Introduction
Endosperm is not just an embryo-nourishing tissue; being the major component of the cereal grain, it nourishes the global population. From an evolutionary perspective, the origin of endosperm (resultant of the second fertilization event) in angiosperms is predicted to have evolved from multiple fertilization events in gymnosperms, leading to polyembryony (Friedman, Reference Friedman1992). In 70% of the angiosperms genome dosage level of the embryo is ‘2n’ with equal genetic contribution from the female (m, maternal) and male (p, paternal) parent, while in the endosperm, it is ‘3n’ (2m:1p) (Raghavan, Reference Raghavan2006). Diversity in endosperm ploidy level of angiosperms broadly ranges from 2n to 15n, especially due to the variability in the genetic contribution of the female parent from 1n to 14n (Friedman et al., Reference Friedman, Madrid and Williams2008). Key genes regulating the cell cycle, mega-sporogenesis or -gametogenesis or endosperm developmental process, including ontogeny and cell numbers are well documented (Motamayor et al., Reference Motamayor, Venzon, Bajon, Sauvanet, Grandjean, Marchand, Bechtold, Pelletier and Horlow2000; Berger et al., Reference Berger, Grini and Schnittger2006; Huh et al., Reference Huh, Bauer, Hsieh and Fischer2007; Fiume and Fletcher, Reference Fiume and Fletcher2012; Batista et al., Reference Batista, Figueiredo, Santos-González and Köhler2019; Huang et al., Reference Huang, Wang, Huang, Wang, Wang, An, Li, Wang and Wu2019; Kirkbride et al., Reference Kirkbride, Lu, Zhang, Mosher, Baulcombe and Chen2019). However, the variability underlying these traits between species and its regulatory mechanism that flexibly accommodates and accepts the variable endosperm dosage level from 2n to 15n, and endosperm cell number in angiosperms is not yet fully understood. The present review highlights the endosperm variability patterns in angiosperms, from variable genome dosage to chlorophyllous endosperm involving carbon fixation; and the potentialities of the genomics tools and techniques to underscore the genetic mechanism for better scientific understanding.
Endosperm variability and development
Endosperm is a tissue in transition across dicot families, while it is the major source of reserves in monocots. However, early endosperm development is remarkably conserved across monocots and dicots (Becraft, Reference Becraft2001). Endosperm development is a dynamic and intricate process involving genome imprinting, dosage balance, positional cues, cell-cycle modifications and programmed cell death (PCD) for a normal ontogeny (Wangenheim, Reference Wangenheim1957; Becraft, Reference Becraft2001; Gehring and Satyaki, Reference Gehring and Satyaki2017; Satyaki and Gehring, Reference Satyaki and Gehring2019). In monocots, where the endosperm is persistent, it is comprised of three cell types, namely starchy endosperm, aleurone layer and transfer cell layers; among these, the aleurone layer retains its viability at seed maturity (Lopes and Larkins, Reference Lopes and Larkins1993; Becraft, Reference Becraft2001; Becraft and Gutierrez-Marcos, Reference Becraft and Gutierrez-Marcos2012). In addition to cell-type variability, the endosperm exhibits variability of inter alia ploidy (both across species and within the seed, as well), chlorophylly and polyendospermy.
Endosperm ploidy variability in sexual and apomictic species
Around 70% of the angiosperms exhibit an endosperm ploidy level of ‘3n’ with two-thirds of the genome dosage contribution from the maternal parent, and the remaining one-third through the paternal parent due to the second fertilization event (Raghavan, Reference Raghavan2006). The maternal to paternal genome ratio of 2:1 is the most balanced dosage levels among parental genomes during sexual reproduction and is postulated as endosperm balance number (EBN) for normal endosperm and seed development (Johnston et al., Reference Johnston, Den Nijs, Peloquin and Hanneman1980). However, in several angiosperms, endosperm ploidy deviates from ‘3n’. A central cell with 1 to 14 nuclei is the source of variability for the endosperm ploidy level, resulting in a genome dosage level of 2n to 15n upon the second fertilization event, reflecting altered maternal dosage levels, ranging from 1m:1p to 14m:1p (Friedman et al., Reference Friedman, Madrid and Williams2008). Genera exhibiting variable endosperm ploidy level due to altered genome dosage levels of maternal parents include Oenothera (Von Wangenheim, Reference Von Wangenheim1962; Haig and Westoby, Reference Haig and Westoby1991), Nuphar (Williams and Friedman, Reference Williams and Friedman2002), Manekia (Arias and Williams, Reference Arias and Williams2008), Gagea (Greilhuber et al., Reference Greilhuber, Ebert, Lorenz and Vyskot2000), Penea, Plumbago, Fritillaria, Plumbagella and Peperomia (Friedman et al., Reference Friedman, Madrid and Williams2008). Invariably, in all these species with seed development through a fertilization event, the paternal genome dosage contribution is uniformly ‘1n’ only.
However, in aposporous apomicts, the paternal genome dosage level could be higher than ‘1n’, at instances where the paternal ploidy level is higher than the maternal one (Haig and Westoby, Reference Haig and Westoby1991; Quarin, Reference Quarin1999; Alves et al., Reference Alves, Carneiro and Araujo2001; Šarhanová et al., Reference Šarhanová, Vašut, Dančák, Bureš and Trávníček2012). In diplosporous apomicts, the maternal dosage alone is inherited with 4n (4m:0p) genome content without the fertilization event (Kollmann et al., Reference Kollmann, Steinger and Roy2000). In certain diplosporous apomictic grass species, such as Elymus, Poa, Eragrostis and Tripsacum, pollination is a necessary event for successful endosperm development – one of the three components of apomictic seed development (Bashaw and Hanna, Reference Bashaw, Hanna and Chapman1990; Kaushal et al., Reference Kaushal, Dwivedi, Radhakrishna, Srivastava, Kumar, Roy and Malaviya2019). For example, Tripsacum with diploids and tetraploids reported to exhibit genome dosage (maternal to paternal) of 8:1 and 8:2 ratios in apomictic plants, but 2:1, 4:1 and 4:2 ratios observed in sexual plants (Grimanelli et al., Reference Grimanelli, Hernández, Perotti and Savidan1997). Paspalum with di-, tri-, tetra-, penta-, hexa- and octaploid species, the endosperm genome dosage level varies widely from 1:1, 2:1, 3:1, 4:1, 4:3, 8:1 to 8:3, with 4:1 genome dosage exhibiting the maximum reproductive efficiency (Burton, Reference Burton1948; Quarin et al., Reference Quarin, Burson and Burton1984; Quarin, Reference Quarin1999; Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015). An AFLP Marker associated with apomictic trait loci in Paspalum has been identified (Labombarda et al., Reference Labombarda, Busti, Caceres, Pupilli and Arcioni2002), its cross-transferability in other related species might be of advantage in differentiating the apomictic from sexual plants. Exceptionally, the occurrence of pseudogamy resulting in the normal EBN genome dosage level of 2:1 is possible only when the reduced sperm cell is fused with the central cell containing a single unreduced polar nucleus as reported in Panicum maximum (Warmke, Reference Warmke1954). However, in general, due to pseudogamy, 4:1 is formed when the two unreduced polar nuclei are fused with one reduced sperm nucleus (Grimanelli et al., Reference Grimanelli, Hernández, Perotti and Savidan1997; Quarin, Reference Quarin1999; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015). Polyploidy and apomixis are reported to be strongly associated, although gametophytic apomixis is reported in diploids (Sharbel et al., Reference Sharbel, Voigt, Corral, Thiel, Varshney, Kumlehn, Vogel and Rotter2009; Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013).
The occurrence of apomixis (gametophytic: apospory and diplospory; sporophytic: adventitious embryony type) is documented in at least 300 species, predominantly from four families, namely Gramineae, Compositae, Rosaceae and Rutaceae, but for more than 35 families in total (Koltunow, Reference Koltunow1993; Khush, Reference Khush1994). As a rule of thumb, gametophytic apomicts (apospory – Cenchrus, Dicanthium, Panicum and Heracium; diplospory – Taraxacum, Ixeris and Antennaria) are polyploid in nature, while the sporophytic ones (adventitious embryony – Citrus) exhibit diploid behaviour (Knox, Reference Knox1967; Young et al., Reference Young, Sherwood and Bashaw1979; Asker and Jerling, Reference Asker and Jerling1992).
From an evolutionary perspective, polyploidization and hybridization events are the drivers of genetic modifications at the genome level for apomixis development (Barke et al., Reference Barke, Daubert and Hörandl2018). Photoperiod (natural) induced apomixis in Dichanthium was the first report documenting the role of the environment, in addition to genetic factors, leading to apomixis (Knox, Reference Knox1967; Rodrigo et al., Reference Rodrigo, Zappacosta, Selva, Garbus, Albertini and Echenique2017). Moreover, experimentally induced apomixis through gamma-ray irradiation has also been documented for maize (Yudin, Reference Yudin1966). Chemically induced apomixis has employed inter alia nitrous oxide, dimethyl sulfoxide, gibberellic acid, 6-benzyl aminopurine, 2,4-dichlorophenoxy acetic acid and zeatin in Datura, Solanum, Ficus, Gossypium and Zea (Montezuma-de-Carvalho, Reference Montezuma-de-Carvalho1967; Arendt, Reference Arendt and Khokhlov1970; Zhou, Reference Zhou1980; Hu et al., Reference Hu, Liang and Wassom1991).
Facultative apomixis – coexistence of apomixis and sexual reproduction – have been demonstrated in Hieracium and Sorghum (Tang, Reference Tang1977; Bicknell and Koltunow, Reference Bicknell and Koltunow2004; Carman et al., Reference Carman, Jamison, Elliott, Dwivedi and Naumova2011). Probably due to the apomictic nature of Hieracium, Gregor Mendel in 1869 obtained contrasting results as compared to Pisum, which he noted as ‘almost opposed behaviour’ at a time when the phenomenon of apomixis was unknown (Savidan, Reference Savidan2000; Bicknell and Koltunow, Reference Bicknell and Koltunow2004). Inheritance of aposporous apomixis has been studied in Pennisetum, Panicum and Brachiaria and reported to be determined by a single dominant locus (Sherwood et al., Reference Sherwood, Berg and Young1994; Valle et al., Reference Valle, Glienke and Leguizamon1994; Savidan, Reference Savidan2000). Genes (protein coding and lncRNA) and epigenetic mechanisms regulating apomixis are well documented (Guerin et al., Reference Guerin, Rossel, Robert, Tsuchiya and Koltunow2000; Albertini et al., Reference Albertini, Marconi, Reale, Barcaccia, Porceddu, Ferranti and Falcinelli2005; Laspina et al., Reference Laspina, Vega, Seijo, González, Martelotto, Stein, Podio, Ortiz, Echenique and Quarin2008; Garcia-Aguilar et al., Reference Garcia-Aguilar, Michaud, Leblanc and Grimanelli2010; Polegri et al., Reference Polegri, Calderini, Arcioni and Pupilli2010; Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013; Hand and Koltunow, Reference Hand and Koltunow2014; Podio et al., Reference Podio, Cáceres, Samoluk, Seijo, Pessino, Ortiz and Pupilli2014a,Reference Podio, Felitti, Siena, Delgado, Mancini, Seijo, González, Pessino and Ortizb; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015; Ortiz et al., Reference Ortiz, Revale, Siena, Podio, Delgado, Stein, Leblanc and Pessino2017; Selva et al., Reference Selva, Siena, Rodrigo, Garbus, Zappacosta, Romero, Ortiz, Pessino, Leblanc and Echenique2017; Tang et al., Reference Tang, Zang, Cheng, Luan, Dai, Xu, Yang, Zhao and Su2017; Bocchini et al., Reference Bocchini, Galla, Pupilli, Bellucci, Barcaccia, Ortiz, Pessino and Albertini2018; Ochogavía et al., Reference Ochogavía, Galla, Seijo, González, Bellucci, Pupilli, Barcaccia, Albertini and Pessino2018; Tang et al., Reference Tang, Xu, Deng, Cheng, Dai, Yang, Liu and Su2019; de Oliveira et al., Reference de Oliveira, Vigna, da Silva, Fávero, de Matta, Azevedo and de Souza2020). However, complete molecular regulatory mechanisms are yet to be revealed and would help fertilize agricultural crops innovatively through fixing the hybrid heterotic vigour in order to propagate indefinitely with enhanced productivity (Dujardin and Hanna, Reference Dujardin and Hanna1983; Khush, Reference Khush1994; Ramulu et al., Reference Ramulu, Sharma, Naumova, Dijkhuis and van Lookeren Campagne1999; Spillane et al., Reference Spillane, Steimer and Grossniklaus2001, Reference Spillane, Curtis and Grossniklaus2004; Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013; Brukhin, Reference Brukhin2017).
Besides, in an inter-specific cross combination, when the ploidy level of the maternal plant (e.g. diploid) is less than that of the paternal plant (e.g. tetraploid); the genome proportion of the paternal plant in the resultant endosperm tissues is greater than ‘1n’, and as per the example, it is ‘2n’ (Quarin, Reference Quarin1999; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015; Batista et al., Reference Batista, Figueiredo, Santos-González and Köhler2019). Evolutionarily, the occurrence of the character – endosperm development in angiosperms – resembles poly-embryonic events (more than one fertilization event leading to embryo formation) in gymnosperms, and later evolved to form endosperm in angiosperms. The genome dosage level variability in the endosperm, from 2n to 15n, might be termed ‘radicals’ or ‘variables’ for the said character, as defined by Vavilov (Reference Vavilov1922).
Polyendospermy and chlorophyllous endosperm
Besides endosperm ploidy level variability (2n to 15n), Arceuthobium americanum Nutt. ex Engelm. has been reported to exhibit polyendospermy – formation of multiple ‘3n’ endosperms (Friedman and Sumner, Reference Friedman and Sumner2009). Non-endospermic species have also been reported, for example in Podostemaceae due to the lack of a central cell (Battaglia, Reference Battaglia1971; Baroux et al., Reference Baroux, Spillane and Grossniklaus2002) and in Trapaceae and Orchidaceae, due to either suppression of primary endosperm nucleus’ cell division or disintegration of nuclei after few cell divisions (Johri et al., Reference Johri, Ambegaokar and Srivastava1992). Very few taxa have been reported to exhibit chlorophyllous endosperm; Amaryllidaceae members are the notable ones (Meerow and Snijman, Reference Meerow and Snijman2001). However, taxa from more than 70 families exhibit chlorophyllous embryos (Johri et al., Reference Johri, Ambegaokar and Srivastava1992) during seed development. However, at maturity, the chlorophyllous nature disappears. The nature or type of photosynthesis, the quantity of carbon being fixed and its relative contribution with respect to the carbon translocated from leaves are open for further scientific research to assess the importance of chlorophyllous seed tissues. Irrespective of the statistical significance of the quantum of carbon fixed through endosperm or embryo, in comparison with translocated carbon, it will provide a greater understanding from a biological and evolutionary viewpoint, if the nature of photosynthesis is different from leaves. Answers from such studies will form a niche for further research questions, especially when the nature of photosynthesis in endosperm and/or embryo is different from leaves, as reported in wheat grain pericarp (Rangan et al., Reference Rangan, Furtado and Henry2016). C4 photosynthesis in wheat grain pericarp is yet an unsettled issue with arguments for and against (Henry et al., Reference Henry, Rangan, Furtado, Busch and Farquhar2017). Species exhibiting variability in photosynthesis type, between leaves and endosperm/embryo parts of a plant might be of much help to throw light on this issue. The importance of non-foliar photosynthesis for yield improvement in crop plants is an untapped potential and is a current prioritized issue globally, on implementing C4 photosynthesis in rice for improved productivity (Normile, Reference Normile2008; Ermakova et al., Reference Ermakova, Danila, Furbank and Von Caemmerer2020; Simkin et al., Reference Simkin, Faralli, Ramamoorthy and Lawson2020). Carbon fixation in non-foliar (reproductive or sink) tissues of crop plants might, potentially, help accelerate grain filling to keep pace with the increase in cell size due to ploidy effects in endosperm tissues and its variability with respect to the cell position in the seed as detailed in the following section.
Endosperm ploidy variability within a seed
The variability in ploidy level of endosperm across species or hybrids derived between parents at different ploidy levels is prominent in crossable species wherein fertile seeds are produced, involving parents differing in their ploidy (Tomaszewska and Kosina, Reference Tomaszewska and Kosina2018). Remarkably, the variability of endosperm ploidy within a seed was reported well before the DNA-double helix structure (Duncan and Ross, Reference Duncan and Ross1950; Swift, Reference Swift1950). First, report to point the variability in the size of cells and nuclei of the young endosperm cells present in the central portion of the endosperm, was made as early as 1931 in maize (Lampe, Reference Lampe1931). However, it was only in 1950 that increased cell and nuclei size were associated with the increased DNA content (Swift, Reference Swift1950) or ploidy levels for those specific cells of enlarged size (Duncan and Ross, Reference Duncan and Ross1950) through quantified DNA content, and identified to be due to the endomitotic process. Genes (like Rhl1) associated with cell size enlargement and higher ploidy were reported much later (Sugimoto-Shirasu et al., Reference Sugimoto-Shirasu, Roberts, Stacey, McCann, Maxwell and Roberts2005). An upsurge of around 64 or 125 and even up to 1000 times the volume of nuclei from the central portion of endosperm, when compared to its peripheral nuclei, was documented in maize (Duncan and Ross, Reference Duncan and Ross1950); the ploidy correlation with DNA content has been reported recently (Santeramo et al., Reference Santeramo, Howell, Ji, Yu, Liu and Kelliher2020). The ploidy level variation of endosperm nucleus within a maize seed, when compared in the aleurone and central regions of the endosperm, was reported to be up to ‘6n’ and ‘24n’, respectively (Swift, Reference Swift1950). Also, the variability in endosperm ploidy among the centrally located endosperm cells was as high as ‘690n’ in maize (Kowles and Phillips, Reference Kowles and Phillips1985). In addition to central region cells of the endosperm, suspensor cells too were reported to undergo endoreduplication events (Lee et al., Reference Lee, Davidson and Duronio2009).
Apparently, the cells present in the central region of the endosperm undergo cell cycle only between S and G phases (endoreduplication) interrupted by gap periods and a doubling time of 24 h (Kowles et al., Reference Kowles, Srienc and Phillips1990, Reference Kowles, Yerk, Srienc, Phillips and Setlow1992b; Schweizer et al., Reference Schweizer, Yerk-Davis, Phillips, Srienc and Jones1995). The Cyclin A gene (CYCA2;3) is one of the key genes regulating the endoreduplication event (Imai et al., Reference Imai, Ohashi, Tsuge, Yoshizumi, Matsui, Oka and Aoyama2006), and the importance of this functionality has been viewed from a broader perspective on overall physiology and development (De Veylder et al., Reference De Veylder, Larkin and Schnittger2011). Besides increased mRNA and protein formation through endoreduplication, it has also been suggested that storing nucleotides by this mechanism might anticipate its use during embryogenesis and germination (Lee et al., Reference Lee, Davidson and Duronio2009). It has been estimated that roughly 3% of the total endosperm cells (positioned in the central region) in maize seed exhibit the variability in ploidy from the normal triploid endosperm cells, and a few cells (approximately 1%) contained around 90 chromosomes (Lin, Reference Lin1977). Ploidy variability, like in maize, has been reported in oats, as well (Tomaszewska and Kosina, Reference Tomaszewska and Kosina2018). Later, endoreduplication in plants was found in cotyledons, roots, cell suspensions, anthers, developing fruits (tomato) and leaves, as well (Joubès and Chevalier, Reference Joubès and Chevalier2000).
Defective kernel (Dek) mutants in maize exhibit reduced mitotic activity and, in turn, the endoreduplication, especially in the central regions of endosperm cell, was found to be controlled by a recessive gene (Kowles et al., Reference Kowles, McMullen, Yerk, Phillips, Kraemer and Srienc1992a) and also lacking an aleurone layer (Becraft and Yi, Reference Becraft and Yi2010). Water deficit (Artlip et al., Reference Artlip, Madison and Setter1995), abscisic acid (Mambelli and Setter, Reference Mambelli and Setter1998), high temperature (Engelen-Eigles et al., Reference Engelen-Eigles, Jones and Phillips2000), parental dosage effect (Kowles et al., Reference Kowles, Yerk, Haas and Phillips1997; Leblanc et al., Reference Leblanc, Pointe and Hernandez2002; Tomaszewska and Kosina, Reference Tomaszewska and Kosina2018) and post-translational modification (Zhao and Grafi, Reference Zhao and Grafi2000) may alter endoreduplication through regulating mitotic cycles in the endosperm, thereby suggesting multiple checkpoints, besides some recessive genes. Key molecular mechanisms involved in endoreduplication and increased ploidy level within the seed were identified as primarily due to the loss of activity of M-phase cyclin-dependent kinase and alterations in S-phase cyclin-dependent kinase (Larkins et al., Reference Larkins, Dilkes, Dante, Coelho, Woo and Liu2001). Post-translational modifications such as hypophosphorylation on high mobility group I/Y protein by CDC2 kinase have been suggested to be associated with endoreduplication events in maize endosperm by alleviating the transcriptional repression by Histone H1 (Zhao and Grafi, Reference Zhao and Grafi2000). Cyclin-dependent kinase inhibitors KRP1 and KRP2 inhibit the cell cycle, leading to the onset of endoreduplication events in the maize endosperm (Coelho et al., Reference Coelho, Dante, Sabelli, Sun, Dilkes, Gordon-Kamm and Larkins2005).
Tolerance to altered genome dosage levels is evident in angiosperms, albeit in a few species only, and in different cells of a tissue, depending on the position (Kowles and Phillips, Reference Kowles and Phillips1985; Joubès and Chevalier, Reference Joubès and Chevalier2000; Friedman et al., Reference Friedman, Madrid and Williams2008). Understanding the linkage between the genetic nature of embryosac variability and endosperm ploidy might help shed light on the genetic regulatory mechanism underlying the ploidy syndrome. The following section associates endosperm ploidy with embryosac variability, using the common genetic factor. However, how far they could be influenced by environmental factors on ploidy variability with respect to the different cell positions within a tissue is yet to be revealed.
Are endosperm ploidy and embryosac variability genetically linked?
Cells destined for endosperm formation are determined during embryosac formation. Later, with the onset of fertilization, the determined central cell, upon second fertilization, gets differentiated into endosperm tissue. To understand the variability in endosperm ploidy level, primarily, the megaspore mother cell (MMC) formation process – its root, needs to be understood. Identification of the diversity in embryosac development might shed some light on the endosperm variability. Normal meiosis of MMC leads to monosporic functional megaspores. The absence of cytokinesis during the second meiotic division or both meiotic divisions yields bi- (two haploid nuclei) or tetra-sporic (four haploid nuclei) functional megaspores. Evolutionarily, bi- and tetra-sporic trait types are derived from the monosporic trait (Arias and Williams, Reference Arias and Williams2008). The single functional megaspore undergoes three mitotic division to yield an eight-nucleate (seven cellulars) mature embryosac (ES), classified as Polygonum type – the most common one across angiosperms (Friedman, Reference Friedman1998). However, mega-gametogenesis devoid of the second or third mitosis, in combination with mono-, bi- or tetra-sporic megaspore, potentially generates 4 or 16 nucleate ES instead of the most frequently found eight-nucleate ES. Structured arrangement of 4 or 8 or 16 nuclei within ES in different species exhibits diverse forms of ES. Structurally (and the number of nuclei as well) differing ES were named upon the taxa from which they were described and reported at first. Documented ES types are Polygonum- (most common), Oenothera-, Allium-, Peperomia-, Penaea-, Drusa-, Fritillaria-, Plumbagella-, Plumbago-, Adoxa-, Butomopsis-, Acalypha indica-, Peperomia hispidula-, Apinagia- and Dicraea types (Maheshwari, Reference Maheshwari1950; Battaglia, Reference Battaglia1971; Friedman et al., Reference Friedman, Madrid and Williams2008). Due to the variability in the number of nuclei of the central cell of ES, accordingly the endosperm ploidy level gets modulated from the normal ‘3n’ endosperm during the second fertilization event.
To obtain an overall understanding on such variable ES for number of nuclei, and, in turn, endosperm genome dosage levels, ES classified on genetic basis can be grouped broadly into seven types (Friedman et al., Reference Friedman, Madrid and Williams2008), namely monosporic 2n (1m:1p), monosporic 3n (2m:1p), bi-sporic 3n (2m:1p), tetra-sporic 3n (2m:1p), tetra-sporic 5n (4m:1p), tetra-sporic 9n (8m:1p) and tetra-sporic 15n (14m:1p). Taxonomically, the order Piperales alone accommodates six of these seven genetic types (Arias and Williams, Reference Arias and Williams2008) excluding the monosporic 2n type, suggesting the possibility for better understanding of the genetic variability of embryosac or endosperm and its development. The law of homologous variation, described by Vavilov (Reference Vavilov1922), might help foresee the endosperm variability in allied taxa as well for the genera reported with variable endosperm ploidy levels. Most of the understanding of the process of organogenesis of MMC and flower or seed development has been reported only in few plant models like Arabidopsis, Medicago, Zea and Nicotiana (Table 1). With the present understanding from these model plants, the genetic mechanism underlying endosperm or ES variability may be revealed.
Table 1. Potential candidate genes involved in regulating plant reproductive development (based on reports of genes/metabolites associated with cell cycle)

Tolerance of altered genome dosage
Several reports have highlighted the importance of balanced genome dosage levels, positional cues, PCD and cell-cycle modifications primarily involved in normal endosperm development and, in turn, seed development and viability (Wangenheim, Reference Wangenheim1957; Becraft, Reference Becraft2001; Satyaki and Gehring, Reference Satyaki and Gehring2019). Whenever there is a deviation from the 2m:1p genome dosage ratio during endosperm formation, either premature cellularization (increased maternal dosage) or lack of cellularization (increased paternal dosage), seed abortion is the most probable result (Scott et al., Reference Scott, Spielman, Bailey and Dickinson1998). However, the evolution (on parsimony basis) of 2m:1p from 1m:1p further from 1m:0p (Cailleau et al., Reference Cailleau, Cheptou and Lenormand2010), and the existence of species accommodating the altered genome dosage, indicates nature's tolerance of genome dosage levels deviating from 2m:1p (Satyaki and Gehring, Reference Satyaki and Gehring2019). Likely, this tolerance mechanism is responsible for the 30% of angiosperms exhibiting altered genome dosage levels, from 1m:1p up to 14m:1p (Raghavan, Reference Raghavan2006; Friedman et al., Reference Friedman, Madrid and Williams2008). It is essential to underpin the genetic regulatory mechanisms of the tolerance of the deviation from the most common 2m:1p, especially so because understanding the genetic regulation and genes involved in endosperm development have been reported mostly in the model plant Arabidopsis (Table 1). However, in well-studied cereal crops like rice, wheat and maize, the initial phase of endosperm development (cell determination phase) is largely unknown (Li and Song, Reference Li and Song2020; Olsen, Reference Olsen2020). Information gained from model plant species may help understand the endosperm development and the genome dosage balance in the species deviating from 2m:1p ratio. Also, methylation patterns involved in parental imprinting to functionalize parent-of-origin effects, when neutralized in both the parents, the impact of the nature of imprinting no longer hinders or alters normal endosperm development (Adams et al., Reference Adams, Vinkenoog, Spielman, Dickinson and Scott2000). Hence, this potentially gives clues on the possible tolerance for the nature's acceptance of deviation from the 2m:1p dosage level.
Roadmap for understanding the endosperm genome variability
Gametophytic mutations (from completion of meiosis until fertilization and maternal control of seed development) affecting mega-gametogenesis follows non-Mendelian segregation (Yadegari and Drews, Reference Yadegari and Drews2004). However, sporophytic mutations affecting mega-gametogenesis (from MMC till completion of meiosis) are yet to be studied. Hence, to sufficiently address this, two approaches may be followed. Firstly, working with the existing knowledge on the reproductive ontogeny in model flowering plants, potential candidate genes could be identified, and further, its validation in the taxa of interest, either through forward or reverse genetics approaches (Jankowicz-Cieslak and Till, Reference Jankowicz-Cieslak, Till, Al-Khayri, Jain and Johnson2015). Alternatively, differential gene expression (RNA-seq) between the two nearest taxa variable for endosperm and embryosac formation may be used to identify and validate the genes or genetic mechanism underlying variability. Irrespective of the approaches taken to identify the candidate genes, validating the identified genes is an important part of the studies for understanding the genome dosage variability. A schematic overview of the roadmap towards understanding the genetic mechanism underlying endosperm or embryosac variability is provided in Fig. 1 and detailed in the following section.
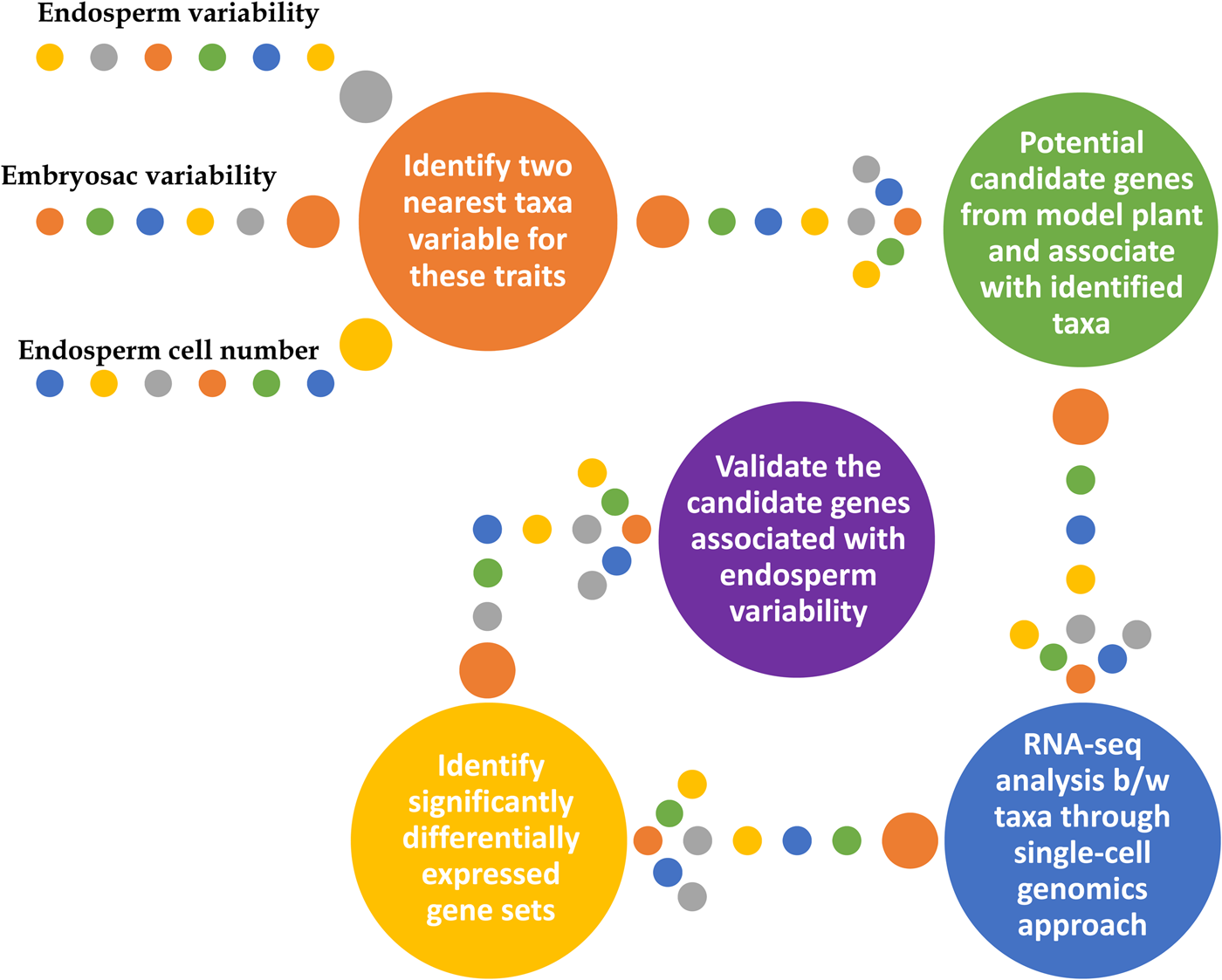
Fig. 1. An overview of the genetic mechanism regulating endosperm and embryosac variability.
Regulators of the cell cycle as potential candidates
With the brief highlights of the genetic or genomic variation in endosperm and embryosac formation, as explained in previous sections, first a framework with identified potential target taxonomical groups for better understanding of the underlying genetic mechanism needs to be made. Secondly, potential candidate genes regulating the cell cycle (mitotic and meiotic) and cytokinetic checkpoints across species are to be identified and charted out (den Boer and Murray, Reference den Boer and Murray2000; Sablowski and Carnier Dornelas, Reference Sablowski and Carnier Dornelas2014; Keçeli et al., Reference Keçeli, De Storme, Geelen and Schmidt2017; Barrada et al., Reference Barrada, Djendli, Desnos, Mercier, Robaglia, Montané and Menand2019; Daigle et al., Reference Daigle, Mazin and Matton2019; Eekhout and De Veylder, Reference Eekhout and De Veylder2019; Lora et al., Reference Lora, Yang and Tucker2019; Lorenzo-Orts et al., Reference Lorenzo-Orts, Witthoeft, Deforges, Martinez, Loubéry, Placzek, Poirier, Hothorn, Jaillais and Hothorn2019; Min et al., Reference Min, Frost and Choi2019; Pinto et al., Reference Pinto, Mendes, Coimbra and Tucker2019; Zühl et al., Reference Zühl, Volkert, Ibberson and Schmidt2019). For easy reference and as a starting point, a list of potential candidate genes (including miRNA and other non-coding RNAs) regulating reproductive development in angiosperms is provided in Table 1. A brief comparison between the identified taxa with its nearest taxa with normal (3n) endosperm, at structural and functional levels for those genes might throw some light on the existing variability and its underlying genetic mechanism.
Epigenetic regulation of genomic imprinting
In addition to genetic mechanisms, the importance of epigenetic mechanisms, especially during ontogeny of endosperm and seed development, is well documented with a role for non-coding small RNAs (Gehring and Satyaki, Reference Gehring and Satyaki2017; Yu et al., Reference Yu, Xu, Wei, Zhang, Chen and Pu2020). Methylation patterns of DNA and various modifications (acetylation and methylation) of histone protein (Song and Chen, Reference Song and Chen2015), in addition to small RNAs (Ng et al., Reference Ng, Lu and Chen2012; Yakovlev et al., Reference Yakovlev, Viejo, Fossdal, Miguel, Dalmay and Chaves2020; Yu et al., Reference Yu, Xu, Wei, Zhang, Chen and Pu2020), are the epigenetic regulators playing key roles in developmental processes. The coordinated act of the Polycomb group (PcG) of proteins and the DNA methylation essentially regulates the epigenetic systems for endosperm development, including the central cell stage of mega-gametogenesis. Gene products of MEDEA (Mea) FERTILIZATION INDEPENDENT SEED DEVELOPMENT (FIS2) and FIS3 (a.k.a: FIE, FERTILIZATION INDEPENDENT ENDOSPERM) classified as PcG type are well-known regulators of the embryo and endosperm development (Köhler et al., Reference Köhler, Hennig, Spillane, Pien, Gruissem and Grossniklaus2003). PcG proteins are well-known transcriptional regulators through histone modifications mediated gene silencing mechanisms, with its homologues present across the animal and plant kingdoms in establishing cell identity and memory (Hsieh et al., Reference Hsieh, Hakim, Ohad and Fischer2003; Di Croce and Helin, Reference Di Croce and Helin2013). The activity of most of the imprinted genes of the endosperm are regulated through epigenetic mechanisms and its complex nature poses difficulties in association with the corresponding phenotype (Pignatta et al., Reference Pignatta, Novitzky, Satyaki and Gehring2018). Besides, the complexities involved in understanding the embryo or endosperm development per se, crosstalk between embryo and endosperm also plays a crucial role in reproductive isolation and seed development. During wide hybridization, such crosstalk induced barriers for genetic exchange lead to hybrid incompatibilities (Fishman and Sweigart, Reference Fishman and Sweigart2018; Roth et al., Reference Roth, Florez-Rueda and Städler2019). Importantly, crosstalk between embryo and endosperm in mature seed during germination is also crucial for successful germination through the supply of seed reserves for the germination processes (Yan et al., Reference Yan, Duermeyer, Leoveanu and Nambara2014; Doll et al., Reference Doll, Royek, Fujita, Okuda, Chamot, Stintzi, Widiez, Hothorn, Schaller and Geldner2020).
Although methylation patterns generally occur in heterochromatic (gene-poor) regions, in transposable element regions, the genic regions appear more methylated than the flanking regions. However, when compared between embryo and endosperm for genome-wide methylation patterns, it was reported that the DEMETER (DME) gene product demethylates the genes of transposable elements in the central cell nuclei of maternal origin before fertilization (Gehring et al., Reference Gehring, Bubb and Henikoff2009). This highlights the role of epigenetic regulation in enforcing the parent-of-origin effect on the endosperm development. In maize endosperm, through genome-wide methylation pattern analysis, it has been reported that there is a reduction of 13–34% methylation when compared to embryo or leaf tissues (Lauria et al., Reference Lauria, Rupe, Guo, Kranz, Pirona, Viotti and Lund2004; Wang et al., Reference Wang, Xia, Zhang, Zhao, Zhao, Hou, Li, Li, Ma and Wang2015). DNA glycosylases (DME) and RNA-directed DNA methylation (RdDM) through siRNA pathways are the two major epigenetic mechanisms regulating plant development with the former for imprinting female gametogenesis and the latter for vegetative tissues (Law and Jacobsen, Reference Law and Jacobsen2010). The importance of Histone H1.2 in regulating DME for its downstream epigenetic regulation to impart an imprinting effect has been identified (Rea et al., Reference Rea, Zheng, Chen, Braud, Bhangu, Rognan and Xiao2012). Also, histones modified by PcG proteins are well-documented regulators at the transcriptional level involved in the normal embryo and endosperm development (Schubert et al., Reference Schubert, Clarenz and Goodrich2005; Moreno-Romero et al., Reference Moreno-Romero, Del Toro-De León, Yadav, Santos-González and Köhler2019). Possibly, this corroborates the reports on endoreduplication in endosperm cells of the central region to have a reduced H1/DNA ratio and thereby enhancing the transcription and translation towards grain filling in endosperm cells (Zhao and Grafi, Reference Zhao and Grafi2000; Larkins et al., Reference Larkins, Dilkes, Dante, Coelho, Woo and Liu2001). This highlights the importance of epigenetic regulation through silencing as well as expression (through negative repression) for normal endosperm and embryo development.
There are very few reports that underscore the association between epigenetic regulation and ploidy level at the species level but none to the author's knowledge with reference to endosperm or seed development. Primarily, a non-linear relationship between the DNA methylation pattern and ploidy level was documented (Li et al., Reference Li, Hu, Xue, Chen, Wang, Song, Chen and Wang2011), when compared between species with different ploidy (di-, tri- and tetra-). Altered ploidy might potentially alter the epigenetic silencing mechanism and thereby altering the normal developmental processes (Scheid et al., Reference Scheid, Jakovleva, Afsar, Maluszynska and Paszkowski1996).
Role of genomics on understanding the endosperm variability
The candidate gene approach is the most traditional, long-standing and widely used methodologies in identifying genes associated with the trait of interest, particularly when the trait is of a quantitative or complex nature (Pflieger et al., Reference Pflieger, Lefebvre and Causse2001; Tabor et al., Reference Tabor, Risch and Myers2002; Zhu and Zhao, Reference Zhu and Zhao2007). However, preliminary knowledge of the trait is required for the identification of the genes underlying it. Also, contrasting genotypes for the trait of interest may be identified at first for generating a QTL map (Wayne and McIntyre, Reference Wayne and McIntyre2002; Zhu and Zhao, Reference Zhu and Zhao2007). With the availability of computational facilities and robust sequencing technologies, the importance of genomics for the identification of genes and pathways is undisputed. Notably, genome-wide scanning approaches and digital candidate gene identification approaches, such as QTL, LD mapping, GWAS, GBS, and functional annotation, either alone or in combination with traditional candidate gene approaches are now frequently used (Mackay and Powell, Reference Mackay and Powell2007; Götz et al., Reference Götz, García-Gómez, Terol, Williams, Nagaraj, Nueda, Robles, Talón, Dopazo and Conesa2008; McCarthy et al., Reference McCarthy, Abecasis, Cardon, Goldstein, Little, Ioannidis and Hirschhorn2008; Chen et al., Reference Chen, Bardes, Aronow and Jegga2009; Zhang et al., Reference Zhang, Ersoz, Lai, Todhunter, Tiwari, Gore, Bradbury, Yu, Arnett and Ordovas2010, Reference Zhang, Tamba, Wen, Li, Ren, Ni, Gao and Zhang2020; Bush and Moore, Reference Bush and Moore2012; Glaubitz et al., Reference Glaubitz, Casstevens, Lu, Harriman, Elshire, Sun and Buckler2014; Torkamaneh et al., Reference Torkamaneh, Boyle, St-Cyr, Légaré, Pomerleau and Belzile2020). The following three subsections provide an overview of the strength of genomics, with special reference towards understanding endosperm variability (both across species and within the seed).
Differential gene expression studies
Significant progress has been made on the genetic regulation of endosperm formation that enhances our understanding of endosperm ontogeny (Table 1). Most of the studies involved the model plant Arabidopsis thaliana to understand the basic genetic mechanism underlying endosperm ontogeny (Table 1). However, genetic regulation underlying the genome-scale variability in endosperm ontogeny between closely related taxa (within Piperales) are yet to be uncovered. Differential gene expression (RNA-seq) in combination with single-cell genomics (scRNA-seq) might be of huge potential to address this. Additionally, for apomixis developmental processes, candidate genes like SERK and APOSTART have been identified (Albertini et al., Reference Albertini, Marconi, Reale, Barcaccia, Porceddu, Ferranti and Falcinelli2005; Podio et al., Reference Podio, Felitti, Siena, Delgado, Mancini, Seijo, González, Pessino and Ortiz2014b). Since apomixis is reported to be tightly associated with the polyploidy mechanism (Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013), species within a genus (or variable cytotypes within a species) exhibiting variable ploidy level with apomictic behaviour are ideal models to identify the molecular mechanisms underlying the apomictic phenomena (Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015; Ochogavía et al., Reference Ochogavía, Galla, Seijo, González, Bellucci, Pupilli, Barcaccia, Albertini and Pessino2018).
Comparison between nearest taxa variables for apomictic behaviour helped identify a set of genes including AGO9 (Olmedo-Monfil et al., Reference Olmedo-Monfil, Durán-Figueroa, Arteaga-Vázquez, Demesa-Arévalo, Autran, Grimanelli, Slotkin, Martienssen and Vielle-Calzada2010), CASEIN KINASE (Depetris et al., Reference Depetris, Acuña, Pozzi, Quarin and Felitti2018), LORELEI (Felitti et al., Reference Felitti, Seijo, González, Podio, Laspina, Siena, Ortiz and Pessino2011), a MAP3K coding gene QUI-GON-JINN (Mancini et al., Reference Mancini, Permingeat, Colono, Siena, Pupilli, Azzaro, de Alencar Dusi, de Campos Carneiro, Podio and Seijo2018), THAUMATIN-LIKE, COPIA, CCD, LEISHMANOLYSIN-LIKE PEPTIDASE and FAR1-RELATED (Ortiz et al., Reference Ortiz, Revale, Siena, Podio, Delgado, Stein, Leblanc and Pessino2017), ORC3 (Siena et al., Reference Siena, Ortiz, Calderini, Paolocci, Cáceres, Kaushal, Grisan, Pessino and Pupilli2016), DORN1 and eATP pathway (Choi et al., Reference Choi, Tanaka, Cao, Qi, Qiu, Liang, Lee and Stacey2014; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015), AMP SYNTHASE, EF-1α, COP9 SIGNALOSOME and ACETOLACTATE SYNTHASE (Cervigni et al., Reference Cervigni, Paniego, Pessino, Selva, Díaz, Spangenberg and Echenique2008), long non-coding RNAs like N13 (Ochogavía et al., Reference Ochogavía, Galla, Seijo, González, Bellucci, Pupilli, Barcaccia, Albertini and Pessino2018) and QGJ (Mancini et al., Reference Mancini, Permingeat, Colono, Siena, Pupilli, Azzaro, de Alencar Dusi, de Campos Carneiro, Podio and Seijo2018), and small RNAs like ATHILA, LINE and ATLANTYS (Olmedo-Monfil et al., Reference Olmedo-Monfil, Durán-Figueroa, Arteaga-Vázquez, Demesa-Arévalo, Autran, Grimanelli, Slotkin, Martienssen and Vielle-Calzada2010), involved in the regulatory processes for apomictic expression. In addition, some recent reports have identified apomixis-related genes in Paspalum (de Oliveira et al., Reference de Oliveira, Vigna, da Silva, Fávero, de Matta, Azevedo and de Souza2020) and Boehmeria (Tang et al., Reference Tang, Xu, Deng, Cheng, Dai, Yang, Liu and Su2019), which provide insight in genetic control of apomeiosis and polyploidy events leading to apomixis behaviour (Savidan, Reference Savidan2000; Hand and Koltunow, Reference Hand and Koltunow2014). Also, regulatory mechanisms modulated at the transcriptional, translational and post-translational levels dissecting the apomictic trait, including epigenetic regulation, signal transduction and evolutionary aspects have been covered in detail (Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013; Brukhin, Reference Brukhin2017; Schmidt, Reference Schmidt2020). Notably, most of the genetic or molecular mechanisms associated with apomictic traits have been reported for the genus Paspalum (Quarin, Reference Quarin1999; Labombarda et al., Reference Labombarda, Busti, Caceres, Pupilli and Arcioni2002; Laspina et al., Reference Laspina, Vega, Seijo, González, Martelotto, Stein, Podio, Ortiz, Echenique and Quarin2008; Polegri et al., Reference Polegri, Calderini, Arcioni and Pupilli2010; Ortiz et al., Reference Ortiz, Quarin, Pessino, Acuña, Martínez, Espinoza, Hojsgaard, Sartor, Cáceres and Pupilli2013, Reference Ortiz, Revale, Siena, Podio, Delgado, Stein, Leblanc and Pessino2017; Podio et al., Reference Podio, Cáceres, Samoluk, Seijo, Pessino, Ortiz and Pupilli2014a; Felitti et al., Reference Felitti, Acuña, Ortiz and Quarin2015; Siena et al., Reference Siena, Ortiz, Calderini, Paolocci, Cáceres, Kaushal, Grisan, Pessino and Pupilli2016; Bocchini et al., Reference Bocchini, Galla, Pupilli, Bellucci, Barcaccia, Ortiz, Pessino and Albertini2018; Depetris et al., Reference Depetris, Acuña, Pozzi, Quarin and Felitti2018; Mancini et al., Reference Mancini, Permingeat, Colono, Siena, Pupilli, Azzaro, de Alencar Dusi, de Campos Carneiro, Podio and Seijo2018; Ochogavía et al., Reference Ochogavía, Galla, Seijo, González, Bellucci, Pupilli, Barcaccia, Albertini and Pessino2018; de Oliveira et al., Reference de Oliveira, Vigna, da Silva, Fávero, de Matta, Azevedo and de Souza2020). These details will be of significant help in mining the genes associated with apomictic behaviour in other species as well, based on homology, using sequences obtainable through next-generation sequencing (NGS) tools, followed by further functional validation.
Differential gene expression in combination with digital candidate gene identification approaches using functional annotation tools is a potential method to identify candidate genes underlying endosperm variability and apomixis. This is easily doable in qualitative traits and is feasible in quantitative traits when the quantum of expression results in a differential expression pattern associated with the contrasting phenotypic traits (Rangan et al., Reference Rangan, Furtado and Henry2020a,Reference Rangan, Furtado, Henry, Gaikwad, Smithers and Knoerzerb). On these advantages, differential gene expression or RNA-seq with functional annotation is a robust upcoming tool for candidate gene identification. While this approach could be used directly for endosperm ploidy variability across species, its utility for endosperm ploidy variability within a seed, and apomictic developmental processes will require additional single-cell sequencing methods (Wagner et al., Reference Wagner, Regev and Yosef2016; Tanay and Regev, Reference Tanay and Regev2017).
Single-cell sequencing for ploidy and epigenetic identification
Single-cell sequencing to understand ploidy level (aneuploidy) and its effect on development are well documented in animal and cell models, and upcoming in plant systems. The techniques in the whole genome, epigenetic and transcriptome studies have been applied to identify differences underlying cancer cells and regular ones (Ferrarini et al., Reference Ferrarini, Forcato, Buson, Tononi, del Monaco, Terracciano, Bolognesi, Fontana, Medoro, Neves, Möhlendick, Rihawi, Ardizzoni, Sumanasuriya, Flohr, Lambros, de Bono, Stoecklein and Manaresi2018). Gene regulation and genome dosage imbalance due to aneuploidy in humans for trisomy dependent regulation, studied using RNA from skin fibroblast cells through single-cell RNA-seq (scRNA-seq) approaches have been reported.
Utilization of single-cell genomics in generating a cell-type atlas, cell-type specificity, resolving molecular relations and functional genomics, and cell differentiation for tissue specificity underlying biological or analytical problems are well documented (Efroni and Birnbaum, Reference Efroni and Birnbaum2016; Tanay and Regev, Reference Tanay and Regev2017; Ryu et al., Reference Ryu, Huang, Kang and Schiefelbein2019; Rich-Griffin et al., Reference Rich-Griffin, Stechemesser, Finch, Lucas, Ott and Schäfer2020). Such studies might potentially be of help as starting material to plan for combining RNA-seq with single-cell genomics – scRNA-seq (Ryu et al., Reference Ryu, Huang, Kang and Schiefelbein2019), to study ploidy variability of individual cells within a seed or endosperm tissues.
For this, NGS and single-cell sequencing methodologies (Nawy, Reference Nawy2014), with the current understanding of the endosperm ontogeny in model plants (Table 1) might be of great help in understanding genome-scale variability of the endosperm. This may provide deeper insights in sporogenesis, gametogenesis, endosperm ontogeny and its variability across species from evolutionary, breeding and crop improvement perspectives. Gene regulatory network relationships and molecular interaction could very well be revealed using the scRNA-seq approaches. The peculiarities of seed resource allocation between maternal and paternal parents are not well-founded on hypotheses explaining the variability (Cailleau et al., Reference Cailleau, Cheptou and Lenormand2010). Such studies will help underscore the genetic mechanisms underlying the biology of dosage compensation, genome imprinting and maternal/paternal effect with special reference to seed development.
Single-cell genomics in combination with RNA-seq and functional annotation approaches might be a robust tool to study and elucidate the underlying genetic events affecting the sporophytic and gametophytic mega-gametogenesis and, in turn, fertilization and seed developmental processes. This will also help in identifying the key candidate genes or molecular events responsible for the variability in the genome/nuclear content of ES from 1n to 14n. Additionally, this may also provide a better understanding of the genetic mechanism that favours seed resource allocation that begins with the occurrence of meiosis and completes before fertilization in cycads and ginkgoes whereas the same begins only after fertilization in angiosperms, with conifers in between (Cailleau et al., Reference Cailleau, Cheptou and Lenormand2010). It will also be of use in endoreduplication events occurring in the central region of endosperm tissue leading to higher ploidy in those specific cell types as compared to the ones at the periphery (Duncan and Ross, Reference Duncan and Ross1950; Swift, Reference Swift1950; Kowles and Phillips, Reference Kowles and Phillips1985; Larkins et al., Reference Larkins, Dilkes, Dante, Coelho, Woo and Liu2001).
Merits and challenges of single-cell RNA-sequencing
Key merit is the availability of techniques and tools reported in model organisms or cells or tissue types, which can be applied with minor modifications to plants (Efroni and Birnbaum, Reference Efroni and Birnbaum2016; Rich-Griffin et al., Reference Rich-Griffin, Stechemesser, Finch, Lucas, Ott and Schäfer2020). However, the presence of cell walls and various secondary metabolites in plants warrants for protocol standardization. Great advantage and strength are expected for studying cell-type or single-cell systems in plants, including the availability of laser-assisted microdissection tools to isolate what we actually see (Kehr, Reference Kehr2003; Nelson et al., Reference Nelson, Tausta, Gandotra and Liu2006; Brandt et al., Reference Brandt, Mascher, Thiel and Murray2018; Sakai et al., Reference Sakai, Taconnat, Borrega, Yansouni, Brunaud, Paysant-Le Roux, Delannoy, Magniette, Lepiniec and Faure2018; Florez-Rueda et al., Reference Florez-Rueda, Waser, Grossniklaus and Bayer2020). With these strategies, it is quite possible to achieve the isolation of genetic material from the cell lines or cell types of interest in plants, especially from endosperm during seed development. Once the genetic material is isolated; then, it is equal to any other system wherein common tools and techniques available for single-cell sequencing are directly applicable. Differential expression through RNA-seq might help identify the key candidate genes involved in endosperm ploidy variability.
In spite of these merits and potential possibilities of the applicability of scRNA-seq methods to understand endosperm ploidy variability, one should be cautious on the possible challenges that might be faced during the process: (1) epigenetic regulation including parent-of-origin and endoreduplication mechanisms are modulated through cell cycle regulators and isolating cell types for comparison might end up in different results; (2) sufficient quantity of homogenous cell types at the same cellular stage within the group is also an important criterion. It could potentially affect the results, and hence, uniformity of samples is imperative; (3) requirement of sufficient biological replicates of the same cellular stage and developmental level for higher confidence scores when the results are subjected to statistical analyses. Addressing these challenges might help save time and resources with proper planning towards understanding the genetic mechanism modulating endosperm variability within the seed, and across species, and their differences.
Conclusion
Understanding the overall seed development is important for seeds that are not just the fulcrum for the survival of many life forms but also are the carriers of genetic imprints across generations, which helps pass through the successful speciation events in the evolutionary timeline. Sporophytic (Mendelian segregation pattern) mutations and their molecular interactions affecting mega-gametogenesis are known to affect fertilization events and, at times, leads to ovule abortion yielding fewer seeds. Variability in endosperm ploidy within a seed (between the central region and the periphery) and at species level has been known for some 90 years. However, its underlying (epi)genetic mechanisms linking imprinting and parent-of-origin effects during its ontogeny are not completely understood. Present-day genomic tools like scRNA-seq might help gain a better picture of gametophyte development and fertilization in plants, especially of the ontogeny of embryo and endosperm, with special reference to variability within the seed and across species. Comparative studies between taxa exhibiting variability for ES structure and genome dosage level might shed light on the tolerance for altered genome dosage, imprinting and endosperm development under varied genetic environments, and its mechanism of action from an evolutionary perspective. The role of epigenetic mechanisms (both silencing and expression through negative repression) in combination with varied ploidy level (within the seed and species level) interactively regulating the embryo and endosperm development are yet to be elucidated. Additionally, genetic mechanisms underlying the other forms of variability like the disintegration of endosperm after a few divisions in certain taxa, cell identity for endoreduplication in specific cells from the central region of the endosperm, chlorophyllous endosperm and its importance in contributing to carbon fixation, apomictic endosperm dosage, composite endosperm and polyendospermy would also be revealed. This will potentially shed light on the variability in resource allocation during seed development processes and might help extend the duration of resource allocation in agricultural crop plants for enhanced productivity.
To exploit the triploid nature (3n) of endosperm tissues, endosperm culture (in vitro) is mostly used for generating triploid plants with superior traits like disease resistance, high yield, larger fruits, etc., in species where the seed is not an economic product (including propagation mode), or felt undesirable as in case of banana, grapes or berries (Thomas and Chaturvedi, Reference Thomas and Chaturvedi2008; Miyashita et al., Reference Miyashita, Ohashi, Shibata, Araki and Hoshino2009). Direct somatic embryogenesis or callus mediated in vitro propagation, as reported in many species (Trolinder and Goodin, Reference Trolinder and Goodin1987; Novak et al., Reference Novak, Afza, Van Duren, Perea-Dallos, Conger and Xiaolang1989; Li et al., Reference Li, Traore, Maximova and Guiltinan1998; Rangan et al., Reference Rangan, Venugopalan, Giridhar and Ravishankar2011), might be useful in deriving triploid plantlets with increased vigour using endosperm culture techniques (Sita et al., Reference Sita, Ram and Vaidyanathan1980; Tulecke et al., Reference Tulecke, McGranahan and Ahmadi1988; Gmitter et al., Reference Gmitter, Ling and Deng1990; Sun et al., Reference Sun, Lu, Liang, Guo, Mo and Xie2011; Antoniazzi et al., Reference Antoniazzi, de Faria, de Carvalho, Mikovski, de Carvalho, de Matos, Reis, Viccini, Pinto and Rocha2018; Van Thang et al., Reference Van Thang, Van Viet, Nam, Tung and Nhut2018). In addition, endosperm cells are targeted for improved grain quality and nutrition, like quality protein maize (Gibbon and Larkins, Reference Gibbon and Larkins2005; Vivek et al., Reference Vivek, Krivanek, Palacios-Rojas, Twumasi-Afriyie and Diallo2008) and golden rice (Beyer et al., Reference Beyer, Al-Babili, Ye, Lucca, Schaub, Welsch and Potrykus2002; Paine et al., Reference Paine, Shipton, Chaggar, Howells, Kennedy, Vernon, Wright, Hinchliffe, Adams and Silverstone2005). Generation of soft, friable and off-white to creamy yellow callus from endosperm tissues of cereal crops, in combination with processing (dried and powdered forms), would gear up agriculture towards lab farming that might lead to culture edible endosperm directly on Petri dishes. At the verge of a climate change scenario, transformation and utilization of the understanding of variability towards an entirely new form (petri cultures of endosperm) may also help feed humanity in the future, contributing towards fulfilling the dreams of the great academician N.I. Vavilov: a hunger-free world.
Acknowledgement
The author is thankful to ICAR-NBPGR, New Delhi, for institutional support.





