Introduction
One of the greatest problems of developing countries is the under-nourishment and malnutrition among the children and women of economically backward sections of society. According to the World Health Organization, half of the malnutrition is due to protein energy malnutrition. As animal proteins are expensive, people of developing countries depend upon seed proteins derived from plants. Seeds of cereals and legumes are the major sources of protein in developing countries, which are consumed directly as food and feed by humans and animals, providing more than 70% of the world's caloric intake (Mandal and Mandal, Reference Mandal and Mandal2000). However, these crops are generally deficient in quality proteins and nutritionally balanced amino acids. Hence there is need for isolation and characterization of genes for nutritionally important proteins that can subsequently be transferred to cereals and legumes by genetic engineering for biofortification purposes.
Finger millet is a wonder crop because it is grown under low or limiting nitrogen conditions and has a protein content of 4.9 to 11.3% (McDonough et al., Reference McDonough, Lloyd and Serna-Saldivar2000). This is comparable to that of the major cereals like wheat and rice, which require large amounts of nitrogenous fertilizers. It ranks fourth in production among millets in the world after sorghum, pearl millet and foxtail millet (Upadhyaya et al., Reference Upadhyaya, Gowda and Reddy2007) and serves as a food security crop because of its high nutritional value and excellent storage qualities (Dida et al., Reference Dida, Srinivasachary, Ramakrishnan, Bennetzen, Gale and Devos2007). Finger millet grains have hypocholestrolaemic (Venkateswaran and Vijayalakshmi, Reference Venkateswaran and Vijayalakshmi2010) and hypoglycaemic properties (Shobana et al., Reference Shobana, Sreerama and Malleshi2009). The seed proteins of finger millet occupy a prime place as they are rich in essential amino acids such as lysine (181 mg/g) and tryptophan (191 mg/g) (FAO, 1970; Indira and Naik, Reference Indira and Naik1971). Its seed proteins have been found to be nutritionally better balanced than other cereals and millets (Ravindran, Reference Ravindran1992).
Seed storage proteins are usually classified according to their solubility: albumins (water soluble), globulins (salt soluble), prolamins (alcohol soluble) and glutelins (soluble in diluted acid/base) (Osborne, Reference Osborne1924; Ciccocioppo et al., Reference Ciccocioppo, Di Sabatino and Corazza2005). The major storage proteins in finger millet kernels are those belonging to the prolamin super family, followed by glutelins (Serna-Saldivar and Rooney, Reference Serna-Saldivar, Rooney and Dendy1995). Prolamins present in cereals are monomeric polypeptide chains with molecular weights ranging from 10 to 80 kDa (van Eckert et al., Reference van Eckert, Bond, Rawson, Klein, Stern and Jordan2010). The prolamins are further characterized as alpha, beta and gamma types, out of which alpha-type prolamins (molecular weight ranging from 19 to 22 kDa) appear to be the most abundant (75–85%). Prolamins are deposited in discrete protein bodies in developing starchy endosperm (Arcalis et al., Reference Arcalis, Stadlmann, Marcel, Drakakaki, Winter, Rodriguez, Fischer, Altmann and Stoger2010). α-Prolamin is a member of the prolamin super family and is characterized by the presence of a α-helical globular domain that contains a conserved pattern of six or eight cysteine residues forming three or four intramolecular disulfide bonds (Mills et al., Reference Mills, Jenkins, Alcocer and Shewry2004). There is increased focus on α-prolamin for its use as a biopolymer due to its helical nature, which increases the thermostability and can be used to form biofilm for encapsulation of processed foods and drugs (Riebel and Mankato, Reference Riebel and Mankato2005). On the other hand, beta, gamma and omega prolamins do not possess enough helical structure to form biofilm.
In recent years, significant progress has been made to develop molecular markers after accumulation of sequencing data of millets, especially the foxtail millets, which have opened up opportunities to isolate nutritionally important genes for their use in biofortification purposes (Muthamilarasan et al., Reference Muthamilarasan, Suesh, Pandey, Kumari, Parid and Prasad2014). So far, no work has yet been carried out to identify, study, detect and characterize the gene for Ec-α-prolamin protein, although it is predominantly expressed in finger millet. In this context, the aim of the present study was to isolate and characterize the Ec-α-prolamin protein and its gene from finger millet, as this will lead to introgression of this gene to other major food crops, such as cereals and legumes for biofortification programmes.
Materials and methods
Experimental material
Eleusine coracana genotype GE-3885 (13.76% protein) was grown in a polyhouse at 25°C/18°C over a 12 h/12 h day/night schedule, and developing spikes were harvested at S1, S2, S3 and S4 stages. Anthesis of finger millet starts from 60 to 80 days after sowing (DAS) and the S1, S2, S3 and S4 stages correspond to spike emergence (80 DAS), pollination stage (80–85 DAS), dough stage (85–92 DAS) and maturation stage (92–102 DAS), respectively (see supplementary figure).
Extraction of seed prolamins
Sequential extraction of seed storage proteins from finger millet. Sequential extraction of the seed storage proteins including total protein and prolamin proteins from different developmental stages (S1, S2, S3 and S4) of high protein-containing genotype GE-3885 (13.76% protein) was carried out by a slight modification of the method by Tatham et al. (Reference Tatham, Fido, Moore, Kasarda, Kuzmicky, Keen and Shewry1996).
SDS-PAGE electrophoresis for isolation of different types of Ec-α-prolamin. The total protein and prolamin protein fractions, isolated from GE-3885 genotype of finger millet by the sequential extraction procedure was analysed by SDS-PAGE (Laemmli, Reference Laemmli1970). The alpha-prolamin fraction was obtained and purified from total prolamin by gel filtration chromatography according to the procedure previously reported for the extraction of rye prolamins (Charbonnier et al., Reference Charbonnier, Terce-Laforgue and Mosse1981). A UV spectrophotometer (UV-1700; Shimadzu, Japan) was used to detect different fractions by recording the absorbance at 280 nm. Eluted protein samples were confirmed by SDS-PAGE.
Generation of monospecific polyclonal antibodies against Ec-α-prolamin
New Zealand White albino rabbits were used for the immunization experiment. The extracted Ec-α-prolamin protein fraction was quantified by Bradford's method (Bradford, Reference Bradford1976). A purified α-prolamin protein fraction (500 ng) was dissolved in phosphate-buffered saline (PBS) at pH 7.4 and emulsified with Freund's complete adjuvant for primary and Freund's incomplete adjuvant for subsequent booster doses. First booster was given after 7 days of primary injection and subsequent booster doses were given at an interval of 7 days. After the fifth booster, a blood sample was collected and serum containing polyclonal antibodies against Ec-α-prolamin was separated. The immunoreactivity of generated antibodies was checked by dot blot analysis.
Expression study of Ec-α-prolamin western blot analysis
Western blotting was used to study the presence of α-prolamin in total protein extracted from different developmental stages of GE-3885 genotype of finger millet. SDS-PAGE was performed and separated proteins were transferred from gel to nitrocellulose membrane (NC) using a semi-dry transfer system (SCIE-PLAS, Cambridge, UK) at 2 mA/cm2 for 90 min. Anti-α prolamin antibody was used at a dilution of 1:250 in PBS buffer containing 5% powdered milk and incubated for 2 h at 37°C with constant shaking. Secondary antibody was goat anti-rabbit ALP conjugate which was incubated at 37°C for 45 min with constant shaking. Membranes were developed colorimetrically by adding BCIP/NBT substrate.
Design of primers for isolation and expression analysis of Ec-α-prolamin gene
In the absence of any knowledge about conserved α-prolamin genes across the cereals and millet, the alpha-zein of maize was used to identify the EST accession number CX265222 of finger millet to design the primers. The identified EST was used to design a pair of primers for isolation of a partial sequence of the Ec-α-prolamin gene, which was subsequently used to design different pairs of primers for performing 3′ RACE and expression analysis of Ec-α-prolamin gene of finger millet. The details of the primer are given in the supplementary Table S1.
RNA isolation and cDNA preparation
The freshly collected developing spikes from S1, S2, S3 and S4 stages were directly used for isolation of total mRNA by using the IRIS system (developed by Institute of Himalayan Bioresource Technology, Palampur). RNA preparations were subjected to DNase digestion according to the manufacturer's instruction (Fermentas) and the purity and integrity of the RNA were checked by performing NanoDrop Spectroscopy and agarose gel electrophoresis. A total RNA (2 µg) sample was used to synthesize cDNA by using oligo(dT)18 primer with RevertAid H Minus Reverse Transcriptase (RT) (Fermentas). The cDNA was checked by performing PCR using tubulin primers. 25 µl PCR reaction mixture contained 1x KCl buffer (Fermentas), 0.2 mM dNTPs, 30 ng of each tubulin primer, 1.5 mM MgCl2, 0.8 U Taq DNA polymerase (Fermentas) and 100 ng of cDNA. Amplification was carried out according to the following temperature profile: 3 min initial denaturation at 94°C, 32 cycles of 94°C for 1 min, 60°C for 30 s and 72°C for 45 s, followed by a final extension of 7 min at 72°C.
Isolation of full-length cDNA clones of Ec-α-prolamin gene from E. coracana
A semi-quantitative RT-PCR-based approach, as per the above reaction, was utilized to amplify partial cDNA of Ec-α-prolamin from the S3 stage of the developing spike of finger millet. The amplified fragment was cloned into pGEM-T Easy vector (Promega, USA) and sequenced. The partial sequence carried an ATG initiation codon in the 5′ region, while the 3′ region was lacking. In order to acquire 3′ region, three forward primers were designed from the 3′ end of the partial sequence. Finally, 3′ RACE was performed using a 3′ RACE kit (second generation, Roche).
Phylogenetic and in silico analysis
In order to detect similarities in amino acid sequence position, the translated amino acid sequence (http://web.expasy.org/translate) of Ec-α-prolamin was aligned using CLUSTAL_W (MEGA) with the sequence obtained from the NCBI database of the grass subfamily. Further, a phylogenetic tree was constructed by using MEGA version 5 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011). Physiochemical properties were deciphered by online tools (http://web.expasy.org/cgi-bin/protparam/protparam). The secondary and tertiary structures were predicted using GOR secondary structure prediction method version IV and Bhagirathi H (Jayaram et al., Reference Jayaram, Dhingra, Lakhanib and Shekhar2012), respectively. Further refinement of the tertiary structure was performed using Motif and domain analysis, which were performed using SMART/MEME/MAST online tools (Letunic et al., Reference Letunic, Doerks and Bork2012).
Expression of Ec-α-prolamin by real-time PCR analysis
Real-time PCR was carried out by designing the primer from the partial sequence of Ec-α-prolamin and as reference primer tubulin from finger millet (see supplementary table). Real Master Mix SYBR ROX (Eppendorf India Limited, Chennai, India) was obtained according to the manufacturer's instructions. The thermo cycler was an Eppendorf thermo cycler eprealplex-4. The following amplification programme was used: 95°C for 10 min; 40 cycles at 95°C for 15 s; 60°C for 30 s; 72°C for 30 s; 60°C for 15 s; and 95°C for 15 s. All samples were amplified in duplicate, and the expression values were calculated.
Results
SDS-PAGE analysis of total seed storage proteins from finger millet
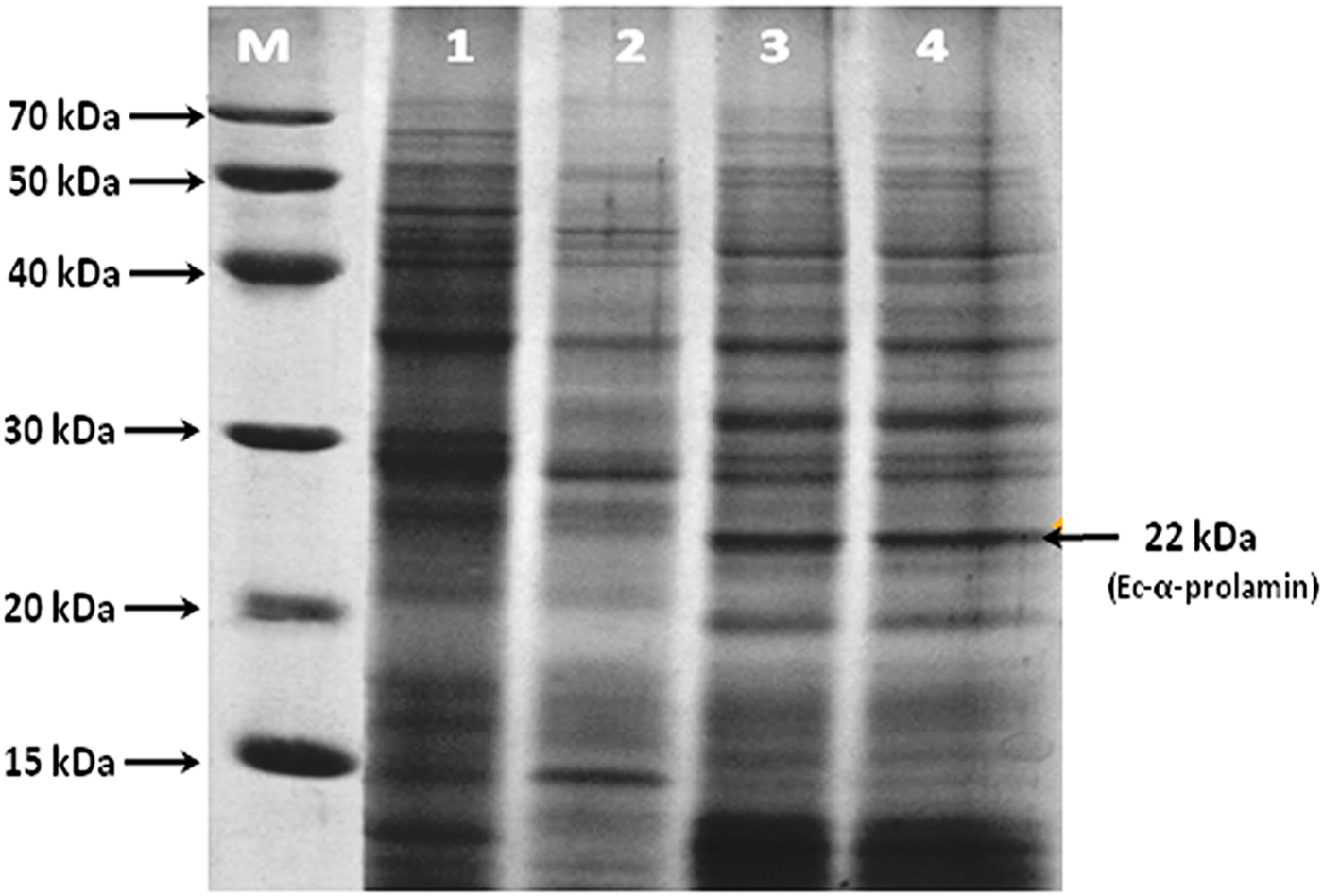
SDS-PAGE analysis of total seed storage proteins extracted from different stages (S1, S2, S3 and S4) of developing spikes was carried out from high protein grain genotype (GE-3885) of finger millet. On the basis of molecular weight, total proteins were characterized and identified as albumins, globulins, glutelins and prolamins. The protein bands falling in the molecular weight range 25–70 kDa were identified as globulins and albumins, while those in the range 15–22 kDa were identified as prolamins. The protein band of 22 kDa was identified as Ec-α-prolamin on the basis of earlier reports. Its expression was observed to be higher in S3 and S4 stages of developing spikes (Fig. 1.).
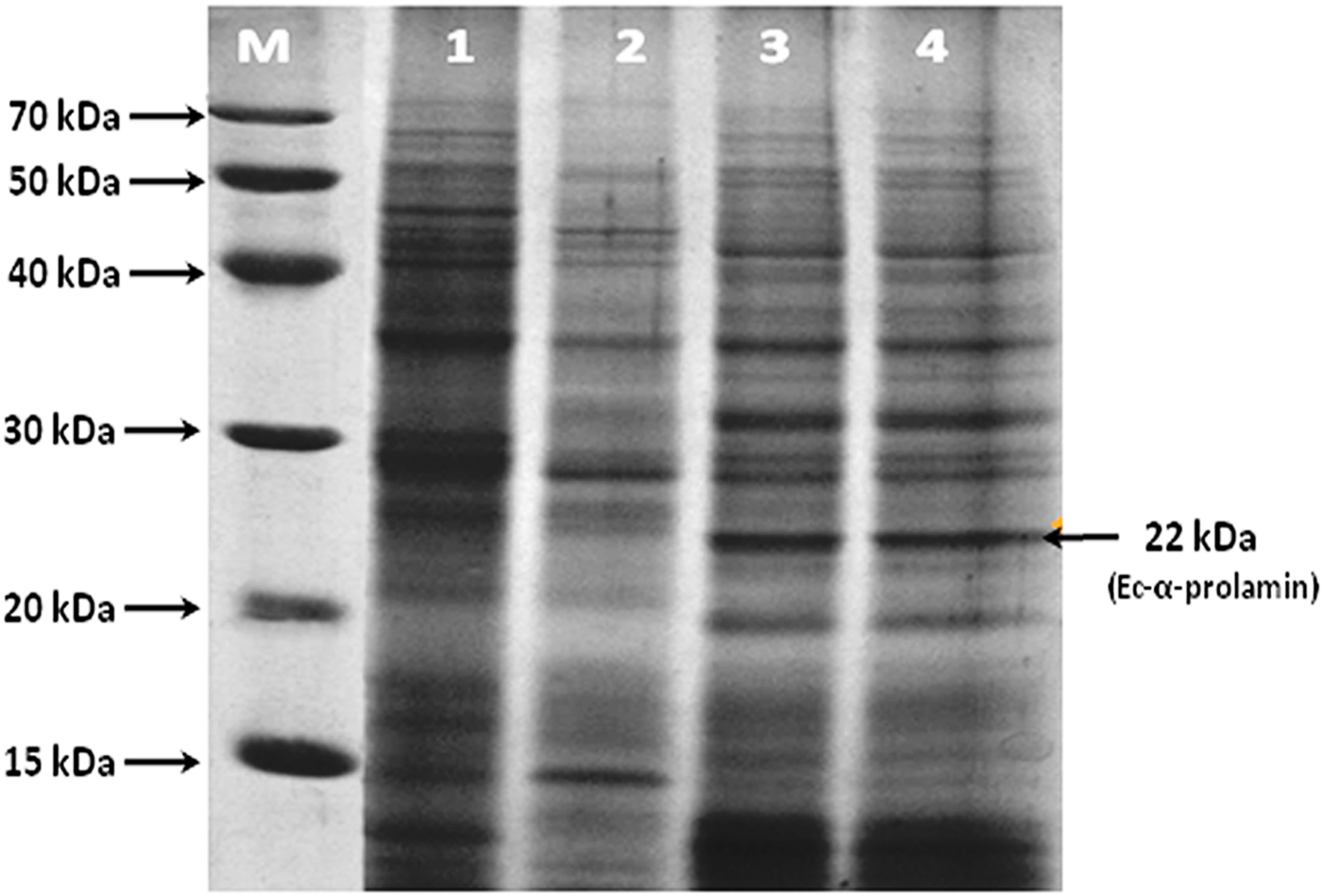
Figure 1. SDS-PAGE analysis of total protein isolated from S1, S2, S3 and S4 stages of finger millet (GE-3885, Brown genotype) showing the expression of the 22 kDa EC-α-prolamin protein. Lane M = medium range protein marker, lane 1 = initial stage (S1) lane 2 = fertilization stage (S2), lane 3 = milky stage (S3) and lane 4 = before harvesting (S4) stage of developing spikes.
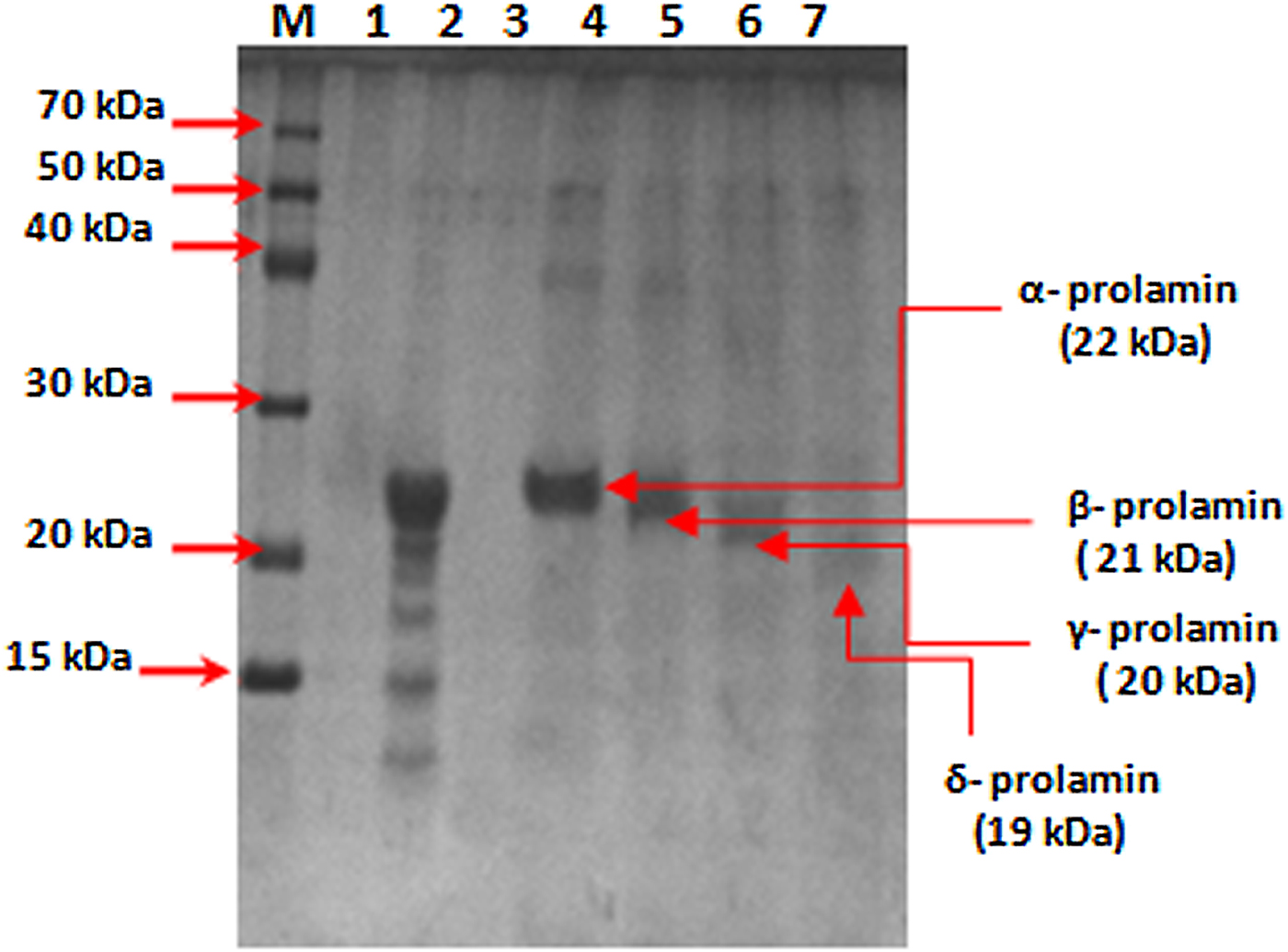
SDS-PAGE analysis of total prolamin proteins from mature seeds of finger millet showed four major bands: α, β, γ and δ prolamin proteins with molecular weights of about 22, 21, 20 and 19 kDa, respectively. The protein band corresponding to 22 kDa was interpreted as α-prolamin (Fig. 2) on the basis of earlier reports (Shewry, Reference Shewry, Belton and Taylor2002). The high band intensity of Ec-α-prolamin (22 kDa) stimulated our interest to characterize this protein in detail at the protein and gene level in Eleusine coracana to explore its nutritional significance. Therefore, the α-prolamin (22 kDa) fraction was purified from total proteins of mature seeds by gel filtration chromatography and analysed by SDS-PAGE (Fig. 2, lane 4). Antisera were then generated against it and its specific reactivity with Ec-α-prolamin protein was checked by a dot blot technique.
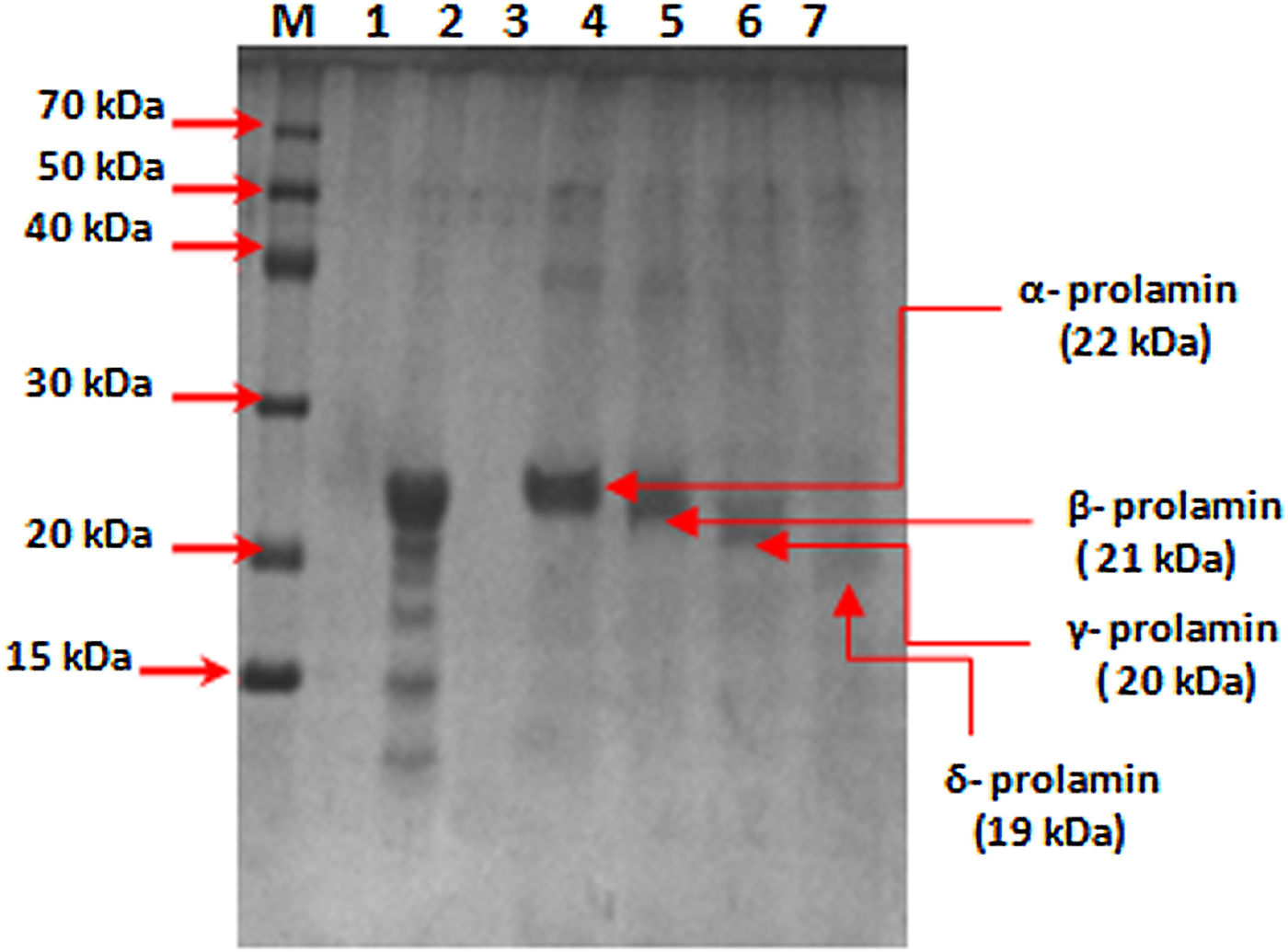
Figure 2. SDS-PAGE analysis of prolamin protein of finger millet (GE-3885, Brown genotype) purified by gel filtration chromatography. Lane M = medium range marker, lane 2 = total prolamin protein, lane 4 = purified alpha-prolamin protein, lane 5 = purified beta-prolamin, lane 6 = purified gamma-prolamin, lane 7 = purified delta-prolamin.
Using western blot analysis, the single Ec-α-prolamin-specific band of approximate 22 kDa was observed in the total protein isolated from different stages of developing spikes in the high protein GE-3885 genotype of finger millet. As a visible band of Ec-α-prolamin was observed in the S3 and S4 developmental stages (Fig. 3), it suggests that α-prolamin synthesis and its accumulation start in the S3 and S4 developmental stages of finger millet.
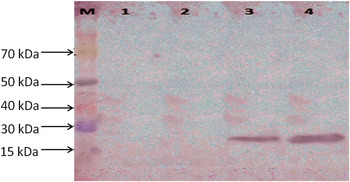
Figure 3. Western blotting of total protein isolated from S1, S2, S3 and S4 stages of finger millet (GE-3885, Brown genotype) showing detection of Ec-α-prolamin protein in S3 and S4 stages. Lane M = medium range protein marker, lane 1 = initial stage (S1), lane 2 = fertilization stage (S2), lane 3 = milky stage (S3) and lane 4 = before harvesting stage (S4) of developing spikes.
Isolation, nucleotide sequencing and characterization of Ec-α-prolamin
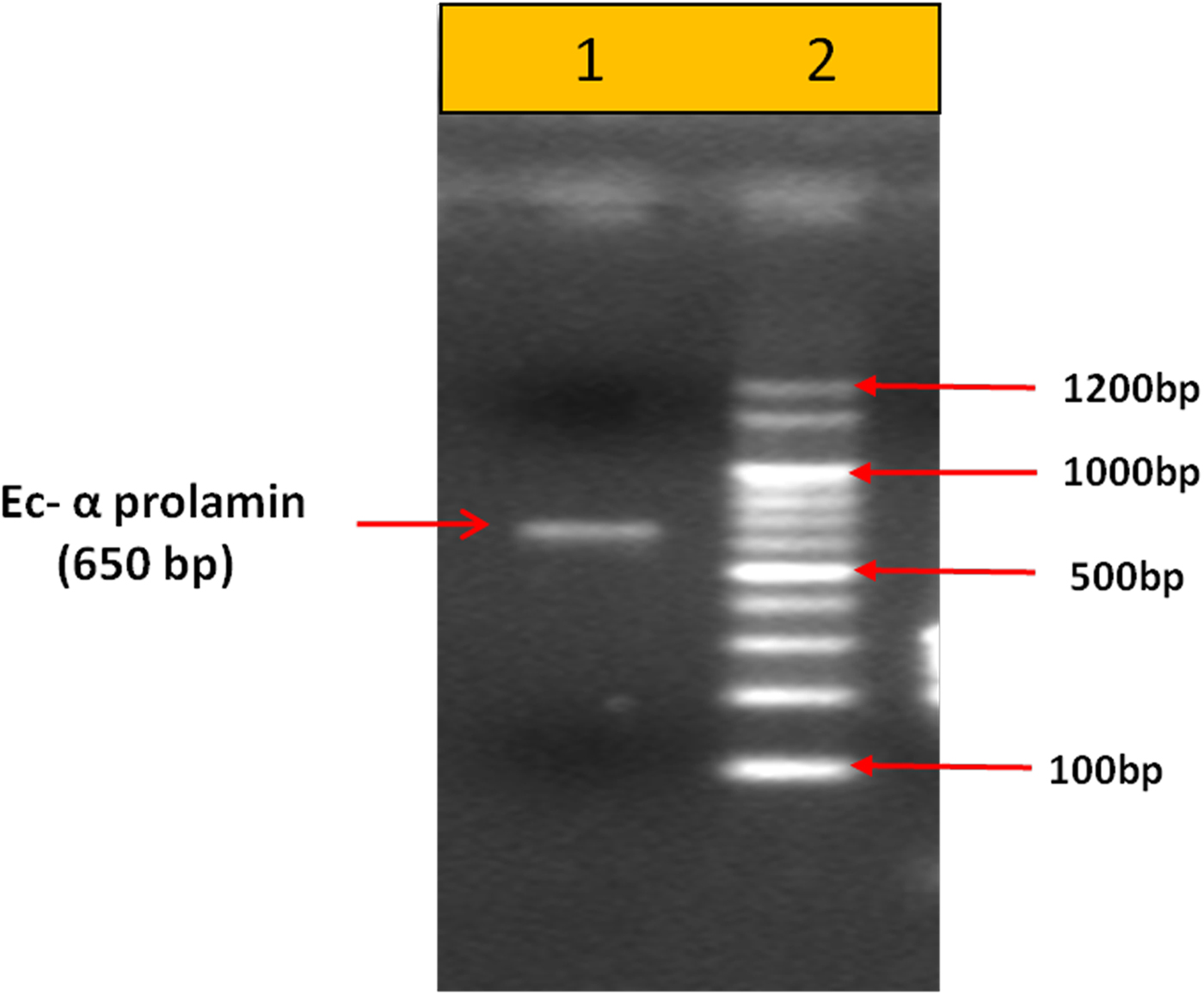
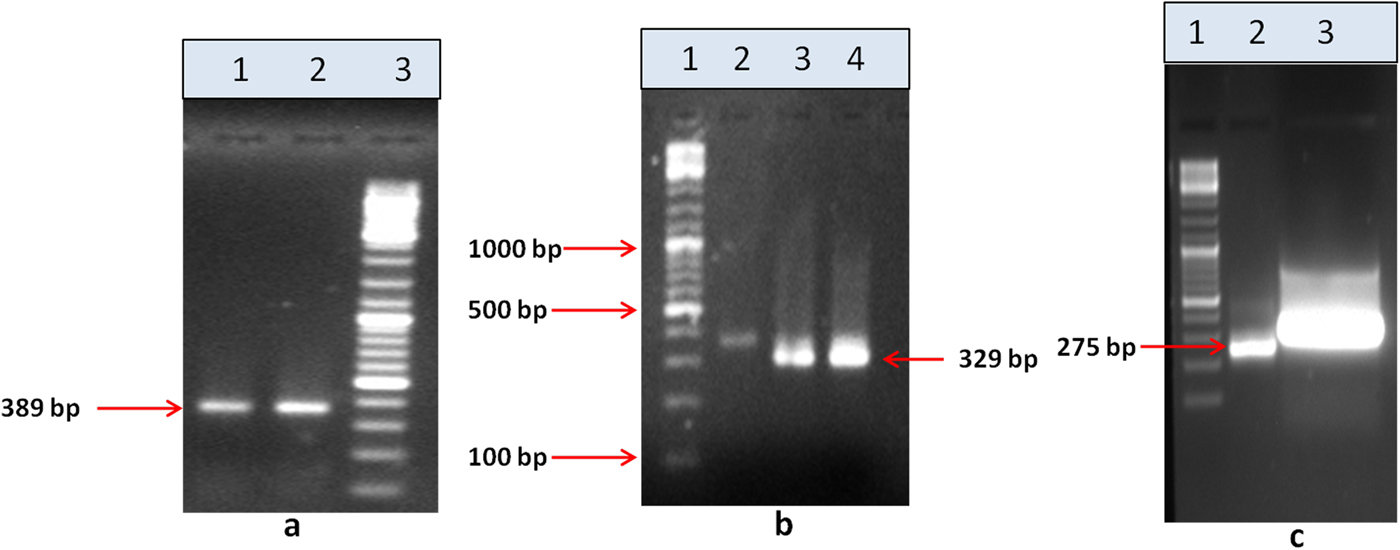
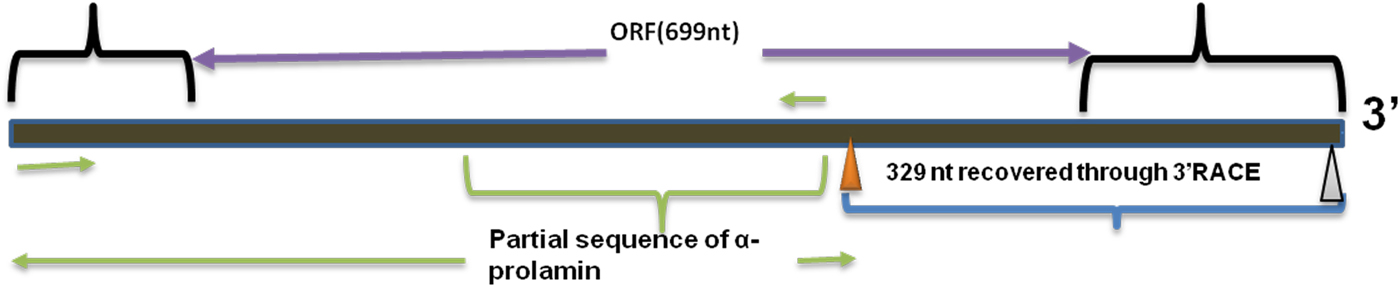
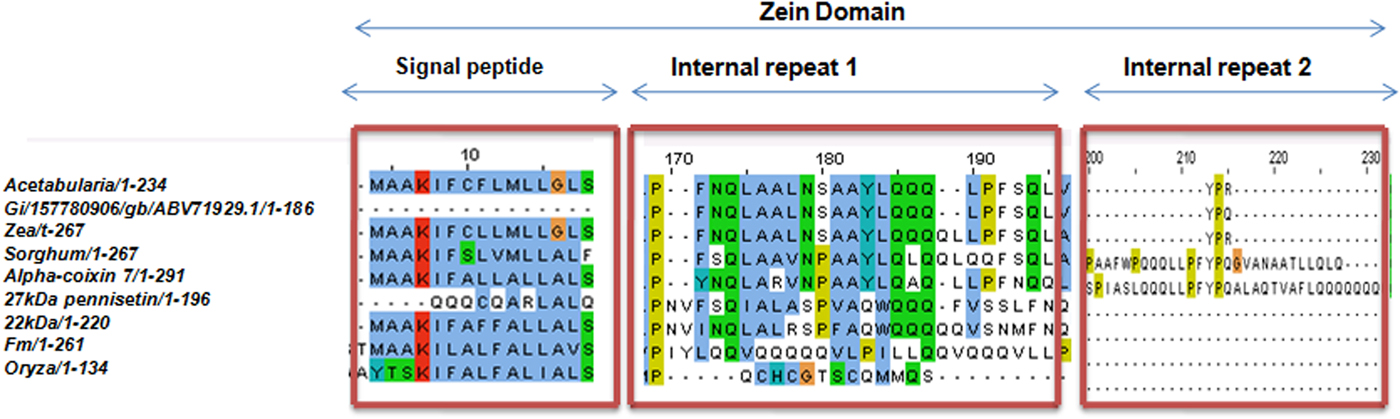
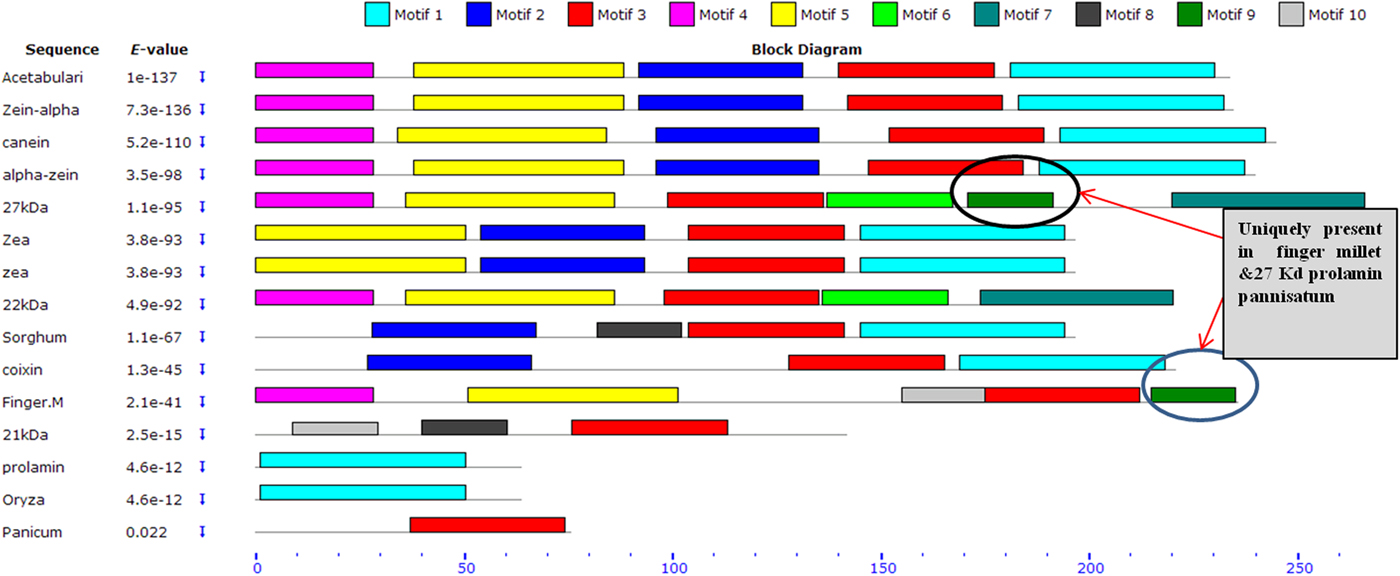
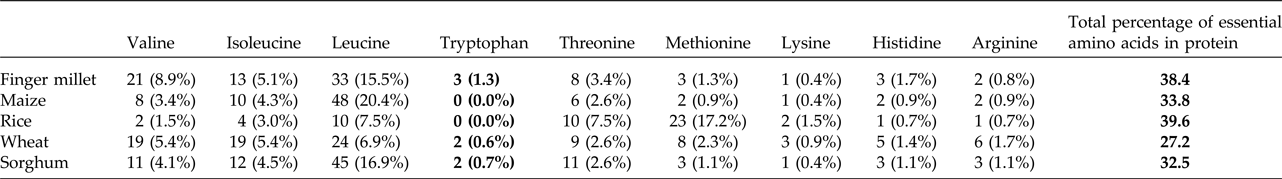
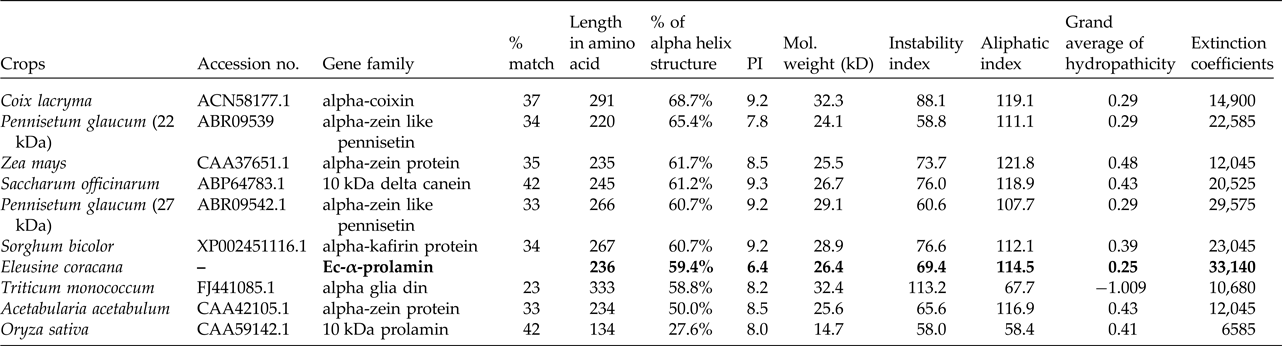
Upon semi-quantitative RT-PCR using primers designed from an EST of finger millet, homologous to alpha zein protein of maize, a 650 bp partial sequence of Ec-α-prolamin was amplified from the GE-3885 genotype of Eleusine coracana (Fig. 4). The amplicon was eluted from the gel, sequenced and subjected to BLASTn, where it was found to match the sequence of alpha zein. The pairwise alignment of the partial sequence of Ec-α-prolamin with alpha zein protein of maize revealed that the 5′ end of the ORF containing the initiation codon (ATG) was present, but the 3′ end was missing. The missing 3′ end of approximately 329 bp containing polyA tail was amplified by 3′ RACE and sequenced (Fig. 5a–c). The 5′ and 3′ ends of ORF were assembled to obtain a full-length sequence of 798 bp encoding 236 amino acids of the Ec-α-prolamin gene along with 209 bp 5′ UTR and 71 bp 3′ UTR (Fig. 6). The nucleotide sequence of the Ec-α-prolamin gene was translated into the amino acid sequence and its amino acid composition was compared with prolamin-related proteins of other cereals and millets. The Ec-α-prolamin protein of finger millet contained 38.4% of essential amino acids, with tryptophan at the highest level (1.3%) among prolamin-related proteins of other cereals and millets (Table 1). Upon BLAST, the Ec-α-prolamin amino acid sequence showed highest homology (42%) with the 10 kDa prolamin protein of Oryza sativa followed by Zea mays (35%) and least homology (23%) with α-gliadin of Triticum monococcum (Table 2). Ec-α-prolamin was found to contain a zein domain that is divided into one signal peptide and two internal repeat sequences (Fig. 7). This conserved domain was also detected in other prolamin-related proteins such as the 19 kDa alpha zein protein of Zea mays, the α-kefrin protein of Sorghum bicolor and the 22 and 27 kDa prolamin proteins of other plants such as Pennisetum glucum, Acetabularia acetabulum and Saccharum officinarum (Fig. 7). Instead of the zein domain, the trypsin-alpha amylase inhibitor domain was observed in the 10 kDa prolamin of Oryza sativa and the α-gliadin of Triticum monococcum. The motif and domain analysis of full-length Ec-α-prolamin by MEME and MAST tools revealed a unique domain (EELFDEFDEIAETSVGEW) present in Ec-α-prolamin of E. coracana and 27 kDa pennisetin (Cenchrus americanus; Fig. 8)
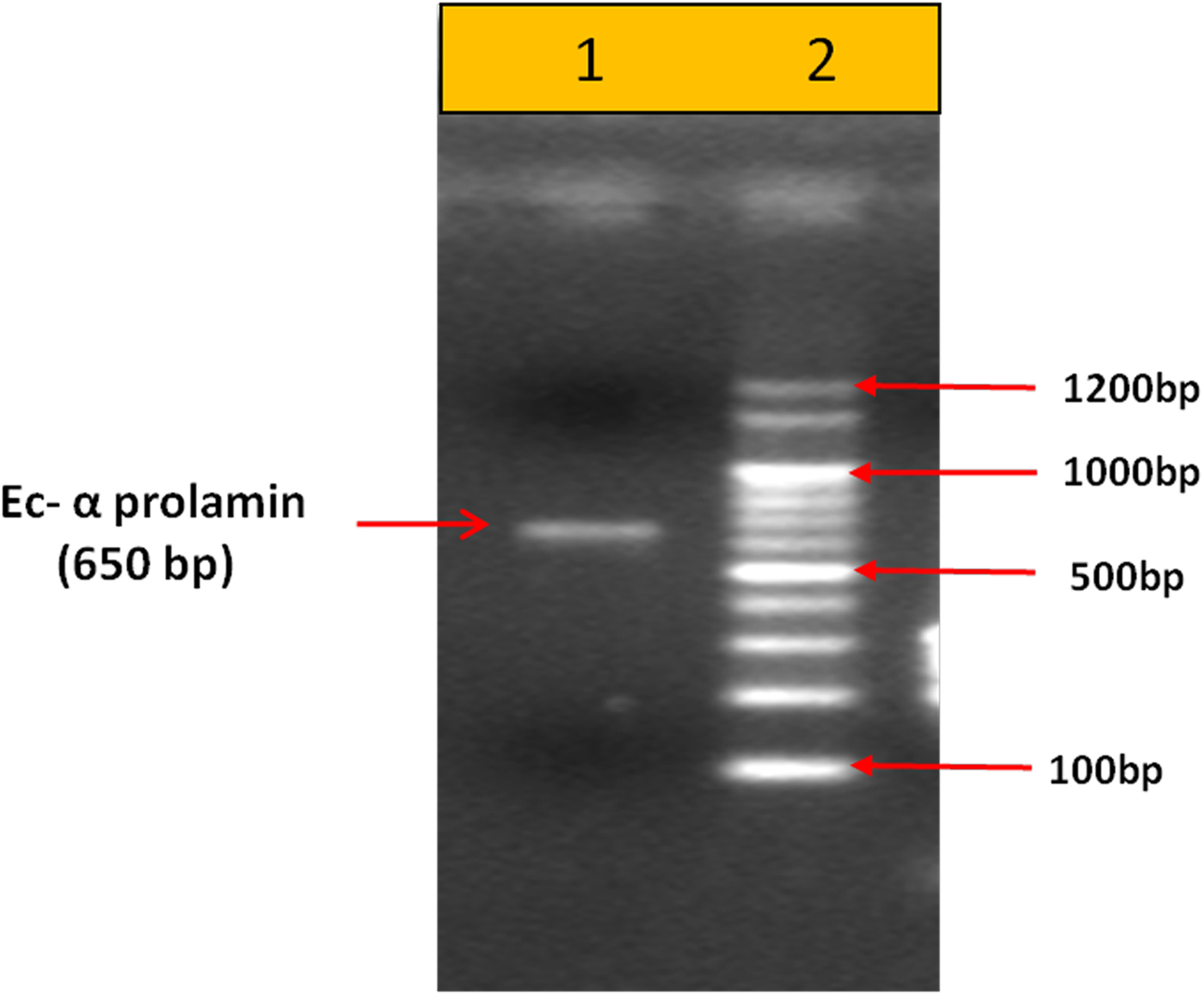
Figure 4. PCR-based amplification of Ec-α-prolamin gene from the GE-3885 genotype of finger millet. Lane 1 = 100 bp DNA ladder; lane 2 = 650 bp amplified gene.
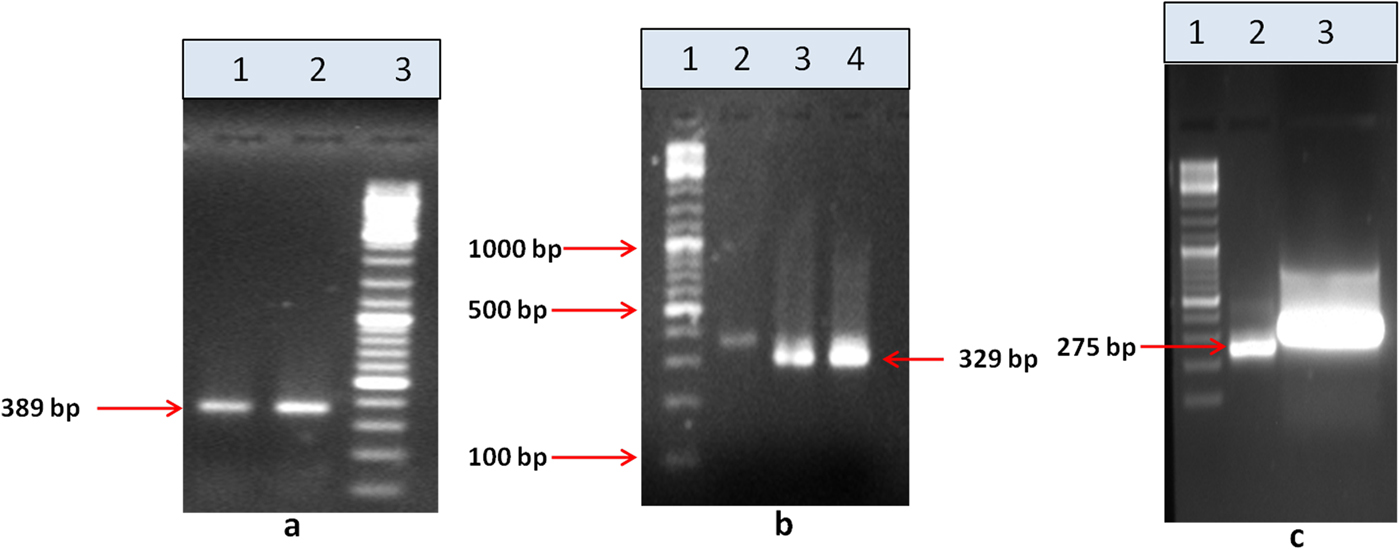
Figure 5. 3′ RACE PCR amplification of the Ec-α-prolamin gene. (a) Amplification of 389 bp by outer primer using cDNA as a template. Lane 1 = Tm −55°C, lane 2 = Tm 60°C, lane 3 = 100 bp DNA ladder. (b) Amplification of 329 bp by inner first primer using PCR product of (a) as template. Lane 1 = 100 bp DNA ladder, lane 2 = first PCR product, lane 3 = amplicon at Tm 55°C, lane 4 = amplicon at Tm 55°C. (c) Amplification of 275 bp by inner second primer using PCR product of (b) as a template. Lane 1 = 100 bp DNA ladder, lane 2 = amplicon, lane 3 = eluted amplicon.
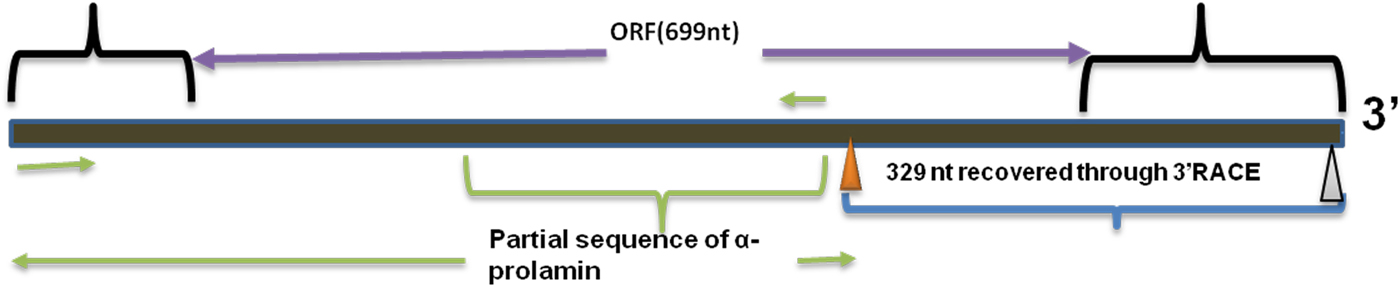
Figure 6. Full-length alpha prolamin gene.
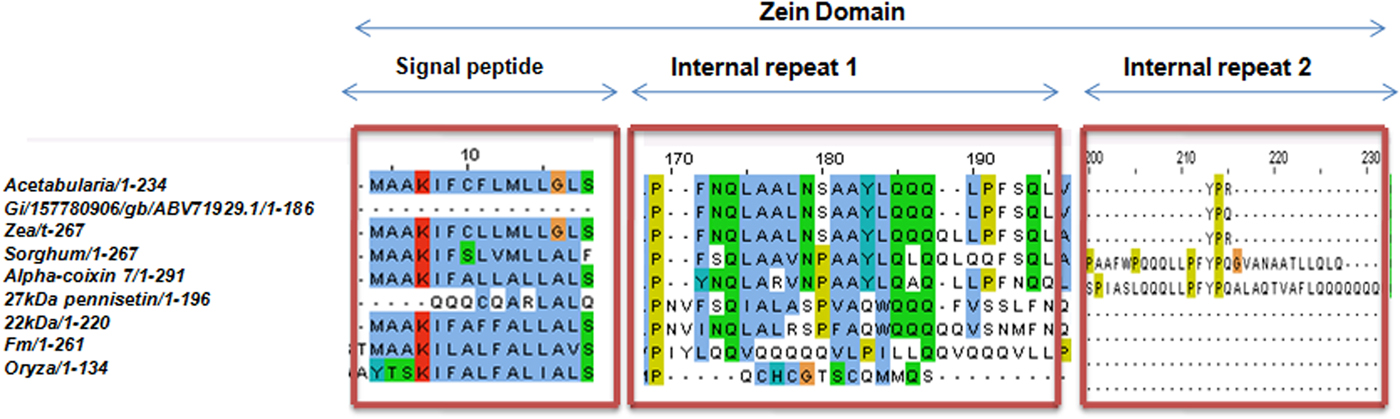
Figure 7. Alignment of amino acid sequence of Ec-α-prolamin with related prolamins of other cereals and millets showing conserved zein domain in all the proteins.
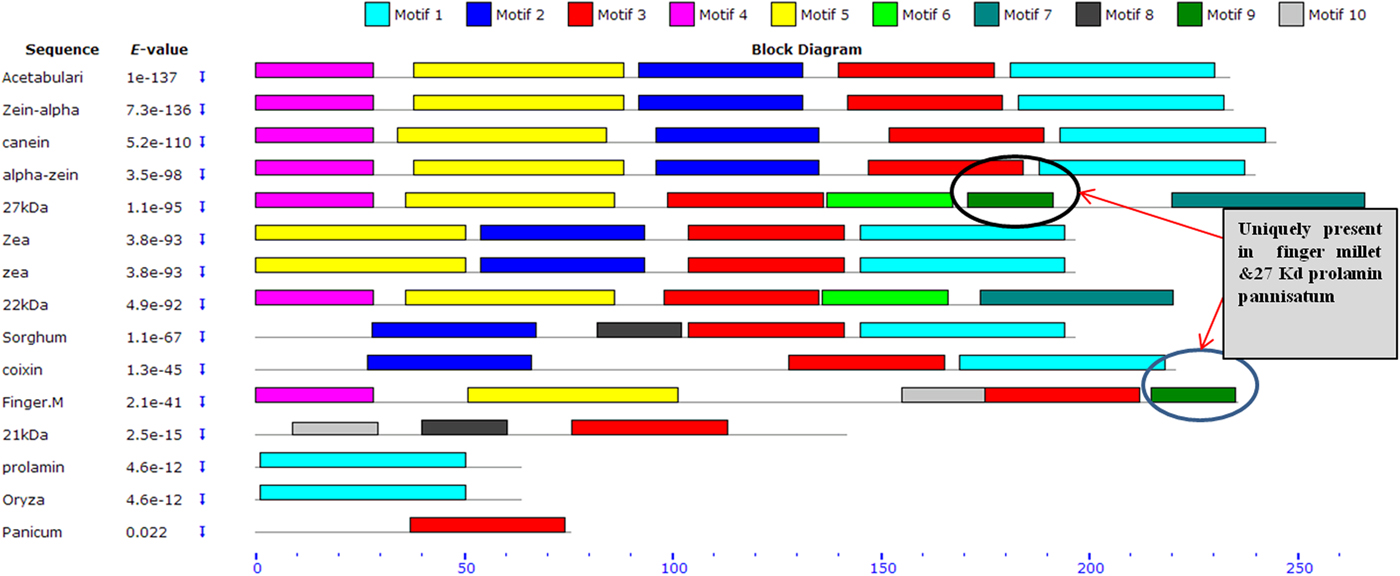
Figure 8. Schematic distribution of respective conserved motifs in Ec-α-prolamin and related prolamins identified by means of MEME software. Arrows indicates unique motif found in prolamin of finger millet and 27 kDa protein of Pennisetin.
Table 1. Comparative analysis of essential amino acids present in cereals and millets of Ec-α-prolamin protein sequence
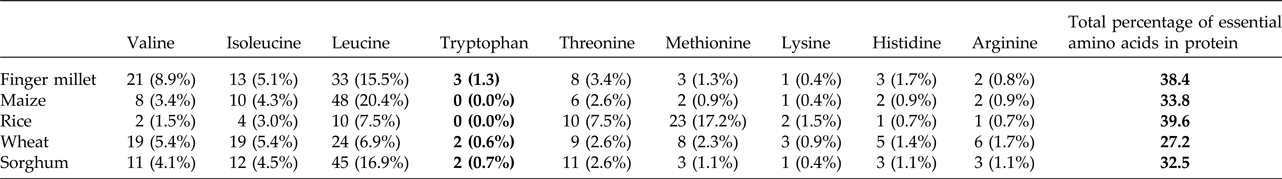
Different values represent the number and percentage of respective essential amino acid out of total number of amino acids in prolamin or prolamin-related proteins in different crops. Bold values in table highlight the Tryptophan conetent found in alpha prolamin of Finger millet compared to that found in other related proteins.
Table 2. Homology of Ec-α-prolamin of Eleusine coracana sequence to other crops and physiochemical properties
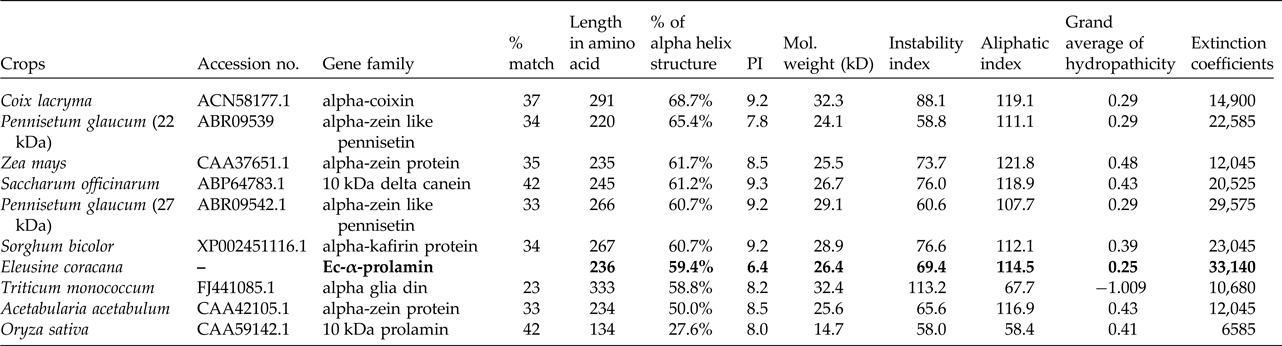
Bold values highlight the properties of EC alpha prolamin (isolated in the present study) relative to those of related proteins in other cereals and millets.
Physiochemical characteristics of Ec-α-prolamin
A comparative study of physiochemical properties of the Ec-α-prolamin of Eleusine coracana and related α-prolamins in different cereals and millet was performed and revealed that proline and glutamine together comprise 28% of the total amino acids in Ec-α-prolamin, whereas their content is only 22% of total amino acids in the prolamin of rice (Yoon et al., Reference Yoon, Lee, Hahn, Kim, Lee, Ji, Kim, Mun, Kim and Kim2012). Thus the prolamin of finger millet is rich in glutamine and proline. The calculated isoelectric point (pI) provides an idea about the solubility of the protein and its mobility in an electric gradient. The pI of Ec-α prolamin is 6.37 (Table 2), at which Ec-α-prolamin proteins are stable and compact. The very high aliphatic index (114.49) of Ec-α-prolamin protein sequences indicates that Ec-α-prolamin protein may be stable at a wide range of temperatures. The instability index value for the Ec-α prolamin was found to be 69.35, which indicates that Ec-α-prolamin is unstable in vitro because proteins with an instability index larger than 40 are considered to be unstable (Guruprasad et al., Reference Guruprasad, Reddy and Pandit1990). The GRAVY index (0.252) of Ec-α prolamin indicates good affinity for interaction with water.
Secondary and tertiary structure of Ec-α-prolamin
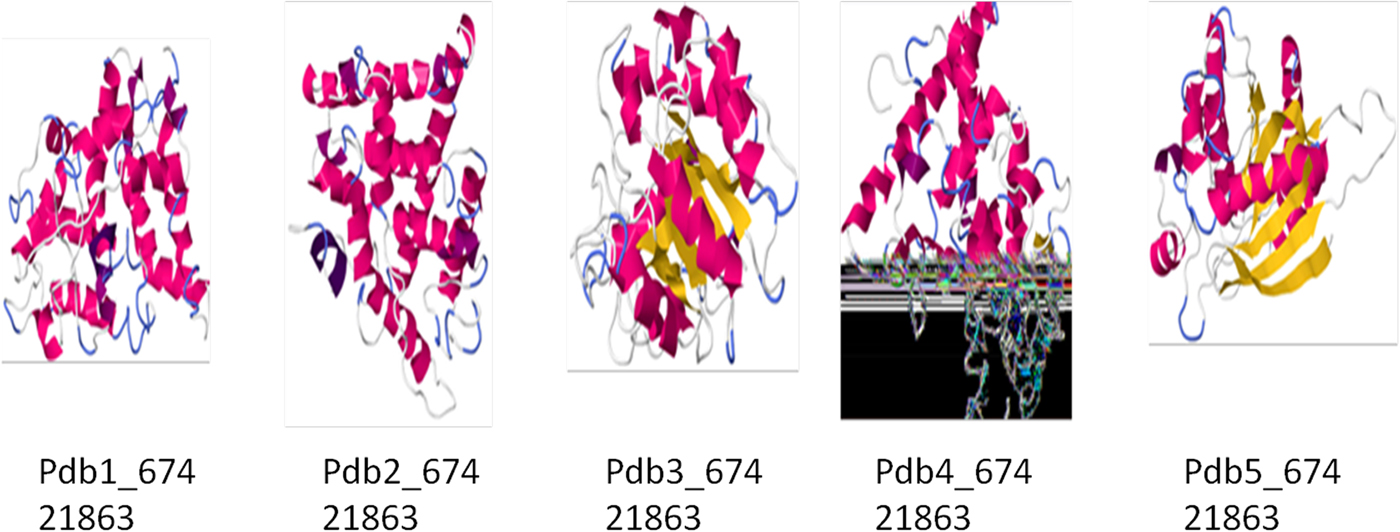
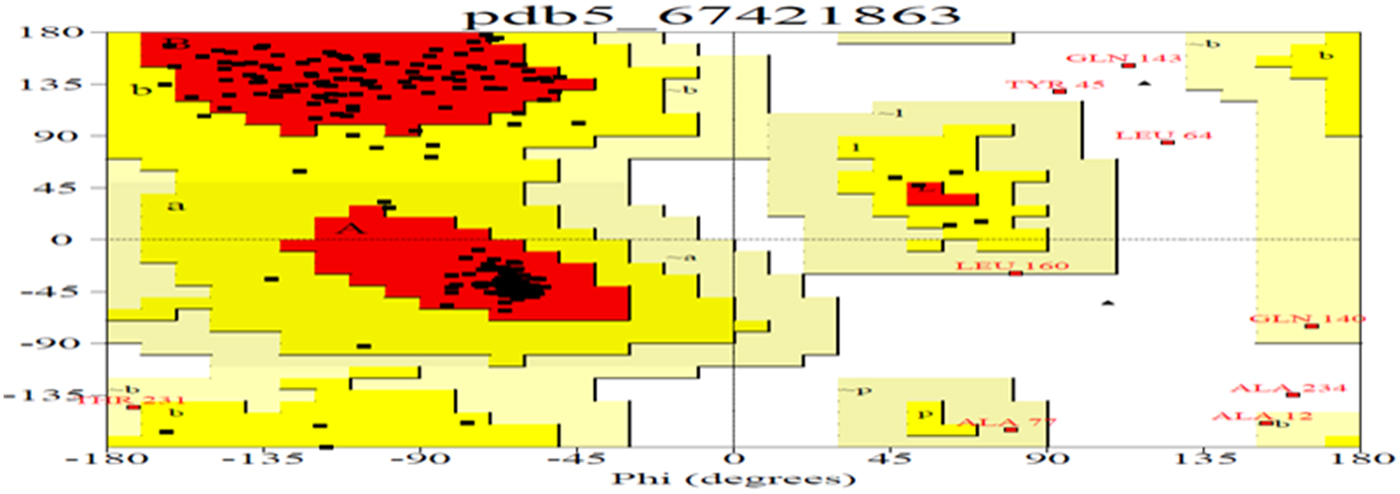
Due to the abundance of alpha helix, this protein could be used to manufacture the best films in terms of clearness, greatest strain at break, highest tensile strength and lowest water vapour permeability. As the helical nature of a protein increases, its chances of cross-linking also increase, and this property can be utilized for formation of biopolymers (da Silva and Taylor, Reference da Silva and Taylor2005; Taylor et al., Reference Taylor, Taylor, Dutton and De Kock2005) in the pharmaceutical industry. Therefore, in order to understand these properties, the secondary and tertiary structures of Ec-α-prolamin were predicted. The Ec-α-prolamin gene has been predicted to encode a 26.4 kDa protein with highest α-helix structure (59.4%) among all reported prolamins (Table 2), which further implies that it can be used to manufacture biofilm and biopolymer (Riebel and Mankato, Reference Riebel and Mankato2005). The tertiary structure of Ec-α-prolamin protein was determined using a homology modelling method. Five tertiary structures could be predicted (Fig. 9), out of which the most accurate structure (pdb5_674) was validated using Ramachandran plotting (Fig. 10). This indicates the maximum number of amino acids of structure falling in the allowed region and only 8% of amino acids fall in disallowed region.

Figure 9. Predicted three-dimensional structures of the Ec-α-prolamin of finger millet. The secondary structure elements are presented as ribbons. Residues nearest to the N-terminus are blue and those nearest to C-terminus are red. Residues in between are assigned other colours spanning the visible spectrum.
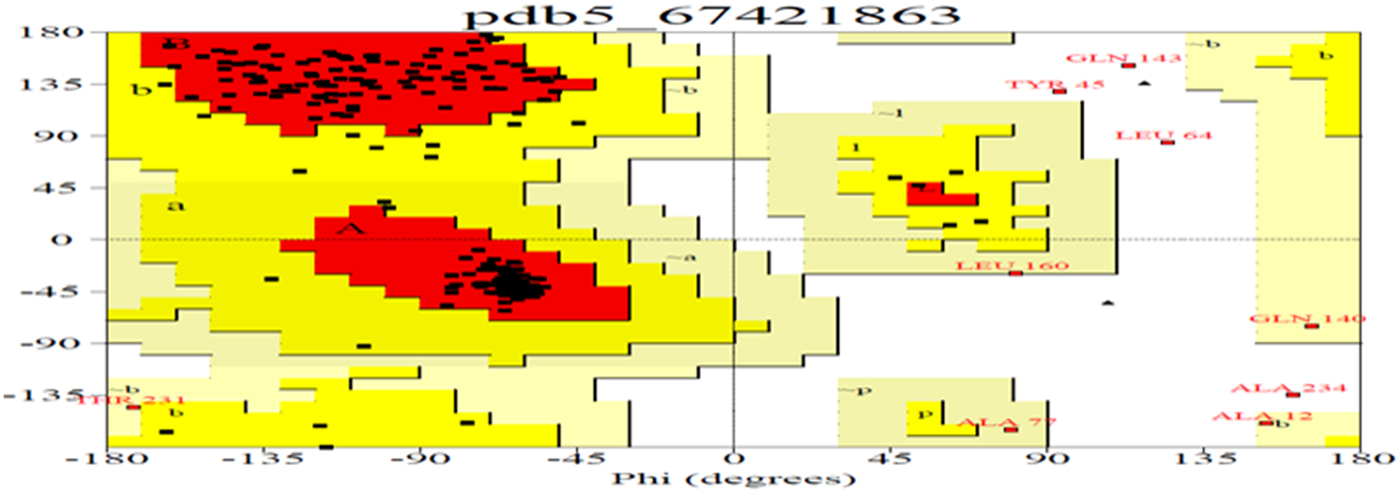
Figure 10. Ramachandran plot analysis for most accurate tertiary structure of Ec-α-prolamin (pdb5674) of finger millet. The plot statistics are: Total number of residues = 205 with 86.9% of residues in the core region (red); 9.3% residues in allowed (yellow); 2.1% in generously allowed (light yellow) and 1.7% residues in the disallowed region (white). Number of glycine residues (labelled as G) is 4, and number of Pro residues is 1.
Synthesis of Ec-α-prolamin in developing spike transcript profiling by qPCR
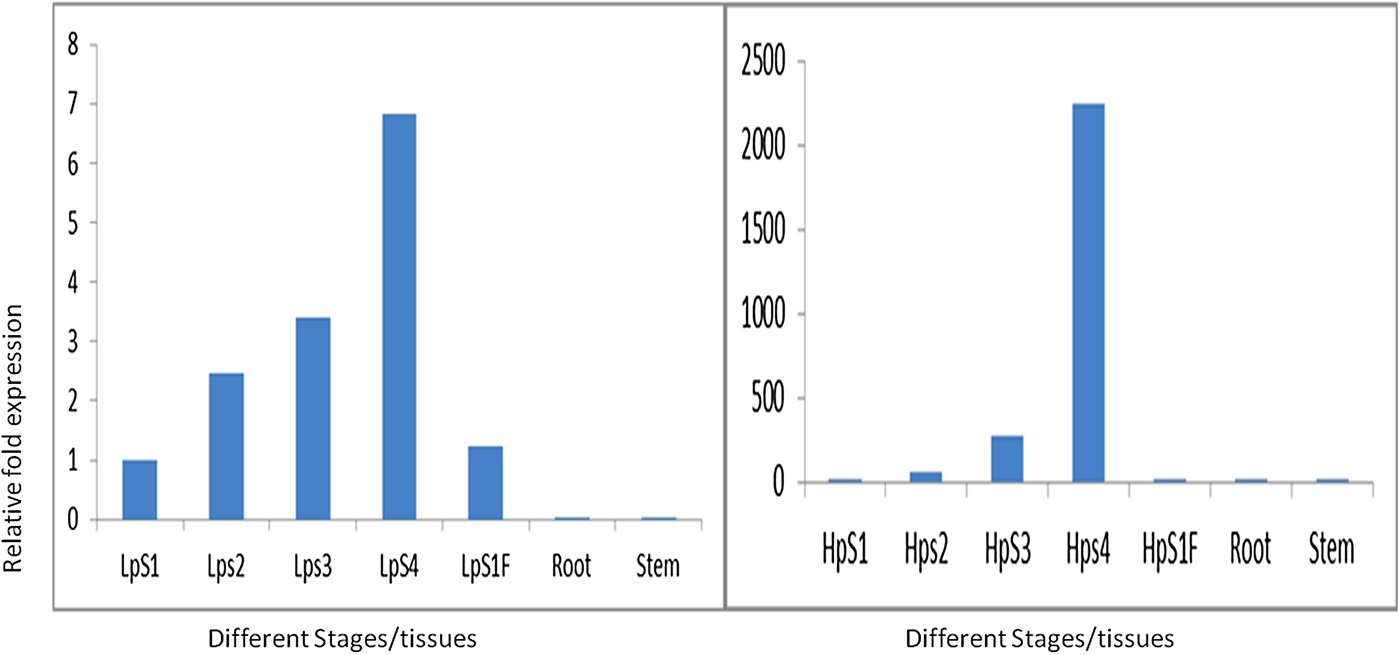
In order to validate the results of western blotting in which the α-prolamins are expressed at the S3 and S4 stages, qPCR was performed at different stages of developing spikes and flag leaf. The expression of Ec-α-prolamin continuously increased over the developing stages of spikes, starting from the date of anthesis to the grain filling stage (Fig. 11). In the case of S1 stage flag leaf (HpS1 F), Ec-α-prolamin expression was detected as very low.
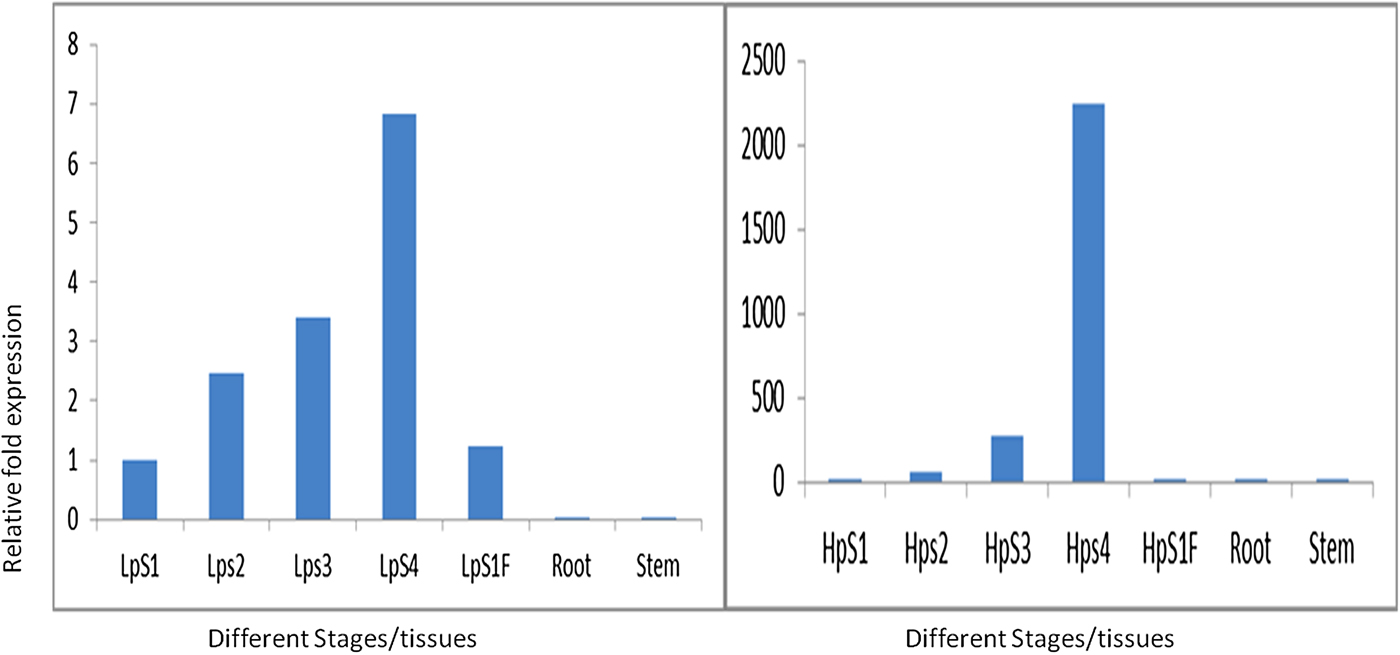
Figure 11. Real-time PCR analysis of Ec-α-prolamin in two contrasting plant genotypes. Left: low protein (GE-1435); right: high protein genoptype (GE-3885) at S1, S2, S3 and S4 stages of developing spikes and in leaf, root and stem. Note the different scales on the y-axis.
Discussion
Isolation and characterization of nutritionally rich proteins and their genes have become important for genetic engineering-based biofortification of cereal staple crops and development of functional foods to solve the problem of malnutrition. In the present investigation, it was revealed that 22 kDa α-prolamin, a seed storage protein of finger millets, predominantly expressed at the S3 and S4 stages of spike development. Its most abundant fraction among the prolamin proteins was sequentially extracted from total proteins of finger millet. In order to explore if it is the major protein contributing to high nutritional value of finger millet, attempts were made to isolate the gene for Ec-α-prolamin of finger millet. GE3885, a high protein genotype of finger millet, is expected to contain a higher abundance of Ec-α-prolamin gene transcripts than the low protein genotype which was used as source for isolation of the gene. It was observed that the isolated Ec-α-prolamin gene-encoded protein has nutritionally a better balanced essential amino acid content than prolamin-related proteins in other cereals and millets. However, the total protein amount was comparable to other cereals and millets (Table 1). Ec-α-prolamin was found to contain the highest amount of tryptophan among the prolamin-related proteins of other cereals and millets. Thus this protein is nutritionally superior to other seed storage proteins and could serve as an attractive target for biofortification programmes.
The precise mechanism of the regulation of amino acid composition is still unknown, but it is assumed that the primary control of the prolamin genes take place at the transcription level (Sørensen et al., Reference Sørensen, Cameron-Mills and Brandt1989) with the help of a regulatory –300 element present upstream of the promoter, which is known as P-box (Forde et al., Reference Forde, Heyworth, Pywell and Kreis1985). This box is known to consist of two conserved motifs, the E motif (TGTAAAGT) and the GLM motif [G(A/G)TGAGTCAT] (also called nitrogen element, or N motif) (Müller and Knudsen, Reference Müller and Knudsen1993). The corresponding two transcription factors, PBF and OPAQUE2, bind to the P-box and E motif, respectively, to regulate prolamin gene expression and its amino acid composition of total grain protein. This observation was based on the report that in the opaque2 mutant, the lysine and tryptophan content were increased nearly twofold in proteins accumulated in the endosperm of maize, compared with the wild-type by a decrease in the Lys ketoglutarate reductase, saccaropine dehydrogenase and tryptophan amino transferase enzymes involved in the downstream catabolism of lysine and tryptophan (Mertz et al., Reference Mertz, Bates and Nelson1964). As the amino acid composition of the Ec-α-prolamin protein is dependent upon the OPAQUE2 transcription factor, which is the same in high and low protein genotypes (L. Kumar et al., unpublished results), it is expected that the Ec-α-prolamin genes of both genotypes may have a similar balanced amino acid composition.
The Ec-α-prolamin gene encodes a 26.36 kDa protein and its secondary structure demonstrates a high proportion of alpha helix structure (59.4%) (Table 2), which increases its prospects to be utilized as biodegradable biopolymer by the pharmaceutical industry (da Silva and Taylor, Reference da Silva and Taylor2005). Ec-α-prolamin has high thermal resistance and the ability to undergo disulfide bond-mediated cross-linking in response to heating with microwave energy. It also induces changes in its secondary structure from α-helical to β-sheet conformation to form oligomers of kafirin/prolamin (Byaruhanga et al., Reference Byaruhanga, Emmambux, Belton, Wellner, Ng and Taylor2006). These properties make it ideal for preparation of bioplastic film with high tensile strength. Besides this, it could also be exploited for the development of functional foods for diabetics because alpha-prolamin proteins form a water-tight matrix that makes starch granules less degradable in the rumen environment. This study can help in development of food with high prolamin content; such food will allow only slow digestion of starch, which on consumption will be of help to the diabetic patient (Hoffman, Reference Hoffman2009).
Alignment of the sequence of Ec-α-prolamin of finger millet with prolamins of other related cereals and millets indicated the presence of a conserved signal peptide at the N-terminal (Fig. 7). As the prolamin protein is tissue specific, the function of the signal peptide is to target the prolamin protein from the ER to protein bodies of the developing endosperm. Ec-α-prolamin has two internal repeat sequences in its Zein domain, which is not observed in other cereals and millets. Prolamins from most cereal crops have Zein domains but others possess lipid transfer protein/seed storage protein/trypsin-alpha amylase inhibitor domain families. It has been reported that proteins such as α-gliadin of Triticum monococum and the10 kDa protein of Oryza sativa possess trypsin-alpha amylase inhibitor domains, responsible for anti-nutritional properties. However, no such domain was found in Ec-α-prolamin, further underscoring its nutritionally superiority over other prolamins. Motif analysis by MEME also indicated that prolamins from different cereals and millets are diverse, as they contained different sets of motifs. However, a unique motif (EELFDEFDEIAETSVGEW) was found in Ec-α-prolamin and in a 27 kDa pennisetin (Cenchrus americanus), a function of which remains unknown.
The expression of Ec-α-prolamin during the developing stages of spikes differing in seed protein contents indicates that the accumulation of seed storage proteins like Ec-α-prolamin occurs at a relatively late stage, which was supported by western blot analysis (Fig. 3). A similar expression pattern of α-prolamin was observed in the low protein genotype of finger millet (L. Kumar et al., unpublished results). Western blotting suggested that the monospecific antiserum against α-prolamin is highly specific as it did not show any reactivity with other types of prolamins. Hence it can be used for the development of an indirect ELISA for specific detection of α-prolamins. Ec-α-prolamin protein accumulation starts from the S3 stage of grain filling. Regulatory controls of prolamin primarily occur at the transcriptional level (Sørensen et al., Reference Sørensen, Cameron-Mills and Brandt1989). The transcription factors OPAQUE2 containing a bZip domain and PBF containing a zinc-finger domain interact with each other, along with an interaction with the upstream sequence of the promoter of the Ec-α-prolamin to up-regulate the protein content during the early S2 grain filling stage of development (L. Kumar et al., unpublished results). As the expression of opaque2 has been found to decrease during the later S3 and S4 stages of seed development, the relatively constant increase in Ec-α-prolamin content may be hypothesized to be dependent on the PBF transcription factor. This clearly indicates that the expression of Ec-α-prolamin is higher due to higher expression of these transcription factors.
Acknowledgements
Technical assistance by Dr Vikram Singh Gaur is also acknowledged.
Financial support
The research described in this manuscript was financially supported by Jawaharlal Nehru memorial fund fellowship and a grant from the Department of Biotechnology.
Conflicts of interest
Authors have no conflicting interests.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S096025851700023X.