hydroxyphenylpyruvate dioxygenase (HPPD) inhibiting herbicides, while topramezone belongs to the pyrazole subclass of HPPD-inhibiting herbicides.
Conclusions
Several weed populations around the world have evolved resistance to herbicides by metabolizing herbicide active ingredients to non-phytotoxic metabolites. Characterization of metabolic resistance mechanisms and underlying biochemical and molecular regulation is a difficult and arduous process. However, recent strides made through the use of contemporary approaches and procedures have made it possible to delineate roles of enzyme systems such as CYPs, GSTs, and GTs in metabolic and multiple resistance to herbicides in plants, including both crops and weeds, as outlined in this review. These biochemical and molecular mechanisms conferring resistance to multiple herbicides with different sites of action indicate weeds possess multiple genes encoding diverse metabolic enzymes, which ultimately result in complex, herbicide-dependent, cross- or multiple-resistance patterns. Cross- and multiple herbicide resistance could pose serious challenges for weed management in the future, especially if these resistance mechanisms do not confer a fitness cost to the plant in the absence of herbicide (Délye Reference Délye2013; Yu and Powles Reference Yu and Powles2014). Additionally, herbicide resistance due to rapid herbicide metabolism has the potential to confer resistance not only to existing commercial herbicides, but also to new or yet-to-be-discovered active ingredients (Yu and Powles Reference Yu and Powles2014). A better understanding of the biochemical, molecular, and genetic mechanisms conferring metabolic resistance to multiple herbicides provides insights into evolving weed populations in response to selection pressures and development of innovative and integrated resistant weed management strategies and demonstrates an urgent need for discovery of a new herbicide site of action (Duke Reference Duke2012).
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/wsc.2018.88
Acknowledgments
VKN gratefully recognizes funding from the Weed Science Society of America toward organizing a symposium at the 2018 WSSA annual meeting in Arlington, VA. DER acknowledges expert technical assistance from Anatoli V. Lygin, Mayandi Sivaguru, and Yousoon Baek, University of Illinois–Urbana. RE acknowledges the joint support of the Biotechnology and Biological Sciences Research Council (grant BB/L001489/1) and Agriculture and Horticulture Development Board (RD-2012-3807). No conflicts of interest have been declared.