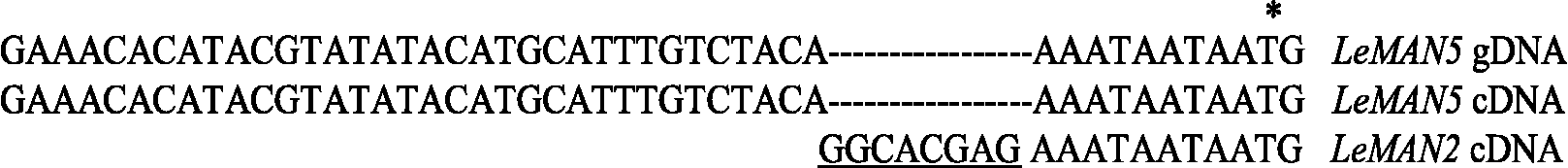Introduction
The cell walls of higher plants contain varying amounts of mannan-type polysaccharides (i.e. mannans, glucomannans, and galacto[gluco]mannans). At least 81 families with soft, watery or fleshy galactomannan-containing endosperms and 19 having hard galactomannan-containing endosperms have been identified (Reid and Meier, Reference Reid and Meier1970). In many seeds, such as those of endospermic legumes, e.g. carob or locust bean (Ceratonia siliqua), fenugreek (Trigonella foenum-graecum) and guar (Cyamopsis tetragonoloba); and of Datura ferox, lettuce (Lactuca sativa), and tomato (Solanum lycopersicum), which do not store appreciable amounts of starch, these polysaccharides are restricted to the endosperm and are responsible for the thickening of its cell walls. They serve as storage carbohydrates. After seed germination, they are degraded by several hydrolases to provide nutrients for the growth of the seedling.
Galactomannans are polysaccharides with β-1,4-linked mannose as a backbone to which a number of α-1,6-linked galactose side-chains are attached. In seeds, those polysaccharides with fewer than 10% galactose substitution of the backbone are often defined as ‘mannans’, while ‘galactomannans’ have more galactose side-chains (Bewley and Reid, Reference Bewley, Reid, Dey and Dixon1985). The two types of polysaccharides are quite different in their physical properties. Galactomannans are highly hydrophilic ‘seed mucilages’ and are present in some endospermic legumes, e.g. carob (McCleary and Matheson, Reference McCleary and Matheson1975), Chinese senna (Cassia tora) and fenugreek (Reid, Reference Reid1971). Mannans are water-insoluble crystalline substances that can confer hardness upon the tissues that contain them, such as seeds of date palm (Phoenix dactylifera) and coffee bean (Coffea arabica) (Bewley and Reid, Reference Bewley, Reid, Dey and Dixon1985). Seed glucomannans contain a backbone of β-1,4-linked glucose and mannose residues. They are sparsely galactose-substituted and resemble the mannans in their physical properties (Bewley and Reid, Reference Bewley, Reid, Dey and Dixon1985). The endosperm cell walls of lettuce (Halmer et al., Reference Halmer, Bewley and Thorpe1975) and tomato (Groot et al., Reference Groot, Kieliszewska-Rokicka, Vermeer and Karssen1988) seeds are composed of large quantities of mannose and smaller quantities of galactose and glucose, suggesting that a large portion of the cell wall polysaccharides is of the mannan type. In lettuce, cell walls of the micropylar and lateral endosperm regions have different polysaccharide compositions; the former has a higher proportion of arabinose (Dutta et al., Reference Dutta, Bradford and Nevins1994).
After seed germination, the galactomannan reserves are mobilized to support the growing seedling. Three enzymes, endo-β-mannanase (EC 3.2.1.78), β-mannosidase (EC 3.2.1.25) and α-galactosidase (EC 3.2.1.22), have been identified as being involved in the hydrolysis of endosperm cell walls. Endo-β-mannanase randomly hydrolyses the β-1,4 mannose backbone chains of mannans and galactomannans to produce mannobiose and mannotriose; α-galactosidase removes the galactose residues from the side-chains of galactomannans; and finally mannotriose and mannobiose are hydrolysed into mannose by β-mannosidase (Reese and Shibata, Reference Reese and Shibata1965; Reid and Meier, Reference Reid and Meier1973; McCleary and Matheson, Reference McCleary and Matheson1975). In different seeds, the syntheses of the three hydrolytic enzymes occur in different places, and mobilization of galactomannan occurs in different ways. In some seeds, e.g. fenugreek, crimson clover (Trifolium incarnatum) (Reid and Meier, Reference Reid and Meier1972) and guar (McClendon et al., Reference McClendon, Nolan and Wenzler1976), the mobilization of galactomannans in the endosperm requires the participation of an aleurone layer. In fenugreek, during galactomannan mobilization, a dissolution zone initially forms in the endosperm next to the aleurone layer and progressively increases in size, inwards, towards the cotyledons (Reid, Reference Reid1971). Unlike in fenugreek, there is no specialization of the carob seed endosperm into storage tissue and aleurone layer. The hydrolytic enzymes endo-β-mannanase, α-galactosidase and β-mannosidase are produced within the endosperm cells themselves, which remain living (Seiler, Reference Seiler1977). After germination, a small amount of wall degradation occurs throughout the endosperm, but the major galactomannan breakdown then proceeds from the embryo outwards. In some seeds, e.g. asparagus (Asparagus officinalis) (Goldberg and Roland, Reference Goldberg and Roland1971) and date (Keusch, Reference Keusch1968), the mobilization of the endosperm begins adjacent to the haustorial cotyledon and then advances outwards toward the testa. A dissolution zone is soon observed around the haustorium as the endosperm cell walls are hydrolysed. It has been suggested that hemicellulases are produced in, and secreted by, the haustorial cotyledon, which remains within the endosperm following germination. However, the endosperm cells are living and are capable of enzyme synthesis (Keusch, Reference Keusch1968; Sekhar and DeMason, Reference Sekhar and DeMason1990; Williams et al., Reference Williams, Bewley, Greenwood, Bourgault and Mo2001).
Endo-β-mannanase is the key enzyme in the mobilization of cell wall mannan polymers, and it has been studied most extensively in seeds, but also in non-seed tissues, e.g. during flower development (Filichkin et al., Reference Filichkin, Leonard, Monteros, Liu and Nonogaki2004) and fruit ripening (Bourgault and Bewley, Reference Bourgault and Bewley2002). There are many reports of the presence of endo-β-mannanase and its isozymes and isoforms in seeds containing galactomannan reserves during and after germination (Halmer et al., Reference Halmer, Bewley and Thorpe1976; Dirk et al., Reference Dirk, Griffen, Downie and Bewley1995; Nonogaki et al., Reference Nonogaki, Nomaguchi and Morohashi1995; Voigt and Bewley, Reference Voigt and Bewley1996; Sánchez and de Miguel, Reference Sánchez and de Miguel1997; Marraccini et al., Reference Marraccini, Rogers, Allard, André, Caillet, Lacoste, Lausanne and Michaux2001; Williams et al., Reference Williams, Bewley, Greenwood, Bourgault and Mo2001; Homrichhausen et al., Reference Homrichhausen, Hewitt and Nonogaki2003; Da Silva et al., Reference Da Silva, Toorop, van Aelst and Hilhorst2004; Gong et al., Reference Gong, Bassel, Wang, Greenwood and Bewley2005). They show spatio-temporal and tissue specificities as well as hormonal control of expression (Kontos and Spyropoulos, Reference Kontos and Spyropoulos1996; Toorop et al., Reference Toorop, Bewley and Hilhorst1996; Voigt and Bewley, Reference Voigt and Bewley1996; Wang et al., Reference Wang, Wang, Ren, Gong and Bewley2005). In fenugreek endosperms, the endo-β-mannanase protein is present before the enzyme activity is detected, suggesting that an inactive form of the enzyme is produced initially (Malek and Bewley, Reference Malek and Bewley1991; Gong, personal communication). The enzyme has been completely or partially purified from seeds of endospermic legumes (McCleary, Reference McCleary1978, Reference McCleary1988), lettuce (Dulson and Bewley, Reference Dulson and Bewley1989; Halmer, Reference Halmer1989), tomato fruit, from which it has been crystallized and its structure determined (Pressey, Reference Pressey1989; Bourgault et al., Reference Bourgault, Oakley, Bewley and Wilce2005), and tomato seeds (Nonogaki et al., Reference Nonogaki, Nomaguchi and Morohashi1995). The genes encoding this enzyme have been isolated and characterized from several species, such as seeds of coffee (Marraccini et al., Reference Marraccini, Rogers, Allard, André, Caillet, Lacoste, Lausanne and Michaux2001; Eira et al., Reference Eira, da Silva, de Castro, Dussert, Walters, Bewley and Hilhorst2006), carrot (Daucus carota) (Homrichhausen et al., Reference Homrichhausen, Hewitt and Nonogaki2003) and lettuce (Wang et al., Reference Wang, Li and Bewley2004), and from seeds and fruits of tomato (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997; Nonogaki et al., Reference Nonogaki, Gee and Bradford2000; Bourgault and Bewley, Reference Bourgault and Bewley2002).
In some seeds, e.g. tomato (Groot and Karssen, Reference Groot and Karssen1987; Haigh and Barlow, Reference Haigh and Barlow1987), Datura ferox (Sánchez and de Miguel, Reference Sánchez and de Miguel1997) and white spruce (Picea glauca) (Downie et al., Reference Downie, Hilhorst and Bewley1997), an increase in the endo-β-mannanase activity is associated with the weakening of the micropylar endosperm (endosperm cap). This weakening is proposed to be the mechanism that facilitates the completion of germination in these seeds (Sánchez et al., Reference Sánchez, Sunell, Labavitch and Bonner1990; Sánchez and de Miguel, Reference Sánchez and de Miguel1997; Nonogaki et al., Reference Nonogaki, Gee and Bradford2000). Consequently, studies have focused on the role of endo-β-mannanase activity in the weakening of the micropylar endosperm cell walls surrounding the radicle and its association with radicle protrusion; tomato seeds, in particular, have been used as the ‘model system’.
Tomato seeds have several advantages as an experimental organism for studying germination and endo-β-mannanase function. The seed is sufficiently large to facilitate its dissection and separation into specific seed tissues, e.g. embryo, endosperm and seed coat. Synthesis of endo-β-mannanases within the endosperm of tomato seeds is promoted by gibberellic acid (GA) and inhibited by abscisic acid (ABA) (Groot et al., Reference Groot, Kieliszewska-Rokicka, Vermeer and Karssen1988). Various mutants of tomato (GA-deficient, ABA-deficient, auxin-insensitive and ethylene-insensitive) are available for studying the roles of these hormones in regulating germination (Kelly and Bradford, Reference Kelly and Bradford1986). Although generally not dormant, tomato seeds can exhibit dormancy under some conditions, and germination is sensitive to far-red light (Downie et al., Reference Downie, Gurusinghe and Bradford1999) and water potential (Dahal and Bradford, Reference Dahal and Bradford1990). The effects of environmental or hormonal regulation on gene expression, therefore, can be compared to their corresponding effects on germination. In the tomato seed, the endosperm surrounding the embryo physically inhibits protrusion of the radicle (Bewley and Black, Reference Bewley and Black1994). When embryos in tomato gib-1 seeds are removed from their dormancy-imposing surrounding endosperm tissues, they complete germination on water despite their inability to produce GA (Groot and Karssen, Reference Groot and Karssen1987). Thus, a weakening of the cell walls in the micropylar endosperm coincidental with an increase in the endo-β-mannanase activity is suggested to permit germination (Bewley, Reference Bewley1997; Bradford et al., Reference Bradford, Chen, Cooley, Dahal, Downie, Fukunaga, Gee, Gurusinghe, Mella, Wu, Yang, Yim, Black, Bradford and Vázquez-Ramos2000). The walls of the lateral endosperm are mobilized following germination as an early source of carbohydrate for the growing seedling (Bewley, Reference Bewley1997).
Genes encoding endo-β-mannanases in tomato
Five genes (LeMAN1, LeMAN2, LeMAN3, LeMAN4 and LeMAN5) encoding endo-β-mannanases in tomato have been isolated and characterized, although there are two forms of LeMAN4 (LeMAN4a and LeMAN4i) (Fig. 1). These genes from tomato are expressed at different developmental stages, from seed germination to fruit ripening (Table 1).

Figure 1 (a) Alignment of amino acid sequences of tomato post-germinative (LeMAN1, GenBank AAB87859), germinative (LeMAN2 AF184238), predominantly post-germinative (LeMAN3 AF290893), fruit (LeMAN4a AY046588), and flower (LeMAN5 AY102168) endo-β-mannanases. (b) Alignment of amino acid sequences of LeMAN1, SGN-U335863, SGN-U316912, SGN-U316863 and SGN-U335864. (c) Alignment of amino acid sequences of LeMAN4a and SGN-U334573; *indicates an identical sequence, : indicates a conservative substitution. Amino acids identical in three or more of the sequences are highlighted by dark shading in A and B; grey shading indicates an uncertain degree of homology.
Table 1 Location and times of expression of endo-β-mannanase genes in tomato

LeMANs 1, 3 and 5, for which the genomic sequences are known, all contain four introns of varying lengths. The encoded amino acid sequences on both sides of the introns are conserved in these genes, as well as in those from Arabidopsis, rice and poplar (Yuan et al., Reference Yuan, Yang, Lai, Lin, Cheng, Nonogaki and Chen2007). The genes of these other species have four or five introns, four of which are highly conserved and the fifth, when present, is less so. Analysis of the intron/exon structure and phylogenetic analysis suggests that both intron gain and loss have played roles in the evolution of endo-β-mannanase genes (Yuan et al., Reference Yuan, Yang, Lai, Lin, Cheng, Nonogaki and Chen2007).
Endo-β-mannanase encoded by LeMAN1 in seeds
LeMAN1 (GenBank accession no. AAB87859) was the first endo-β-mannanase cDNA identified in tomato (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997), with a reported nucleotide sequence of 1307 bp, and an open reading frame encoding 369 amino acids. More recently, a missing motif of 28 amino acids was identified in the N-terminus of this enzyme (GenBank AF017144). The transcripts of LeMAN1 are expressed in the lateral endosperm after germination, and its protein product functions as a post-germinative hydrolase to mobilize galactomannan in the cell walls to support the growth of the seedling (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997; Nonogaki et al., Reference Nonogaki, Gee and Bradford2000).
Endo-β-mannanase encoded by LeMAN2 in seeds
LeMAN2 (GenBank AF184238) is another endo-β-mannanase cDNA identified in tomato (Nonogaki et al., Reference Nonogaki, Gee and Bradford2000). Its protein shares 79% amino acid sequence similarity with the post-germinative enzyme encoded by LeMAN1. It is expressed exclusively in the micropylar endosperm in germinating seeds, indicating that it is involved in the weakening of the micropylar endosperm to allow radicle emergence to complete germination (Nonogaki et al., Reference Nonogaki, Gee and Bradford2000).
Endo-β-mannanase encoded by LeMAN3 in seeds
LeMAN3 (GenBank AF290893) was the first fully sequenced endo-β-mannanase gene identified from tomato, and its protein product is more identical with LeMAN1 than LeMAN2. Its expression pattern has not been characterized to date. Hence, to determine where mRNA encoded by this gene is expressed in tomato, Northern blot analyses were carried out. Total RNA from germinating and germinated wild-type seed, flower, leaf, root, pericarp of fruit, and stem tissues was hybridized with a LeMAN3 probe; the transcripts of LeMAN3 were detected only in seed tissues (Fig. 2a). Transcripts of this gene were barely detectable late during germination compared to the very large amount detected after this was completed (Fig. 2a). Tomato gib-1 mutant seeds, which are incapable of producing GA and cannot germinate without application of this hormone, were used to investigate hormonal regulation of the expression of this gene. LeMAN3 mRNA was not detected in gib-1 mutant seeds on water, but it was present after 48 h in germinating seeds on GA and in germinated seeds, particularly at 72 h after imbibition (Fig. 2b). mRNAs from gib-1 embryos and endosperms, dissected from seeds imbibed in water or GA for various times, were also subjected to Northern blot analysis. The transcripts of LeMAN3 were detected only in the endosperms dissected from the already germinated mutant seeds imbibed in GA (Fig. 2c). Only by long exposure of the blot were the transcripts detectable before completion of germination (radicle protrusion). There was no expression detected of LeMAN3 mRNA in 100 μM ABA-imbibed wild-type seeds (Fig. 2d), although this could be due indirectly to the failure of the seeds to complete germination.

Figure 2 (a) Northern blot of total RNA from tomato wild-type whole seeds imbibed in water for 48–96 h and from tomato flower (flw), leaf, root, fruit pericarp (peri), and stem tissues. (b) Northern blot of mRNA from tomato gib-1 mutant intact seeds imbibed in water or gibberellic acid (GA) for 24–96 h. (c) Northern blot of mRNA from tomato gib-1 mutant seed parts (embryo, endosperm) that were dissected from seeds imbibed in water or gibberellic acid (GA) for 48–72 h. (d) Northern blot of mRNA from tomato wild-type whole seeds imbibed in water or abscisic acid (ABA, 100 μM) for 24–72 h. rRNA stained with ethidium bromide demonstrates equal loading in each lane of all gels. All blots were hybridized with 32P-labelled LeMAN3 probe. NG, non-germinated; G, germinated.
Endo-β-mannanase encoded by LeMAN4a, LeMAN4i in the fruit
Endo-β-mannanase activity is also present in tomato fruit, mainly in the skin and outer pericarp, where the enzyme exists in an active or an inactive form, depending upon the cultivar (Bewley et al., Reference Bewley, Banik, Bourgault, Feurtado, Toorop and Hilhorst2000; Banik et al., Reference Banik, Bourgault and Bewley2001). Thus, endo-β-mannanase likely plays a role in ripening-associated softening (Pressey, Reference Pressey1989; Bewley et al., Reference Bewley, Banik, Bourgault, Feurtado, Toorop and Hilhorst2000); its activity increases in the outer tissues of the fruit as it ripens from the turning to the red stages. In several non-ripening tomato mutants that fail to soften, there is reduced enzyme activity (Bewley et al., Reference Bewley, Banik, Bourgault, Feurtado, Toorop and Hilhorst2000). Endo-β-mannanase cDNAs (LeMAN4a GenBank AY046588, LeMAN4i GenBank AY046589) were cloned and characterized from ripening tomato cv. Trust fruit, which produces an active enzyme, and from the cv. Walter, which produces an inactive enzyme (Bourgault and Bewley, Reference Bourgault and Bewley2002). Another endo-β-mannanase cDNA, LeMAN4 (GenBank AY034075) was also isolated from tomato cv. Castalia fruit (Carrington et al., Reference Carrington, Vendrell and Domínguez-Puigjaner2002). The amino acid sequence encoded by LeMAN4 is identical with that of LeMAN4i, except for one amino acid difference. They are likely the same gene, and the sequence difference was probably a DNA sequencing error.
Besides its mannan hydrolase activity, the endo-β-mannanase encoded by LeMAN4 from ripe tomato fruit functions as a mannan transglycosylase in the presence of mannan-derived oligosaccharides (Schröder et al., Reference Schröder, Wegrzyn, Sharma and Atkinson2006).
Endo-β-mannanase encoded by LeMAN5 in anthers
Endo-β-mannanase activity is also present in developing tomato anthers (Downie et al., Reference Downie, Hilhorst and Bewley1994) and in pollen (Filichikin et al., Reference Filichkin, Leonard, Monteros, Liu and Nonogaki2004). A novel endo-β-mannanase gene LeMAN5 (GenBank AY102168) was identified in the tomato genome by genome-walking polymerase chain reaction (PCR) (Filichkin et al., Reference Filichkin, Leonard, Monteros, Liu and Nonogaki2004). Its cDNA has 98% identity with LeMAN2 cDNA, and its entire coding region is identical to that of LeMAN2. Transcript expression of LeMAN5 occurs in tomato anthers and pollen. Its putative promoter, driving a GUS-reporter, is activated in anthers and pollen of transgenic Arabidopsis, suggesting that endo-β-mannanase LeMAN5 is associated with anther and pollen development (Filichkin et al., Reference Filichkin, Leonard, Monteros, Liu and Nonogaki2004).
Putative endo-β-mannanases from tomato database
Expressed sequence tags (EST) of five more unigenes, possibly encoding endo-β-mannanases, were found in the tomato EST database by BLASTing the above LeMAN genes in the Sol Genomics Network database (http://www.sgn.cornell.edu/) (Table 1). They share a high similarity with the LeMAN genes. Some ESTs are more identical to LeMAN1 (Fig. 1b) and others more with LeMAN4 (Fig. 1c).
Properties of endo-β-mannanases in tomato
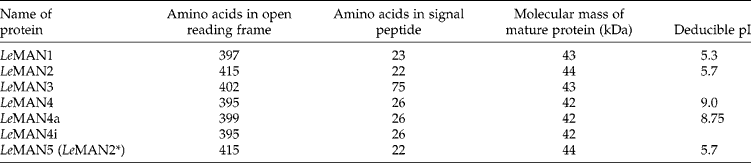
The proteins encoded by the endo-β-mannanase genes share quite high amino acid sequence similarities with each other (Fig. 1). However, there is only about a 30% identity between tomato endo-β-mannanases and those from Aspergillus aculeatus (Christgau et al., Reference Christgau, Kauppinen, Vind, Kofod and Dalboge1994) and Trichoderma reeseii (unpublished data, GenBank L25310). Southern blot analysis suggests that tomato endo-β-mannanases are encoded by a gene family of four genes or more (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997; Nonogaki et al., Reference Nonogaki, Gee and Bradford2000). Signal peptides are located at the N-terminus of these proteins, as would be expected since they are secreted; the molecular mass of the mature proteins is 42–44 kDa. The characteristics of these enzymes are shown in Table 2.
Table 2 Characteristics of endo-β-mannanase proteins in tomato
In tomato seeds, the endo-β-mannanases (LeMANs 1–3) are highly soluble and easily extractable in water or low-salt buffers, as is the enzyme from the anthers and pollen (LeMAN5). In contrast, the enzyme in the fruit skin (outer and inner pericarp) requires a salt-containing buffer for extraction, which is indicative of its tight association with the cell walls. This is the case whether or not the enzyme is active or inactive, as shown by immunolocalization (Bewley et al., Reference Bewley, Banik, Bourgault, Feurtado, Toorop and Hilhorst2000; Banik et al., Reference Banik, Bourgault and Bewley2001).
A two-nucleotide deletion at the 3’ end of tomato fruit LeMAN4i causes an open reading frame shift and a resultant truncated C-terminus in the inactive enzyme, compared with the active enzyme encoded by the LeMAN4a gene (Bourgault and Bewley, Reference Bourgault and Bewley2002). The truncated inactive enzyme lacks a Ser–Lys–Leu–Ser (SKLS) sequence present in the C-terminus of the LeMAN4a, which is suggestive of its importance for endo-β-mannanase activity. Of these four amino acids, leucine (Leu) is the most important one for full enzyme activity. It is likely required for correct protein folding to allow for binding of the galactomannan substrate (Bourgault et al., Reference Bourgault, Oakley, Bewley and Wilce2005). Leu is also present close to the C-terminus in seed-specific LeMAN1 (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997), LeMAN2 (LeMAN5) and LeMAN3 (Nonogaki et al., Reference Nonogaki, Gee and Bradford2000), and in one of the endo-β-mannanases cloned from coffee (CaMANB) (Marraccini et al., Reference Marraccini, Rogers, Allard, André, Caillet, Lacoste, Lausanne and Michaux2001) (Fig. 3). The enzyme C-terminal sequences from coffee (CaMANA), carrot (Homrichhausen et al., Reference Homrichhausen, Hewitt and Nonogaki2003) and lettuce seeds (Wang et al., Reference Wang, Li and Bewley2004) contain only one common Leu (which is not regarded as being important for activity because it is present in LeMAN4i), but there is no Leu closer to the C-terminus (Fig. 3). Despite its low sequence similarity with higher-plant endo-β-mannanases, that of Trichoderma reeseii also possesses a Leu residue in the penultimate position of its C-terminus.

Figure 3 C-terminus amino acid sequences of endo-β-mannanases CaMANA (coffee GenBank AJ293305), CaMANB (coffee AJ278996), DcMAN1 (carrot AF545503), LeMAN1 (tomato), LeMAN2 (tomato), LeMAN3 (tomato), LeMAN4a (tomato), LeMAN4i (tomato) and LsMAN1 (AJ315978). The amino acid leucine (L) that is common to all endo-β-mannanases is highlighted by dark shading. The terminal Leu (L) is boxed.
Characterization of LeMAN promoter sequences and the regulation of gene expression of LeMANs
The PLACE database (http://www.dna.affrc.go.jp/htdocs/PLACE/; Higo et al., Reference Higo, Ugawa, Iwamoto and Korenaga1999) was used to search for cis-acting regulatory DNA elements in the upstream regions of LeMAN1, LeMAN3, LeMAN4a and LeMAN5. The results (Table 3) show that ABA-responsive elements (ABREs, Simpson et al., Reference Simpson, Nakashima, Narusaka, Seki, Shinozaki and Yamaguchi-Shinozaki2003) are in the upstream sequences of LeMAN1 and LeMAN3, and that GA-responsive elements (GAREs, Ogawa et al., Reference Ogawa, Hanada, Yamauchi, Kuwahara, Kamiya and Yamaguchi2003; Sutoh and Yamauchi, Reference Sutoh and Yamauchi2003) are present in the upstream sequences of all the four genes. These promoter sequences also contain several copies of pollen-specific motifs (AGAAA) (POLLEN1LELAT52, Bate and Twell, Reference Bate and Twell1998), a motif associated with seed storage protein genes (EBOXBNNAPA, Stalberg et al., Reference Stalberg, Ellerstom, Ezcurra, Ablov and Rask1996) and copies of a root-specific element (ROOTMOTIFTAPOX1, Elmayan and Tepfer, Reference Elmayan and Tepfer1995).
Table 3 Putative cis-elements in the upstream sequences of endo-β-mannanase genes in tomato

Many experiments have shown that ABA inhibits and GA induces seed germination. The expression of LeMAN2, which is involved in seed germination, is induced by GA in gib-1 mutant seeds (Nonogaki et al., Reference Nonogaki, Gee and Bradford2000), but it is not inhibited by ABA at 100 μM in wild-type seeds. The endo-β-mannanases encoded by LeMAN1 and LeMAN3 (Fig. 2c) are induced by GA in isolated endosperms. However, because expression of LeMAN1 and LeMAN3 is a post-germinative event and ABA inhibits germination, the genes are presumably not expressed in the presence of this inhibitor due to a failure of the seeds to complete germination. The regulation of gene expression by ABA and GA corresponds to the related motifs present in the promoter regions of these genes. Although there are no reports on the regulation of LeMAN4, it is probably also regulated by GA, considering that the GARE motif is present in its promoter sequence.
Comparison of endo-β-mannanase cDNAs from seeds with their genomic sequences
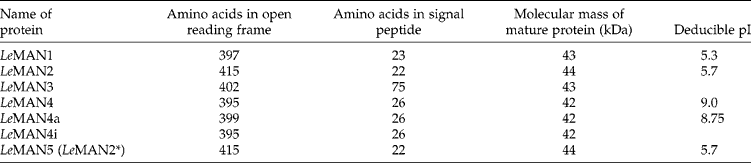
The LeMAN5 cDNA shows a very high identity with LeMAN2 cDNA, and its entire coding region is identical to that of LeMAN2. The only difference between these two cDNAs is an eight nucleotide sequence (GGCACGAG) present in the 5′ untranslated region (5′ UTR) of LeMAN2 cDNA, but not in that of LeMAN5 cDNA or its corresponding genomic DNA (gDNA) (Fig. 4). This extra sequence is likely to be an artefact generated from the adaptor used in the Stratagene Uni-ZAP XR Premade Library. Thus, LeMAN5 and LeMAN2 cDNAs (and genes) can be regarded as being identical. Hence, in consultation with Dr Hiro Nonogaki and his colleagues, who first published data on LeMAN5, it is agreed this gene be renumbered in GenBank as LeMAN2*, to identify its similarity to LeMAN2, but its separate experimental and tissue origin. Using the same Stratagene kit, this eight-nucleotide sequence was also produced at the 5′ UTR of the LeMAN1 cDNA (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997), but it is not present in the genomic sequence.
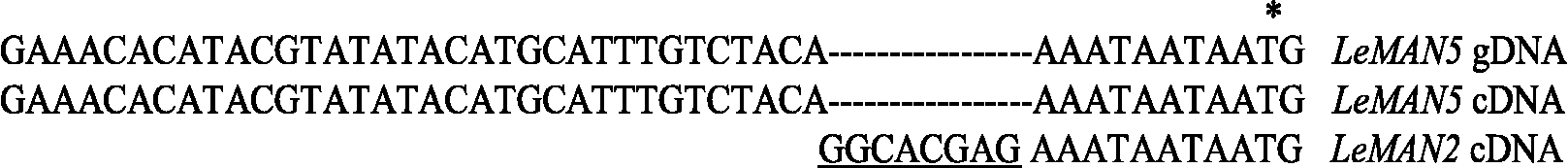
Figure 4 Comparison of the DNA sequences at the 5′ end of LeMAN5 cDNA and its corresponding genomic DNA (gDNA) with that of LeMAN2 cDNA. The first ATG in the coding region is indicated by an asterisk. The eight-nucleotide sequence (GGCACGAG) in the LeMAN2 cDNA is also at the 5′ end of the LeMAN1 cDNA (Bewley et al., Reference Bewley, Burton, Morohashi and Fincher1997), and is an artefact obtained during the synthesis of the cDNA libraries for both clones, which were prepared using the Stratagene Uni-ZAP XR Premade Library Kit.
Summary
According to previous publications and the tomato genomic database, more than 10 genes encoding endo-β-mannanase are present in the tomato genome. Of these, five have been identified and their functions investigated at different developmental stages of tomato: LeMANs 1–3 in the endosperm, LeMAN4 in the fruit pericarp, and LeMAN5 in the flower, although since LeMAN5 and LeMAN2 are the same gene, the former is now designated as LeMAN2*. Genes encoding endo-β-mannanases in other tissues, such as in the leaf (Sol Genomics Network database), have not been investigated. According to the tomato EST database, more than one gene is expressed in the fruit pericarp. They may function redundantly or in different special regions thereof, as do LeMAN1 and LeMAN2 in the seed endosperm. A major challenge in studying these genes is the high similarity of the sequences, making it difficult to distinguish between them. The similar motifs present in the upstream sequences of these genes suggest that they should be expressed in the same tissue at the same developmental stages. But some of them are expressed only at specific stages, in different tissues or in distinct regions of a tissue. The mechanism that regulates differential expression of the LeMAN genes in tomato remains to be determined.
Acknowledgements
This work is supported by an NSERC Postgraduate Scholarship awarded to X.G., and NSERC Discovery Grant 044191 to J.D.B. We are grateful to Dr Hiro Nonogaki for reading the manuscript, for elucidating the likely reason for the differences in sequence between the cDNAs of LeMAN2 and 5, and for suggesting the renumbering of LeMan5.