Introduction
PEPCase (phosphoenolpyruvate carboxylase) (EC 4.1.1.31) in C3 plants is recognized as an important enzyme in the anaplerotic provision of carbon skeletons to the citrate acid cycle (O'Leary et al., Reference O'Leary, Park and Plaxton2011). This enzyme catalyses the irreversible β-carboxylation of phosphoenolpyruvate in the presence of HCO3 − to yield oxaloacetate, which is a precursor of citrate, malate and aspartic acid. The PEPCase enzyme can be allosterically regulated by certain substances, such as malate and glucose-6-phosphate (G6P), in accordance with its phosphorylation status. The physiological role of this enzyme in various plant organs has been investigated by examining its enzymological properties and expression patterns: the relationship between PEPCase and nitrogen metabolism, and the contribution of PEPCase to nitrogen assimilation have been well established by previous studies (Van Quy et al., Reference Van Quy, Foyer and Champigny1991; Sugiharto and Sugiyama, Reference Sugiharto and Sugiyama1992; Van Quy and Champigny, Reference Van Quy and Champigny1992; Huppe and Turpin, Reference Huppe and Turpin1994; Podesta and Plaxton, 1994a, b; Suzuki et al., Reference Suzuki, Crétin, Omata and Sugiyama1994; Duff and Chollet, Reference Duff and Chollet1995; Zhang et al., Reference Zhang, Bin and Chollet1995). Recent molecular approaches have revealed that C3 plants contain several PEPCase isoforms that can be classified into plant and bacterial types; the bacterial-type PEPCase isoforms lack the distinctive N-terminal serine-phosphorylation site, which is seen in plant-type PEPCase (Sánchez and Cejudo, Reference Sánchez and Cejudo2003; Sullivan et al., Reference Sullivan, Jenkins and Nimmo2004; Mamedov et al., Reference Mamedov, Moellering and Chollet2005; Gennidakis et al., Reference Gennidakis, Rao, Greenham, Uhrig, O'Leary, Snedden, Lu and Plaxton2007; Igawa et al., Reference Igawa, Fujiwara, Tanaka, Fukao and Yanagawa2010).
A physiological role of PEPCase in immature seeds has been investigated and has been speculated to include support of the biosynthesis of storage compounds. In castor oil seeds, two PEPCases were characterized, and their possible functions in storage protein and fatty acid synthesis were discussed (Blonde and Plaxton, Reference Blonde and Plaxton2003). A PEPCase in Vicia faba L. seems to be involved in amino acid synthesis in cotyledons (Golombek et al., Reference Golombek, Heim, Horstmann, Wobus and Weber1999). We have shown a correlation between nitrogen content and PEPCase activity in some soybean cultivars (Sugimoto et al., Reference Sugimoto, Tanaka, Monma, Kawamura and Saio1989). We also observed an increase in PEPCase activity in developing rice seeds in response to nitrogen fertilization in the flowering period, causing simultaneous high nitrogen accumulation in rice seeds (Sugimoto et al., Reference Sugimoto, Sueyoshi, Oji, Ando, Fujita, Mae, Matsumoto, Mori and Sekiya1997). However, it remains unclear whether PEPCase contributes to nitrogen accumulation in seeds.
Rice is one of the major crops in the world and the nitrogen content in its seeds is a crucial key to its quality (Yan-Lin et al., Reference Yan-Lin, Jing-Feng, Shao-Hong and Ren-Chao2007). Rice seeds are considered to provide 20% of dietary protein intake in all developing countries (International Year of Rice Secretariat, 2004). In contrast, the protein content in rice grains influences the texture of cooked rice (Tamaki et al., Reference Tamaki, Ebata, Tashiro and Ishikawa1989; Hamaker and Griffin, Reference Hamaker and Griffin1990; Okadome et al., Reference Okadome, Kurihara, Kusuda, Toyoshimna, Kim, Shimotsubo, Matsuda and Ohtsubo1999; Champagne et al., Reference Champagne, Bett-Garber, Thomson and Fitzgerald2001; Martin and Fitzgerald, Reference Martin and Fitzgerald2002; Derycke et al., Reference Derycke, Veraverbeke, Vandeputte, De Man, Hoseney and Deleour2005; Xie et al., Reference Xie, Chen, Duan, Zhu and Liao2008) and flavour (Onate et al., Reference Onate, del Mundo and Juliano1964; Juliano et al., Reference Juliano, Onate and del Mundo1965; Park et al., Reference Park, Kim and Kim2001; Champagne et al., Reference Champagne, Bett-Garber, McClung and Bergman2004, Reference Champagne, Bett-Garher, Grimm and McClung2007). Low nitrogen content is also a critical factor of brewer's rice seed quality (Furukawa et al., Reference Furukawa, Tanaka, Masumura, Ogihara, Kiyokawa and Wakai2006). Although it also increases the nitrogen content in rice seeds (Perez et al., Reference Perez, Juliano, Liboon, Alcantara and Cassman1996), nitrogen fertilization in the flowering stage is an effective technique to increase the yield.
Six PEPCase isoforms have been found in the rice genome: five plant-type isoforms and a bacterial type having an expression pattern with distinctive features (Sánchez and Cejudo, Reference Sánchez and Cejudo2003; Lin et al., Reference Lin, Wei, Jiang, Li, Qian, Attia and Yang2004; Masumoto et al., Reference Masumoto, Miyazawa, Ohkawa, Fukuda, Taniguchi, Murayama, Kusano, Saito, Fukayama and Miyao2010). The putative polypeptide length diverges slightly among them, ranging from 92.5 to 103.6 kDa. At the protein level, four PEPCase subunits have been detected in leaves by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting (Matsuoka and Hata, Reference Matsuoka and Hata1987). Although such molecular-level information has been accumulated for rice PEPCase, there has been no report on PEPCase in developing rice seeds.
In this study, we cloned PEPCase Osppc1 (MSU ID: LOC_Os02g14770, assigned at the Rice Genome Annotation Project: http://rice.plantbiology.msu.edu/) as a candidate gene working in developing rice seeds. This full-length cDNA was isolated from a cDNA library of developing rice seeds with its isoform Osppc3. Sequence analysis revealed that the translation product of Osppc1 seemed to have PEPCase enzymatic characteristics similar to a C3-type isoform, and recombinant protein expression analysis confirmed its predicted enzymatic properties. Osppc1 mRNA in seeds was increased after the flowering stage and peaked in the middle of the grain-filling stage, when the rate of nitrogen accumulation in seeds was rapid. On the other hand, Osppc3 mRNA was expressed strongly during the early part of the grain-filling stage. Osppc1 showed a co-expression pattern with the two aminotransferases that are located downstream of the PEPCase reaction in the metabolic pathway. Osppc1 may work in supporting biosynthesis of amino acids that are consumed in storage protein synthesis in rice seeds, by the provision of oxaloacetate (OAA).
Materials and methods
Plant material
Rice (Oryza sativa, L. cv. Nipponbare) plants for DNA and RNA experiments were grown in an experimental greenhouse at Kyoto Prefectural University under normal conditions in 1999. The flowering date was marked for each hull. Developing rice seeds were collected at 4, 7, 10 and 14 d after flowering and immediately frozen in liquid nitrogen. The samples were stored at − 80°C until use. Developing rice seeds for protein analysis were grown in a paddy field at Kobe University in 1997 and 1998. Developing rice seeds at approximately 10 d after flowering were collected and frozen in liquid nitrogen. The samples were stored at − 20°C until use.
cDNA cloning
We first prepared cDNA probes obtained by reverse transcription-polymerase chain reaction (RT-PCR) to screen PEPCase cDNA from developing rice seed-cDNA libraries. Total RNA of developing rice seeds at 7 d after flowering was extracted as described elsewhere (Masumura et al., Reference Masumura, Hibino, Kidzu, Mitsukawa, Tanaka and Fujii1990), and poly(A)+ RNA was purified from the total RNA using a polyAT tract system (Promega, Madison, Wisconsin, USA). The poly(A)+ RNA was reverse-transcribed using Superscript II (Life Technologies, Carlsbad, California, USA) following the manufacturer's instructions. RT-PCR reactions were performed using rTaq polymerase (Toyobo, Osaka, Japan). The primers for RT-PCR were designed based on the nucleotide sequences of a maize C3 PEPCase, a maize C4 PEPCase and three rice expression sequence tag sequences (Genbank ID: AU069759, C27144 and C72139) showing homology to the PEPCase nucleotide sequences in C3 plants (Table 1). The RT-PCR parameters for the different primer sets were as follows: (i) for the primer set ZmppcCF and Zmppc4R, initial denaturation at 94°C for 2 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 90 s, and a final extension at 72°C for 7 min; (ii) for Zmppc3F and Zmppc3R, initial denaturation at 94°C for 2 min followed by 5 cycles of 94°C for 30 s, 47°C for 30 s and 72°C for 90 s and 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 90 s, and a final extension at 72°C for 7 min; (iii) for ZmppcCF and Zmppc3R, initial denaturation at 94°C for 2 min followed by 5 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 90 s and 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 90 s, and a final extension at 72°C for 7 min; (iv) for AU069759F and C72139R, initial denaturation at 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 90 s, and a final extension at 72°C for 7 min. The amplified fragments were collected separately after agarose gel electrophoresis and analysed using a BigDye Terminator Cycle Sequencing Ready Reaction Kit and ABI-PRISM 310 Genetic Analyzer (Perkin-Elmer, Heidelberg, Germany) to confirm that their sequences were partial fragments of PEPCase.
Table 1 Primer sequences for RT-PCR

Next, we screened PEPC cDNAs from a cDNA library of developing rice seeds at 7 d after flowering using RT-PCR-amplified cDNA probes as probes for the plaque hybridization. This cDNA library was constructed from developing rice seeds at 7 d after flowering using a λZiplox cDNA cloning kit (Life Technologies) following the manufacturer's instructions. The cDNA library was comprised of 1.3 × 106 independent clones. Positive phage clones were isolated from the libraries by plaque hybridization with the probes incorporating [32P]dCTP (32P-labelled deoxycytidine triphosphate) by using a BcaBEST DNA labelling kit (Takara BIO, Otsu, Japan), and their cDNA inserts were separately excised and converted into pZL-1 plasmid clone in DH10B using the λZIPLox system (Life Technologies). The obtained plasmid clones were classified into groups according to the obtained EcoRI restriction digestion patterns. A representative clone that seemed to have the longest insert for each group was sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction Kit and ABI-PRISM 310 Genetic Analyzer. The sequences of the two representative clone inserts were deposited separately in GenBank on 24 May 2000 and 26 January 2001, respectively (GenBank ID Nos. AF271995 and AB055074).
Phylogenetic tree construction
A phylogenetic tree of PEPCase was constructed using GENETYX software (GENETYX, Tokyo, Japan). The polypeptide sequences were corrected based on the results of homology search of Osppc1 and Osppc3 against non-redundant protein database ‘nr’. The top 100 sequences for each search were collected from GenBank and integrated into a non-redundant sequence set. After selecting sequences of seven monocotyledonous species (Sorghum bicolor, Zea mays, Hordeum vulgare, Brachypodium distachyon, Saccharum sp., Phalaenopsis amabilis, Phalaenopsis equestris, Echinochloa crus-galli) and five dicotyledonous species (Arabidopsis thaliana, Glycine max, Ricinus communis, Medicago truncatula, Populus trichocarpa), the multiple alignment of the selected sequences was carried out with rice PEPCase sequences. The alignment data were used for constructing a phylogenetic tree with the neighbour-joining method. Bootstrap testing was conducted with 1000 resamples (Saitou and Nei, Reference Saitou and Nei1987). Because the Osppc-b sequence, which encodes the bacterial type PEPCase in rice, showed quite low conservation with the other rice PEPCase sequences, it was omitted on this tree.
Northern hybridization
The total RNA of developing rice seeds at 4, 7, 10 and 14 d after flowering was extracted by the method described previously (Masumura et al., Reference Masumura, Hibino, Kidzu, Mitsukawa, Tanaka and Fujii1990) and approximately 20 μg of total RNA from rice seeds was subjected to electrophoresis on 1.2% agarose gels containing 1.8% formaldehyde, and transferred to a nitrocellulose membrane (Hybond-C, Amersham Biosciences, Piscataway, New Jersey, USA) with a solution of 0.1 N NaOH/3 M NaCl. The membrane was baked at 80°C for 2 h to allow linking of the RNA to the membrane. [32P]dCTP-labelled cDNA probes were prepared by using a BcaBEST DNA labelling kit. The PCR products for gene-specific regions of the two isolated cDNAs, positions 3061–3272 of the longer cDNA clone insert and positions 742–991 of the other cDNA clone insert, were used as templates. The membrane was prehybridized for 5 h at 42°C in hybridization solution [6 × saline sodium phosphate EDTA buffer (SSPE), 5 × Denhardt's reagent, 50% (v/v) formamide, 0.1% (w/v) SDS, 20 mg ml− 1 denatured salmon sperm DNA]. Then the membrane was hybridized at 42°C for approximately 16 h in hybridization buffer with the cDNA probe. After hybridization, the membrane was washed twice with wash buffer [0.1 × saline sodium citrate buffer (SSC), 0.1% (w/v) SDS] at 42°C for 15 min, and exposed to X-ray film at − 80°C. The expression of Actin1 was monitored as an internal control.
Heterologous expression of PEPCase protein
The full-length cDNA for Osppc1 was cloned into a NotI site of the pET-30b (+) His-tag vector and transformed into Escherichia coli JM109 (DE3) pLysS (Novagen, Madison, Wisconsin, USA). The transformed E. coli cells were grown in Luria–Bertani (LB) media with 50 μg ml− 1 of kanamycin at 37°C until its middle growth-phase (OD600= 0.6), and production of Osppc1 protein was induced by incubation with 0.6 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 6 h.
Enzyme activity assay
PEPCase activity was assayed by coupling with the malate dehydrogenase reaction according to the method of Echevarria et al. (Reference Echevarria, Pacquit, Bakrim, Osuna, Delgado, Arrio-Dupont and Vidal1994) with modifications. This assay was performed in 1 ml of a medium containing 50 mM Tris–HCl, 5 mM MgSO4, 0.15 mM NADH, 5 mM KHCO3, 5 mM PEP (cyclohexylammonium salt), 4 mM dithiothreitol (DTT) and 1.5 U pig heart malate dehydrogenase at 30°C at pH 8.3. The decreasing rate of the absorbance at 340 nm was measured. One unit of enzyme activity was defined as the amount of PEPCase catalysing the production of 1 μmol of oxaloacetate per minute at 25°C. The assay was carried out three times for each sample, and the average of the measured values was adopted. The protein concentration was determined by the method of Bradford (Reference Bradford1976).
Purification of recombinant Osppc1 protein
Recombinant protein was extracted from the E. coli cells as follows. The bacterial culture was centrifuged to collect the E. coli cells. The obtained bacterial pellet was suspended in extraction buffer [50 mM sodium phosphate (pH 7.8), 300 mM NaCl], frozen in liquid nitrogen and defrosted by dipping the centrifuge tube into water. The E. coli cells were shaken vigorously. This frozen–defrost–shaking procedure was repeated ten times. After centrifuging the suspension, the supernatant was collected as a crude extract.
Purification of the recombinant PEPCase protein was carried out in the following steps at 4°C by chasing PEPCase activity. At the first step, the crude extract was brought to 20% (w/v) ammonium sulphate, and the supernatant was collected by centrifugation. The supernatant was brought to 50% (w/v) ammonium sulphate, and the precipitate was collected by centrifugation. The protein pellet was suspended in buffer A [50 mM Hepes/KOH, pH 7.8, 10 μM leupeptin, 100 μM phenylmethanesulphonyl fluoride (PMSF) and 20% (w/v) (NH4)2SO4] and loaded on to Butyl-TOYOPEARL 650M (Tosoh, Tokyo, Japan) equilibrated in buffer A. The Butyl-TOYOPEARL 650M column was washed with an equal volume of equilibration solution, and the PEPCase protein was eluted with a linear gradient of 100–0% buffer A simultaneously with 0–100% buffer A without (NH4)2SO4. The fractions exhibiting PEPCase activity were collected and subjected to diafiltration against buffer B [50 mM sodium phosphate (pH 7.8), 300 mM NaCl] overnight and loaded on to TALON Metal Affinity Resin (Clontech Laboratories, Mountain View, California, USA) equilibrated in buffer B. The resin column was washed with a tenfold volume of buffer B. PEPCase was eluted with buffer B supplemented with 150 mM of imidazole and collected. The TALON Metal Affinity chromatography step was repeated once, and the obtained purified fraction was brought up to 50% (v/v) glycerol and stored at − 20°C. SDS-PAGE analysis was performed by the method of Laemmli (Reference Laemmli1970).
Purification of PEPCase from developing rice seeds
All procedures for PEPCase purification were performed at 4°C. PEPCase was extracted from developing rice seeds at 10 d after flowering. The frozen seeds were ground by a homogenizer in extraction buffer C [100 mM Tris–HCl, pH 7.8, 1 mM EDTA, 10 mM 2-mercaptoethanol, 10 mM l-malate and 20% (w/v) glycerol] with Complete Protease Inhibitor Cocktail (F. Hoffmann-La Roche, Basel, Switzerland). The homogenate was filtered through two layers of gauze and centrifuged to collect the supernatant liquid.
PEPCase protein was purified from the supernatant by four steps of column chromatography. The crude extract was brought to 20% (w/v) ammonium sulphate, and the supernatant was collected by centrifugation. Next, the supernatant was loaded on to Butyl-TOYOPEARL 650M equilibrated in buffer D [100 mM Tris–HCl (pH 7.8), 1 mM EDTA, 1 mM 2-mercaptoethanol, 5 mM l-malate, 1 mM PMSF, 5 μg ml− 1 chymostatin and 10% (w/v) glycerol] with 20% (w/v) (NH4)2SO4. The Butyl-TOYOPEARL 650M column was washed with an equal volume of the equilibration solution, and PEPCase protein was eluted with a linear gradient of 100–0% buffer A with 20% (w/v) (NH4)2SO4 and a linear gradient of 0–100% buffer D simultaneously. The fractions including PEPCase activity were collected and subjected to diafiltration against buffer C overnight and loaded on to DEAE-TOYOPEARL 650M equilibrated in buffer C. The DEAE-TOYOPEARL 650M column was washed with an equal volume of the equilibration solution, and PEPCase was eluted with a linear gradient of 100–0% buffer D and a linear gradient of 0–100% buffer D with 0.5 M NaCl simultaneously. The eluted fractions exhibiting PEPCase activity were collected, diafiltered against buffer E [50 mM sodium phosphate (pH 7.8), 1 mM EDTA, 1 mM 2-mercaptoethanol, 5 mM l-malate, 1 mM PMSF, 5 μg ml− 1 chymostatin and 10% (w/v) glycerol] and loaded on to a hydroxyapatite column equilibrated with buffer E. The hydroxyapatite column was washed with an equal volume of the equilibration solution, and PEPCase was eluted by a linear gradient of 100–0% buffer E and a linear gradient of 0–100% buffer E with 400 mM NaCl simultaneously. The eluted fractions exhibiting PEPCase activity were collected, diafiltered against buffer D and loaded on to a Mono-Q chromatographic column (Pharmacia Biotech, Piscataway, New Jersey, USA). The same procedure was carried out using a DEAE-TOYOPEARL 650M column chromatography to obtain the purified PEPCase fraction.
Mining of co-expressed genes with Osppc1
We downloaded 624 rice gene expression profiles from a public database, the Gene Expression Omnibus (GEO) monitored by the Affymetrix rice genome array platform GPL2025 (Barrett et al., Reference Barrett, Troup, Wilhite, Ledoux, Evangelista, Kim, Tomashevsky, Marshall, Phillippy, Sherman, Muertter, Holko, Ayanbule, Yefanov and Soboleva2010). The raw data were pre-processed using GCRMA routine in R/BioConductor (http://www.bioconductor.org/) to the log2-transformed gene expression level (Irizarry, Reference Irizarry2003). After excluding the data of indica rice, callus, inoculated samples and seeds, the remaining 38 profiles were used to calculate the Pearson correlation efficiency between Osppc1 and the other genes (supplementary Table S1, available online). Positive Pearson correlations were statistically validated by a test of non-correlations. Spearman correlation coefficients were calculated to extract reliable co-expression relationships. The functional annotations of the rice genes were obtained from OryzaExpress (Hamada et al., Reference Hamada, Hongo, Suwabe, Shimizu, Nagayama, Abe, Kikuchi, Yamamoto, Fujii, Yokoyama, Tsuchida, Sano, Mochizuki, Oki, Horiuchi, Fujita, Watanabe, Matsuoka, Kurata and Yano2011).
Results
cDNA cloning of PEPCase Osppc1 and Osppc3
We isolated PEPCase cDNAs from developing rice seeds at 7 d after flowering and obtained 3.3 kb of a full-length cDNA and 1.1 kb of a partial-length cDNA of PEPCase. By searching against the rice whole-genome sequence, we mapped these cDNA sequences into the corresponding genes that were annotated as PEPCase, MSU ID: LOC_Os02g14770 and LOC_Os01g55350, respectively, at the Rice Genome Annotation Project. These genes were described as Osppc1 and Osppc3 by Masumoto et al. (Reference Masumoto, Miyazawa, Ohkawa, Fukuda, Taniguchi, Murayama, Kusano, Saito, Fukayama and Miyao2010). According to the genomic sequence of the latter gene, we amplified the corresponding full-length cDNA from developing rice seeds by RT-PCR using a 5′-UTR and 3′-UTR sequence primer set (5′-UTR–GGGAAAAGGAGTCGGCTTTC-, 3′-UTR–CCATAACACACCATCAGAAC-) and cloned it into pGEM-T Easy vector (Promega). The insert sequence was confirmed to be identical to the sequence of the partial cDNA clone and the predicted coding sequence in the rice genome.
Sequence analysis of PEPCase Osppc1 and Osppc3 cDNA
The nucleotide sequences of the 3308 bp cDNA insert of Osppc1 and 2961 bp cDNA insert of Osppc3 contain the longest open reading frames: 2904 bp in the second frame and 2898 bp in the third frame, respectively (Fig. 1). We noticed that the potential translation initiation codon of Osppc1 could be assigned to either the first ATG at positions 161–163 or the second ATG at positions 188–190 in frame according to the sequence alignment with the maize PEPCase cDNA sequence (AB012228). We designated the second ATG as the translation initiation codon based on the similarity to the maize gene sequence (Dong et al., Reference Dong, Masuda, Kawamura, Hata and Izui1998). The deduced polypeptide sequences from Osppc1 and Osppc3 showed typical conserved features of plant PEPCase (Kai et al., Reference Kai, Matsumura and Izui2003). These sequence regions included the consensus sequences for the PEPCase motifs; phosphorylation site, catalytic bases, G6P binding site, hydrophobic pockets, PEP binding site, tetramer formation, Mg2+ binding site, HCO3 − binding site, PEP and aspartate binding site (see supplementary Fig. S1, available online). Both sequences possess an alanine residue at position 778/775 which is equivalent to position 774 of the C4 PEPCases of Flaveria (Svensson et al., Reference Svensson, Bläsing and Westhoff1997). This implies that Osppc1 and Osppc3 encode a PEPCase having substrate affinities similar to those of the non-photosynthetic type PEPCase (Bläsing et al., Reference Bläsing, Westhoff and Svensson2000).

Figure 1 Graphical representation of primary structures of Osppc1 and Osppc3. Open reading frames of Osppc1 and Osppc3 are shown. The black bar shows the longest open reading frame and the hatched bar represents poly(A). Sequence alignment of the deduced polypeptide of the 5′ region of Osppc1 and maize root-type PEPCase is shown in the box.
We constructed a phylogenetic tree of Osppc1, Osppc3 and other PEPCases from 51 polypeptide sequences deposited in GenBank to investigate the molecular evolutionary relationship among them (Fig. 2). Osppc1 and Osppc3 were clustered into subfamilies comprising genes of monocotyledonous plant species and gramineous plant species genes, respectively. In our phylogenetic tree, Osppc1 is likely to be the orthologue of the maize root-type gene (NP_001105503.1), whereas Osppc3 is likely to be the orthologue of the sugarcane PEPCase (P29193.1) argued as a housekeeping isoform (Albert et al., Reference Albert, Martin and Sun1992). The same results were also obtained in the case of phylogenetic analysis by Kimura's 2-parameter model (data not shown). Osppc1 shares significant homology with maize root-type PEPCase (95% identity, 98% similarity), and Osppc3 shares significant homology with the sugarcane PEPCase (92% identity, 98% similarity). These results suggest the following: (1) the gene family including Osppc1 and Osppc3 evolved from a common ancestor with monocotyledonous and dicotyledonous plant species before the divergence of C3, C4 and CAM plant species; (2) Osppc1 might encode PEPCase with enzymatic properties similar to those of the maize root-form PEPCase; (3) the gene subfamily including Osppc3 was evolved specifically in gramineous plant species; and (4) Osppc3 might encode a housekeeping PEPCase.

Figure 2 A phylogenetic tree of PEPCases. The tree was constructed from 51 polypeptide sequences including Osppc1, Osppc2a, Osppc2b, Osppc3 and Osppc4.
Gene expression of Osppc1 and Osppc3 in developing rice seeds
To monitor the gene expression pattern of Osppc1 and Osppc3 in developing rice seeds, we performed Northern blot analysis by using gene-specific probes corresponding to the 3′-UTR sequences. We detected expression of these genes throughout the grain-filling stages (Fig. 3). The expression patterns of Osppc1 and Osppc3 were slightly different from each other; the expression of Osppc1 was highest at 7 d after flowering (DAF) (early–middle stage), and the expression of Osppc3 was highest at 4 DAF (early stage), although the mRNA accumulation of both genes decreased toward the end of the maturation stage. These decreasing trends are likely to be consistent with the pattern of PEPCase activity in developing rice seeds (Sugimoto et al., Reference Sugimoto, Sueyoshi, Oji, Ando, Fujita, Mae, Matsumoto, Mori and Sekiya1997).

Figure 3 (colour online) Gene expression of Osppc1 and Osppc3 in developing rice seeds.
Enzymatic properties of Osppc1
In order to obtain biochemical information about Osppc1, we expressed the recombinant Osppc1 protein in E. coli in order to characterize its kinetics. The recombinant protein from the cells was purified by monitoring PEPCase activity, as summarized in Table 2. The relative PEPCase activity of the purified recombinant protein was 20.3 units (mg protein)− 1, which was 41.5-fold that of the crude extract. The detected polypeptide showed mobility similar to a molecular weight marker of 130 kDa, which was consistent with the expected size of the deduced polypeptide comprised of 1028 amino acids, including His-tagged sequences (Fig. 4). We confirmed that a His-tagged protein showing mobility identical to the polypeptide in SDS-PAGE was detected in Western blotting analysis using an anti-His-tag antibody (data not shown).
Table 2 Purification of recombinant Osppc1 protein


Figure 4 (colour online) SDS-PAGE analysis of recombinant Osppc1 protein. The fraction showing PEPCase activity from each purification step was electrophoresed and stained with Coomassie brilliant blue. The size of molecular mass markers is indicated on the left. (A) Lanes: 1, molecular mass marker; 2, crude extract (no insert); 3, crude extract; 4, (NH4)2SO4 fractionation; 5, butyl-TOYOPEARL; 6, TALON Metal (first). (B) Lanes: 1, molecular mass marker; 2, TALON Metal (second).
The enzymatic parameters of the Osppc1 protein were measured and compared with those of other PEPCase proteins characterized previously (Dong et al., Reference Dong, Masuda, Kawamura, Hata and Izui1998; Masumoto et al., Reference Masumoto, Miyazawa, Ohkawa, Fukuda, Taniguchi, Murayama, Kusano, Saito, Fukayama and Miyao2010). The pH optimum of Osppc1 was determined to be 9.0, and the relative activity at pH 7.0 was decreased to approximately 75% of that at pH 9.0. The fundamental parameters shown in Table 3 are similar to those of the maize root-type PEPCase. Nevertheless, the regulatory properties appeared to be different with respect to the inhibitory effect of malate, which is one of the allosteric effectors of PEPCase (Table 4). Compared to other rice PEPCase enzymes, the I 50 (malate) of Osppc1 was higher. This suggests that Osppc1 works more actively in an intracellular environment close to pH 7.0 than the other PEPCases listed. The influences of G6P and glycine on the inhibitory effect of malate seem to be similar to those of maize root-type PEPCase.
Table 3 Fundamental kinetic parameters of Osppc1 protein

Table 4 Effect of G6P and glycine on I 0.5 * (l-malate)

* I 0.5 (mM) was determined as the concentration of l-malate in the reaction buffer to reduce PEPCase activity by half. The concentration of both glucose-6-phosphate (G6P) and glycine was 10 mM. Parameters of recombinant maize root-type PEPCase are from Dong et al. (Reference Dong, Masuda, Kawamura, Hata and Izui1998). Parameters of recombinant Osppc2a and Osppc4 are from Masumoto et al. (Reference Masumoto, Miyazawa, Ohkawa, Fukuda, Taniguchi, Murayama, Kusano, Saito, Fukayama and Miyao2010).
Detection of the PEPCase protein in developing rice seeds
We prepared a partially purified PEPCase from rice seeds in the middle of the maturation stage. The relative activity was 35.6 U (mg protein)− 1. In the partially purified PEPCase fraction of developing rice seeds, we detected two major polypeptides in the 97–116 kDa mass range (Fig. 5A). These two polypeptides were also immunologically detected using a polyclonal antibody against a soybean mature seed PEPCase with the same mobility (data not shown). The estimated molecular weights of the polypeptides were 109.5 and 103.5 kDa, respectively. These values were quite close to the molecular weights of the deduced proteins of Osppc1 (109.2 kDa) and Osppc3 (105.3 kDa), respectively (see supplementary Table S2, available online). Two polypeptides were also detected in both embryos and endosperms with aleurone layers (Fig. 5B).

Figure 5 (A) SDS-PAGE analysis of purified PEPCase from developing rice seeds. Lanes: 1, molecular weight marker; 2, PEPCase fraction. (B) Immunological detection of PEPCase polypeptides in developing rice seeds using a polyclonal antibody of soybean seed PEPCase (Sugimoto et al., Reference Sugimoto, Kawasaki, Kato, Whittier, Shibata and Kawamura1992). Embryos and endosperms were prepared from developing rice seeds at 10 d after flowering. Crude extract proteins from them were separated on SDS-PAGE and analysed by Western blotting. Lanes: 1, embryo; 2, endosperm.
Co-expressed genes of Osppc1 in rice plants
In order to investigate whether Osppc1 is co-regulated with any other metabolic gene in amino-acid synthetic pathways, we mined co-expressed genes with Osppc1 from DNA microarray data of the Affymetrix rice gene chip. Under the threshold of 1% statistical significance for Pearson correlation, we found that 47 putative metabolic genes showed a strong positive correlation with Osppc1. This gene list was narrowed down to 28 genes to eliminate apparent false-positive correlations by checking the Spearman correlation efficiency at the threshold of 0.7 (Table 5). Out of the 28 genes, 22 genes were annotated to encode enzymes, including three aminotransferase genes: aspartate aminotransferase (AST), alanine aminotransferase (ALT) and aminotransferase of class IV family. Enolase 2 (ENO2), succinate dehydrogenase flavoprotein subunit (SdhA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cysteine synthase were also listed.
Table 5 Metabolic genes co-expressed with Osppc1 in rice vegetative organs

The co-expressed genes that were assigned as the three aminotransferases, the cysteine synthase, the two glycolytic pathway enzymes and the TCA cycle enzyme protein mentioned above were reannotated by evaluating their protein sequences to clarify whether these genes could work coordinately with Osppc1. LOC_Os01g55540, annotated as AST, was also described as a cytoplasmic AST (Song et al., Reference Song, Yamamoto, Shomura, Yano, Minobe and Sasaki1996), the enzyme of which catalyses the reversible reaction between OAA and aspartate. The AST, which was called OsAAT2, was overexpressed in rice, and enhanced seed nitrogen metabolism (Zhou et al., Reference Zhou, Cai, Xiao, Li, Zhang and Lian2009). LOC_Os10g25130, annotated as ALT, seemed to be a cytosolic enzyme because no signal, mitochondrial targeting or chloroplast transit peptides were found in iPSORT prediction (http://ipsort.hgc.jp/). Kikuchi et al. (Reference Kikuchi, Hirose, Toki, Akama and Takaiwa1999) observed that ALT was expressed in the maturing endosperm and suggested the involvement in nitrogen metabolism during the maturation of rice seeds. LOC_Os03g24460, annotated as an aminotransferase of class IV family, was also assigned as a branched-chain amino acid aminotransferase (BCAT) in the UniProt database (http://www.uniprot.org/). BCAT catalyses the synthesis or the degradation of leucine, isoleucine and valine (Diebold et al., Reference Diebold, Schuster, Däschner and Binder2002). The intracellular localization of this BCAT protein was predicted as a plastid gene by iPSORT. The ENO2 (LOC_Os03g14450) and the cysteine synthase (LOC_Os03g53650) had no signal, mitochondrial targeting or chloroplast transit peptides in iPSORT prediction. In contrast, the SdhA (LOC_Os07g04240) and the GAPDH (LOC_Os02g38920) had mitochondrial targeting signal sequences. Therefore, given the above facts, the ENO2, the AST and the ALT were mapped near the PEPCase reaction on the metabolic pathway (Fig. 6).
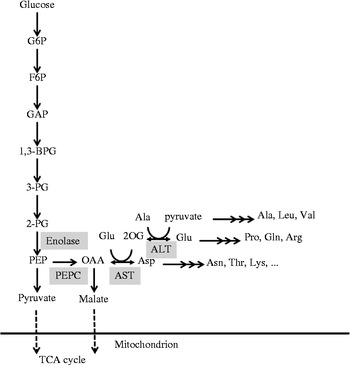
Figure 6 A metabolic pathway map of glycolysis with phosphoenolpyruvate carboxylase (PEPC), aspartate aminotransferase (AST) and alanine aminotransferase (ALT) reactions. The enolase, PEPC, AST and ALT were mapped on the consecutive reactions from 2-phosphoglyceric acid (2-PG) to glutamate (Glu) and pyruvate.
Discussion
In the present study, we isolated two full-length cDNAs of PEPCases Osppc1 and Osppc3 that were expressed in developing rice seeds. The deduced amino acid sequence of Osppc1 was quite similar to that of maize root-form PEPCase and shared the typical conserved features of plant PEPCases. Biochemical assay of the recombinant protein of Osppc1 demonstrated that its kinetic properties were similar to those of the maize PEPCase, except for the inhibition status by malate. We observed that its gene expression was high in the early and the middle stages of grain filling, in which PEPCase activity was also high in parallel (Sugimoto et al., Reference Sugimoto, Sueyoshi, Oji, Ando, Fujita, Mae, Matsumoto, Mori and Sekiya1997). The corresponding PEPCase protein of Osppc1 seemed to accumulate in developing rice seeds at these stages. During these stages, nitrogen accumulation and synthesis of storage proteins in rice grains were notable (Yamagata et al., Reference Yamagata, Sugimoto, Tanaka and Kasai1982; Iwasaki et al., Reference Iwasaki, Mae, Makino, Ohira and Ojima1992). Considering all these results together, we propose that Osppc1 is involved in nitrogen accumulation in developing rice seeds.
Interestingly, co-expression analysis indicated that Osppc1 was coordinately regulated with the two aminotransferases AST and ALT that seem to be involved in seed nitrogen metabolism. These aminotransferase genes were strongly expressed in developing seed tissues (Kikuchi et al., Reference Kikuchi, Hirose, Toki, Akama and Takaiwa1999; Zhou et al., Reference Zhou, Cai, Xiao, Li, Zhang and Lian2009). Therefore, Osppc1, AST and ALT might be involved in the same biological process in immature seeds. On the metabolic map, AST and ALT are located near the PEPCase reaction (Fig. 6). OAA generated from phosphoenolpyruvate could be used for synthesis of aspartic acid (ASP) and 2-oxoglutarate (2-OG) by AST. 2-OG is a substrate of ALT, which generates glutamic acid (Glu) and pyruvate. Asp, Glu and pyruvate are precursors of other amino acids. So, the function of Osppc1 might be to provide OAA which is consumed in amino acid synthesis. Analysis of mutants or transgenic rice lines is expected to verify the physiological role of Osppc1 in nitrogen metabolism.
PEPCase might be one of the essential enzymes to achieve an increased flow rate of the AST reaction. Sentoku et al. (Reference Sentoku, Taniguchi, Sugiyama, Ishimaru, Ohsugi, Takaiwa and Toki2000) reported that the overexpression of proso millet cytoplasmic AST led to the enhancement of PEPC protein expression in transgenic tobacco. This observation suggests that a metabolic change caused by overexpression of the cytoplasmic AST affects the regulation of the PEPCase gene. Enhancement of the AST reaction would induce the consumption of cytosolic OAA and Glu and the production of cytosolic Asp, 2-OG and the derivatives of Asp and 2-OG. According to Zhou et al. (Reference Zhou, Cai, Xiao, Li, Zhang and Lian2009), overexpression of cytoplasmic AST enhanced amino acid and protein content in rice seeds, suggesting that free amino acids were richer during maturation in the transgenic rice. Thus, it seems that PEPCase responds to conditions of low C/N ratio for replenishment of OAA. This concept agrees with the report that RNAi knockdown of the chloroplast PEPCase, Osppc4, suppressed ammonium assimilation and amino acid synthesis by reducing levels of organic acids (Masumoto et al., Reference Masumoto, Miyazawa, Ohkawa, Fukuda, Taniguchi, Murayama, Kusano, Saito, Fukayama and Miyao2010). For production of rice seeds with much higher protein content, Osppc1 might be useful, together with other key genes such as AST and ALT, in combinational genetic transformation, which can enhance the genes of several consecutive enzymes on a metabolic pathway (Zhu et al., Reference Zhu, Naqvi, Breitenbach, Sandmann, Christou and Capell2008).
The existence of PEPCase isoforms in immature seeds has been described in several plant species. We isolated two cDNAs of PEPCase in a developing soybean seed library (Sugimoto et al., Reference Sugimoto, Kawasaki, Kato, Whittier, Shibata and Kawamura1992) and confirmed the accumulation of two PEPCase polypeptides in the seeds (unpublished data). In faba bean, the cDNAs of two PEPCase isoforms were isolated, and their expression patterns differed (Golombek et al., Reference Golombek, Heim, Horstmann, Wobus and Weber1999). Moreover, Blonde and Plaxton (Reference Blonde and Plaxton2003) reported that there were two C3 PEPCase isoforms that were likely to be involved in supporting the synthesis of storage proteins and fatty acids, respectively, in castor bean seeds. With regard to monocotyledonous species, González et al. (Reference González, Osuna, Echevarría, Vidal and Cejudo1998) reported that two main PEPCase polypeptides with apparent molecular masses of 108 and 103 kDa were detected in developing wheat grains. Consistent with these findings, in the present work we isolated cDNAs of two PEPCase isoforms that were expressed in developing rice seeds, and detected two PEPCase polypeptides with apparent molecular masses of 109.5 and 103.5 kDa that seemed to correspond to the cDNAs. As a preliminary result, we observed that two main PEPCase polypeptides were accumulated in rice embryos and endosperms with aleurone layers. In a public gene expression database from microarray and RNAseq (Sato et al., Reference Sato, Takehisa, Kamatsuki, Minami, Namiki, Ikawa, Ohyanagi, Sugimoto, Antonio and Nagamura2013; the Rice Genome Annotation Project: http://rice.plantbiology.msu.edu/), we observed that Osppc1 was strongly expressed in endosperm and embryo. On the contrary, we observed that Osppc2b was expressed moderately in embryo. The expression levels of the other PEPCase isoforms genes were low. Further analysis of the localization of each PEPCase polypeptide would help to clarify the functional partitioning of the isoforms in developing rice seeds.
Our phylogenetic analysis revealed that Osppc1 was in the same gene family as maize root-form PEPCase. The fundamental enzymatic properties of both proteins were similar to each other, as shown by the biochemical results (Table 3). However, the regulatory properties of malate at pH 7.3 diverged (Table 4). The differing regulatory properties might be due to the evolutionary changes among PEPCases at the amino acid sequence level. Among the sequence differences between Osppc1 and maize root-form PEPCase, we found conservation of ten sites between Osppc1 and a PEPCase isoform in Hordeum vulgare (BAJ94338.1), which is in the nearest location in our phylogenetic tree (see supplementary Fig. S2, available online). With regard to eight of these sites, the amino acid changes yielded different characteristics in polarity or charge. Further studies, such as site-directed mutagenesis analyses, will be needed to investigate factors determining malate inhibition.
A possible physiological explanation for the kinetic property of Osppc1 in malate inhibition is that it is a countermeasure against hypoxia. It is well known that in plant root cells under oxygen deprivation the cytosol is acidified (Roberts et al., Reference Roberts, Hooks, Miaullis, Edwards and Webster1992; Xia and Saglio, Reference Xia and Saglio1992; Gout et al., Reference Gout, Boisson, Aubert, Douce and Bligny2001; Tournaire-Roux et al., Reference Tournaire-Roux, Sutka, Javot, Gout, Gerbeau, Luu, Bligny and Maurel2003). Paddy rice plants are occasionally grown under a low oxygen environment. Roberts et al. (Reference Roberts, Hooks, Miaullis, Edwards and Webster1992) showed that the concentration of malate in root cells was immediately increased for alanine synthesis when rice plants were exposed to hypoxia. Rolletschek et al. (Reference Rolletschek, Radchuk, Klukas, Schreiber, Wobus and Borisjuk2005) reported a marked degree of oxygen starvation in developing soybean seeds because of the consumption of ATP for the synthesis of storage compounds such as lipids and proteins. A decrease of pH of barley starchy endosperm during the maturation period has also been observed with an increase in the malate pool (Macnicol and Jacobsen, Reference Macnicol and Jacobsen1992).
We are still conducting our studies on Osppc3. Its deduced amino acid sequence is relatively similar to those of the isoforms Osppc2a and Osppc2b, but its expression seems to be restricted to spikelets (Masumoto et al., Reference Masumoto, Miyazawa, Ohkawa, Fukuda, Taniguchi, Murayama, Kusano, Saito, Fukayama and Miyao2010). Notably, SCPEPCD1, an orthologous gene of Osppc3 in sugarcane, was expressed in several organs, especially stems (Albert et al., Reference Albert, Martin and Sun1992). In order to investigate the biochemical properties of Osppc3, we have established a construct to express the recombinant protein in E. coli. Our in silico analysis using Affymetrix gene chip data in rice might also provide an opportunity to determine the function of Osppc3 in developing rice seeds.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0960258513000354.
Acknowledgements
We appreciate the technical support of Dr Akihiro Kurita and Dr Shigeto Morita of Kyoto Prefectural University in the cloning of PEPCase cDNAs. We also thank Dr Daisuke Shibata at the Kazusa DNA Research Institute for critical suggestions in cDNA analysis of PEPCase.
Conflicts of interest
None.














