Introduction
In response to The European Renewable Energy Directive of 2009 (2009/28/EC) along with the Italian public policy to support biogas, the number of production plants has led to an increase of more than 200 anaerobic digestion plants since 2002, with a logarithmic increase from 2008 to 2013 reaching a total of 1300 plants. In 2015, the total capacity of these plants has been about 2 billion m3 of methane per year (Carrosio, Reference Carrosio2011; Molinari and Donati, Reference Molinari and Donati2015). This growth trend is expected to remain constant until 2030 when about 8.5 billion m3 of methane per year is expected to be produced (Consorzio Italiano Biogas, 2017). Most plants for anaerobic biogas production are associated with intensive agricultural and livestock activity in northern Italy. They represented a solution to the limits imposed by the European Nitrate Directive (91/676/CEE), and further EU updates aiming at protecting water from nitrate contamination through intensive use of fertilizers and intensive livestock farming (Monteny, Reference Monteny2001; Musacchio et al., Reference Musacchio, Re, Mas-Pla and Sacchi2020). With the aim of curbing nitrate pollution, some areas within the regions were classified as nitrate vulnerable zone either because they belonged to the watersheds of river basins or due to their inherently drainable soils which make them prone to nitrate leaching on underlying water tables (Monteny, Reference Monteny2001).
The fast increase of anaerobic digestion plants for biogas production in Northern Italy provided an opportunity for sustainable disposal of manure and drastic reduction of the great environmental risk deriving from that high nitrogen surplus (Carrosio, Reference Carrosio2011). In the meantime, digestates from anaerobic digestion biogas plants associated with agricultural activities, which currently cover 75% of the total national biogas production in Italy, were used as organic amendment and fertilizer in cropland surrounding biogas production in compliance with the limits imposed by the European Nitrate Directive. For years, silage maize (Zea mays L.), livestock manure and energy crops have been the predominant feedstock for biogas anaerobic digesters. As a consequence, maize has been widely cultivated in the Po Valley (northern Italy) until 2018. More recently, in response to the European Renewable Energy Directives and the EU Directive of 2018 (2018/2001) on the promotion of use of energy from renewable sources, Italy promoted a reduction of the use of silage maize as feedstocks for anaerobic digestion by incentivizing the use of crop residues, biomass from cover crops and food-processing waste. Owing to the widespread diffusion of anaerobic digestion plants in the Po Valley, there is significant scope for investigating the effects of digestate on maize, which is the crop predominantly cultivated in these areas.
Although digestates from anaerobic digestion have several known agricultural benefits such as soil amendment for high fertilization properties and the ability to enrich soil with carbon (Tambone et al., Reference Tambone, Scaglia, D'Imporzano, Schievano, Orzi, Salati and Adani2010; Nkoa, Reference Nkoa2014; Chiew et al., Reference Chiew, Spångberg, Baky, Hansson and Jönsson2015; Barzee et al., Reference Barzee, Edalati, El-Mashad, Wang, Scow and Zhang2019), the effect of digestates on soil suppressiveness has yet to be investigated sufficiently. Soil suppressiveness is the natural ability of soil to support optimal plant growth and health. The main basis of this soil state is the microbial component together, of course, with the organic matter content (Broadbent and Baker, Reference Broadbent and Baker1974; Mazzola, Reference Mazzola2002; De Corato, Reference De Corato2020). Soil microbiome has a primary role in controlling soil-borne pathogens through a number of mechanisms such as competition for nutrients, antibiosis and induction of host resistance, which do not permit a pathogen to become established or to persist or for the pathogen to become progressively less severe even though it survives in the soil (Cook and Baker, Reference Cook and Baker1983). The short-term in-pot experiments performed to date to investigate the effects of digestate and sewage sludge application on soil suppressiveness have not always show clear positive effects due to the variability of material of origin, doses, methodology applied and different time intervals between incorporation and soil sampling (Ghini et al., Reference Ghini, Patrício, Bettiol, de Almeida, de H and Maia2007; Franke-Whittle et al., Reference Franke-Whittle, Juárez, Insam, Schweizer, Naef, Topp, Kelderer, Rühmer, Baab, Henfrey and Manici2020). Nevertheless, little has been investigated until now on microbial changes that can increase soil suppressiveness with repeated field digestate applications.
The majority of agricultural soils in the Po Valley are characterized by low soil organic matter content (SOM <2%) and they are subjected to further SOM decline due to climate change and intensive agricultural practices . SOM and biodiversity decline are linked to crop yield decline worldwide in intensive growing regions (Oldfield et al., Reference Oldfield, Bradford and Wood2019). Crop yield decline is an overall reduction of plant vigor and resistance to abiotic stress, due to the loss of soil resilience and microbial diversity, which affect plant production in intensively cultivated areas such as the Po Valley (Manici and Caputo, Reference Manici and Caputo2010). This phenomenon is mainly due to a reduction of rooting ability in the crops caused by a reduced soil microbial activity and diversity with a consequent gradual increase of opportunistic soil-borne pathogens.
In this context, microbial benefits from the incorporation of digestate from anaerobic biogas production were assessed in digestate-amended fields located near three anaerobic biogas production plants associated with agricultural and livestock activity in the Po Valley. The final aims of this study were: (i) evaluating whether and to what extent repeat soil amended with digestates can improve soil suppressiveness and (ii) identifying soil microbial components responsible for soil suppressiveness increase under this experimental set.
Materials and methods
A 2-yr study was performed to compare three digestate-amended soils to the relevant non-amended soils. Microbial benefits (soil suppressiveness) were evaluated as plant growth response with a greenhouse plant growth assays using maize, on which quali-quantitative analysis of root fungal endophytes was performed, whereas in the rhizosphere quantitative changes of actinomycetes and Pseudomonas spp., taken as bacterial indicator of soil suppressiveness, were evaluated.
Experimental fields and soil sampling
The impact of digestate amendment was assessed using soil sampled in three sites located near plants for anaerobic production of biogas which were selected in three representative growing areas across the Po Valley (Fig. 1). They were in the following provinces: Cremona (CR), Modena (MO) and Forli (FL) located, respectively, in the central-west, central and east Po Valley (Fig. 1, Table 1). Two close experimental fields were selected in each of the three sites where soil samples were collected in the consecutive years 2017 and 2018. Each couple of experimental fields was similar for management and crop sequences (maize was the recurrent crop in the last five-teen years, with fewest periodical breaks with triticale or wheat) and both were cultivated with maize at sampling time. However, one field (Digestate) had a history of soil amendment with digestate from a near anaerobic digestion plant, the other (Control) did not (Table 1). The amount of digestate in each site had been applied by not exceeding the annual limits of nitrogen per hectare based on EU-Nitrate Directives for the Nitrate Vulnerable Zones (Monteny, Reference Monteny2001) which were identified in 2006 by the two regions where the experimental fields were located (Lombardy and Emilia Romagna) (Table 1). In the site of Cremona province (CR), located in a nitrate vulnerable zone of the Lombardy region (ERSAF, 2013), separated solid fraction of digestate at a dose estimated to apply a nitrogen rate to not exceeding the annual amount per year, accounting for 170 kg N ha−1 yr−1, was incorporated into the soil three times from fall 2006 to fall 2016 according to the regional directives (Capponi and Barbanti, Reference Capponi and Barbanti2010). In the sites of Modena (MO) and Forlì (FL) provinces, where the experimental sites were out of nitrate vulnerable zone (Emilia Romagna, Reference Emilia Romagna2013), separate digestate fraction and whole digestate fraction were applied at doses estimated to apply a nitrogen rate not exceeding the limits (maximum 340 kg N ha−1 yr−1) and according to the rules for application as imposed by the regional regulation in compliance with the nitrates directive (Capponi and Barbanti, Reference Capponi and Barbanti2010).
Fig. 1. Map of the plants for anaerobic biogas production selected for this study in northern Italy.
Table 1. Main features of sampling sites and main organic sources of the relevant anaerobic digestion plants for biogas production.in all cases previous

a Maize was the recurrent crop in all sampling fields, in all cases they were subjected to periodical (3–4 yr) rotation with sugar beet or wheat or legume crop (alfalfa?).
b Soil texture classified using the Soil Triangle Hydraulic Properties Calculator (Saxton et al., Reference Saxton, Rawls, Romberger and Papendick1986).
c Soil organic matter (%).
d,e Main and secondary waste sources respectively for anaerobic digestion in the close plant.
f Standard deviation near to mean of three replicates.
g Nitrate vulnerable zones (Monteny, Reference Monteny2001).
Soil sampling was performed in the same fields in late May 2017 and 2018 at V6–V10 (six leaves to ten leaves) maize developmental stage (Ciampitti, Reference Ciampitti2016). Five soil subsamples in the 0–25 cm topsoil after having removed the 2 cm superficial layer were collected in five points along the diagonal of each field to obtain a total of 10 kg of soil per field. Subsamples were mixed manually and partially dried for 6 days at room temperature (20–22°C). In 2017, 300 g-sample was taken for physico-chemical analysis.
Soil texture was estimated with the standard pipette method and clay (Cl), silt (Si) and sand (Sa) percentages of USDA classification were determined using the Soil Triangle Hydraulic Properties Calculator (Saxton et al., Reference Saxton, Rawls, Romberger and Papendick1986). Total organic carbon (TOC) content was quantified through dry combustion using a TOC Vario Select analyzer (Elementar, Germany), which conducts a catalytic combustion by high temperatures in an air environment (Vitti et al., Reference Vitti, Stellacci, Leogrande, Mastrangelo, Cazzato and Ventrella2016). TOC of each sample (6) was the mean of three analyzed subsamples.
Weather data
Daily values of air temperatures (°C) and precipitation (mm) for the period from 1st January to 30th May of the years 2017 and 2018 were retrieved by weather stations maintained by the Regional Agencies for Environmental Protection (https://www.arpalombardia.it/; https://www.arpae.it/). Data were recorded electronically every 15 min for temperature and every hour for precipitation, automatically validated and harmonized according to national guidelines for hydro-meteorological data (Barbero et al., Reference Barbero, Zaccagnino and Mariani S2017) and then aggregated to daily values. Such weather stations are located at distances ranging from 3.3 to 8.1 km from the sampled fields. According to Launay and Guérif (Reference Launay and Guérif2003), a 14 km-threshold can be regarded as the ‘maximum distance between fields and stations beyond which it was no longer possible to consider the weather described by the station as being sufficiently representative of that of the field’.
The climate in the three sites is warm temperate with hot summer (Cfa) according to the Köppen-Giger taxonomy (Peel et al., Reference Peel, Finlayson and McMahon2007). Average annual air temperature is about 12.8–13.4°C, ranging from 0 to 6°C in winter months, to peaks over 30°C in summer. Total annual precipitation is in the range of 690–810 mm, with two main rainy periods in spring and fall, and a minimum in July and August.
Plant growth assay
Soil samples from each of six sampled fields were arranged in 18 pots (500 g of soil per pot) to obtain three replicates of six pots each per field; then they were arranged in a greenhouse according to a randomized block design. Each pot was sown with one untreated maize seed and grown in the greenhouse at 24 ± 2°C under natural light for 30 days. Water was regularly supplied to avoid water limitation to plant growth. Being plant growth with the in-pot assay a way to estimate the weight of the soil microbial components on crop health (namely, way to compare soil suppressive ability) (Neiendam Nielsen et al., Reference Neiendam Nielsen, Winding, Svend, Hansen, Hendriksen and Kroer2002), pots were fertilized with ammonium sulfate (1 g per pot, co-responding to a rate of 120 kg N ha−1). This aimed at reducing the effect of different N supply for maize plants in digestate-amended (Digestate) and unamended (Control) soils. At the end of the 30-day period, plants were harvested, carefully separated from soil clod. Then rhizosphere soil adhering to roots was sampled after having gently shaken the roots of each replicate of six plants to remove soil which was not tightly adhering to the roots. In that way, three replicates of rhizosphere soil sample from each of the six treatments were obtained. A subsample of 25 g of soil was taken, air dried at room temperature for 12 h and stored in 50 ml sterile vials at −80°C. Finally, biomass of individual plants was then divided into below and above ground part. Six above ground parts of each replicate were grouped and dried until constant weight at 65°C to determine dry matter production; whereas six roots of each replicate were grouped and processed to analyze root colonization by fungal endophytes.
Root-colonization by fungal endophytes and pathogenicity of test
Roots of each replicate of six plants were washed under running water for 10 min, disinfected for 2 min in 1.5% sodium hypochlorite, rinsed twice with sterile water, dried under sterile air flow for 15 min; then 18 (three per plant) root explants of 0.2–0.3 mm per replicate, for a total of 54 explants per each of six treatments, were processed as described in Manici et al. (Reference Manici, Caputo, Nicoletti, Leteo and Campanelli2018) to estimate colonization frequency and relative species composition of root endophytic fungal communities using culture-based methodology and the most common taxonomy manuals for fungal species identification. Instead, Setophoma terrestris was identified using molecular tools by comparing with BLAST internal transcribed spacer (ITS) the sequences generate according to the method described by Manici and Bonora (Reference Manici and Bonora2007) to the corresponding nucleotide sequences in the GenBank of the National Centre for Biotechnology Information (NCBI), U.S. National Library of Medicine, available on the web at: https://www.ncbi.nlm.nih.gov/.
In July 2018, one isolate of S. terrestris, the root endophyte most abundant in both 2017 and 2018, was subjected to pathogenicity test on maize to estimate its impact on dry matter losses. Fifteen-day-old pure culture grown on potato sucrose agar of the S. terrestris isolate, whose ITS sequence reference number in the GenBank was MG944392, was inoculated on sterile sand–maize meal media into two 800 ml flasks containing 270 g sand, 30 g corn meal and 60 ml water. Two flasks were inoculated with three 6 mm diameter colony disks each and incubated for 28 days at 24°C. A total of 7.2 kg of topsoil taken from the experimental field of Forli was equally divided. One part was amended with the two flasks of maize meal inoculum of S. terrestris (1:4 v/v) and divided into nine pots. Similarly, the other part was amended at the same v/v rate with sterile maize-meal substrate and divided into nine pots to obtain the un-infested treatment. Pathogenicity test was arranged according to a totally randomized design with three replicates of three pots each. Plants were grown for 1 month in July in a shady greenhouse and were regularly irrigated to avoid water stress. Plants were then processed as described for previous in-pot test. Data of above ground part were expressed as dry matter (g) per plant.
Quantification of actinomycetes and Pseudomonas
Total genomic DNA was extracted from 0.25 g of rhizosphere soil (dry weight) using the PowerSoil DNA Isolation kit according to the manufacturer's instructions (MoBio Laboratories, Carlsbad, CA, USA). Quantification and quality control of DNA were performed using Infinite 200 NanoQuant (Trading AG, Switzerland) and DNA was stored at −20°C until use. Three DNA extractions per sample were performed for quantitative polymerase chain reaction (qPCR) and PCR-denaturing gradient gel electrophoresis (DGGE).
Quantification of rhizosphere bacterial DNA was performed by qPCR quantification assays using the primer pairs shown in Table 2. Arthrobacter sialophilus (DSMZ 7306) and Pseudomonas chlororaphis (DSMZ 6508) strains from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) were inserted as reference of Actinomyces and Pseudomonas, respectively. The DNA of the two aforementioned DSMZ bacterial strains was amplified as described in Saccà et al. (Reference Saccà, Manici, Caputo and Frisullo2019). Resulting amplicons were purified using the PureLink Quick PCR Purification kit (Invitrogen) and quantified by Infinite 200 NanoQuant (Trading AG, Switzerland). The gene copy number calculation was obtained using the formula: gene copy/μl = DNA [ng/μl] × 6.02 × 1023/base pairs × 660 × 109. Purified amplicons were serially diluted tenfold and four replicates were used for standard curve generation for quantification of unknown samples (Bustin et al., Reference Bustin, Benes, Garson, Hellemans, Huggett, Kubista, Mueller, Nolan, Pfaffl, Shipley, Vandesompele and Wittwer2009). The slope of the standard curves was used to calculate qPCR reaction efficiency.
Table 2. Description of primer pairs

qPCR assays were carried out using the Rotor-Gene SYBR® Green PCR kit (Qiagen, Hilden, Germany) on a Rotor-Gene 6000 (Corbett Research), according to the manufacturer's instructions. Two technical replicates were performed for two identical independent runs, to assess reproducibility of the assays. Briefly, 1× Rotor-Gene SYBR® Green PCR Master mix was used in a final reaction volume of 25 μl, with a final primer concentration of 1 μM and 2.5 μl of template. After an initial PCR activation step at 95°C for 5 min, cycling conditions consisted of 5 s denaturation at 95°C, and 40 cycles of combined annealing extension at 65°C for 10 s. Post-amplification melting curve analysis was performed to verify specificity and identity of qPCR products, with a ramp from 55 to 99°C, rising by 1°C each step. Results were analyzed with the Rotor-Gene 6000 series software 1.7 program. Sterile water was used as no-template control in each run. The resulting CT values, namely the value inverse to the amount of target nucleic acid in soil samples, were subjected to statistical analysis.
DGGE fingerprinting of actinomycetes and Pseudomonas
Qualitative analysis of actinomycetes and Pseudomonas populations was performed with DGGE analysis using a nested PCR approach and the primer pairs quoted in Table 2 as already described by Saccà et al. (Reference Saccà, Manici, Caputo and Frisullo2019). Two replicates of PCR products per each treatment were loaded on DGGE gel and when analyzing the electrophoretic pattern, a relative density score to each band based on a 1–5 scale (maximum band size 5 = 100%; minimum band size 1 = 20%) was assigned. In that way, taxa-abundance matrices were obtained by making possible comparing alpha diversity.
Data analysis
Data were analyzed with Statgraphics centurion software (2005 STATPOINT Inc., Virginia, USA). Quantitative, or parametric, data were subjected to two-way analysis of variance (ANOVA) and mean separation test using Fisher's least significant difference procedure. All percentage data were ln(x + 1) transformed before running statistical analysis. Multiple Pearson's correlation was used to estimate the relationship between plant growth and all the other estimated variables. Species/abundance data were handled with nonparametric multivariate analysis of variance (NPMANOVA) with Bray–Curtis distance using PAST version 3.24, software for analysis in paleoecology (Hammer et al., Reference Hammer, Harper and Ryan2001). Shannon (H) and Simpson (1-Dominance) biodiversity indices were calculated from the species/abundance of PCR-DGGE matrices using the above-mentioned PAST version. Biodiversity indices were compared with nonparametric Kruskal–Wallis test using the Statgraphics program.
Results
Plant growth response and root fungal colonization
The growth response of maize significantly differed both for site and for treatment in 2017 and 2018 (Table 3). Digestate-amended soil (Digestate) gave an increase in plant biomass accounting for 34% in 2017 and 28% in 2018, respectively, as compared to unamended soil (Control). As shown by the significant interaction site × treatment, in 2017 plant response to amended and unamended soils varied according to the soil origin (Table 3). Plant response to digestate-amended and to corresponding untreated soil (Control) in each site is shown in Figure 2 in which plant response to amended and unamended soils was analyzed within each site. The latter showed that plant biomass in digestate-amended soil largely exceeded the control soil only in the easternmost experimental site (FL) (P < 0.001) in 2017 and in the north western (CR) (P < 0.01) in 2018 (Fig. 2).

Fig. 2. Plant biomass and root colonization frequency recorded at end of in-pot growth assay with maize in digestate-amended and unamended control soils taken from the experimental field in three sites (located in the provinces of CR: Cremona; MO: Modena; FL: Forlì). aPearson correlation (18 counts) between plant biomass and root colonization frequency.
Table 3. Two-way ANOVA for the variables plant biomass (dry matter) and root fungal infection frequency recorded in two subsequent years with a bioassay with maize grown in-pot on soil samples taken from (A) digestate-amended and non-amended fields, in (B) three sites of the Po Valley
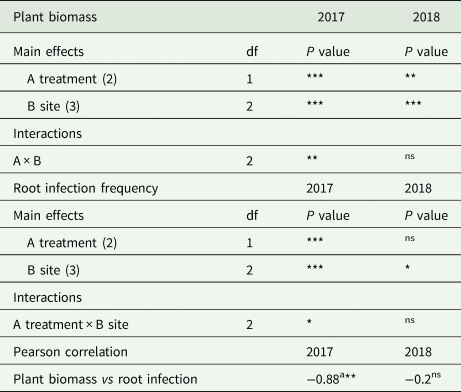
The analysis of root infection frequency was performed on arcsin-transformed data.
***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; nsnot significant.
a r value, 18 counts.
Fungal root colonization frequency was in all cases higher in digestate-amended soils as compared to the corresponding unamended control soils in both 2017 and 2018 (Fig. 2). Nevertheless, root infection frequency by fungal endophytes significantly differed for soil treatment only in 2017 (Table 3). Fungal root colonization frequency always negatively correlated with plant biomass (Table 3); nevertheless, Pearson correlation between those parameters was high and significant (r: −0.88, P > 0.01) in 2017, but not in 2018 (Table 3). The negative correlation between plant biomass production and root colonization frequency indicated that root colonizing fungi acted as root pathogens and digestate-amended soils were able to reduce the fungal root infection responsible of root development reduction (Fig. 3).

Fig. 3. Higher root development of maize grown in pot on digestate-amended soil as compared to unamended control. Soil samples from FL site that in 2017 showed a significant higher biomass development and a lower root fungal infection frequency in digestate-amended as compared to unamended control.
Among the fungal species isolated from maize roots at the end of in-pot test, Fusarium oxysporum and S. terrestris were the two most abundant and widespread fungal species occurring in both 2017 and 2018 (Figs. 4 and 5). F. oxysporum is the most spread soil inhabiting Fusarium sp. worldwide which can vary from neutral to weakly pathogenic on maize like many fusariums; conversely, S. terrestris, which had been already reported as responsible for yield losses in maize in other counties (Chambers, Reference Chambers1987; Newby et al., Reference Newby, Carroll and Whittington1997; Lević et al., Reference Lević, Petrović, Stanković, Krnjaja and Stanković2011), had not been so far reported as important maize root pathogen in Italy; therefore, it was subjected to a pathogenicity test. Quite all the other root colonizing fungi such as Fusarium spp., Helminthosporium turcicum, Macrophomina phaseolina and others (Figs. 4 and 5) which were isolated with lower frequency from maize roots, were all reported as potential root pathogens of maize (Munkvold and White, Reference Munkvold and White2015).

Fig. 4. 2017. Composition of root endophytic fungal communities of maize grown in soil samples from the couple of fields (digestate-amended and unamended control) of each site (located in the provinces of CR: Cremona; MO: Modena; FL: Forlì).

Fig. 5. 2018. Composition of root endophytic fungal communities of maize grown in soil samples from the couple of fields (digestate-amended and unamended control) of each site (located in the provinces of CR: Cremona; MO: Modena; FL: Forlì).
Pathogenicity test indicated S. terrestris as responsible for biomass loss of 69 and 64% in dry and fresh biomass, respectively, and this fungus was re-isolated only from roots of inoculated plants and not from the uninoculated control ones. This finding suggested that S. terrestris isolated from maize in this study acted as root pathogen and was responsible of plant growth reduction.
Weather
The two years considered in this study showed very contrasting values of precipitation, but not of temperature. In 2017, cumulated precipitation was respectively 167 mm in CR, 148 mm in MO and 164 mm in FL in the period January–May, showing a dry trend compared to the seasonal averages of the Po Valley. Year 2018 was more rainy and cumulative precipitation increases ranged from +189% in CR (315 mm), +207% in MO (306 mm) and +248% in FL (406 mm) in the same period (Fig. 6). Temperature at sowing time (day 9, Fig. 6) was above 10°C in 2017 and around 5°C in 2018, without marked differences among the experimental sites. During the period from sowing to V6–V10 maize developmental stage average temperature increased up to 15.6°C in MO, 15.8°C in FL and 18.3°C in CR, with reduced variability between years.

Fig. 6. Cumulated precipitation in the experimental sites in the Cremona (CR), Modena (MO) and Forlì (FL) provinces during the 5 months before soil sampling for the in-pot assay to test soil suppressiveness in 2017 and 2018.
Quantitative analysis of Pseudomonas and actinomycetes
Actinomycetes resulted significantly higher in digestate-treated soil in both the years (Table 4). However, as suggested by the significant (P < 0.01) interaction between the two factors of variability (site × treatment), they showed different response to soil treatment in the three experimental sites. Actinomycetes quantity did not correlate with plant biomass production or with root infection (Table 4).
Table 4. Two-way ANOVA of nucleic acid amount of actinomycetes and Pseudomonas recorded with qPCR
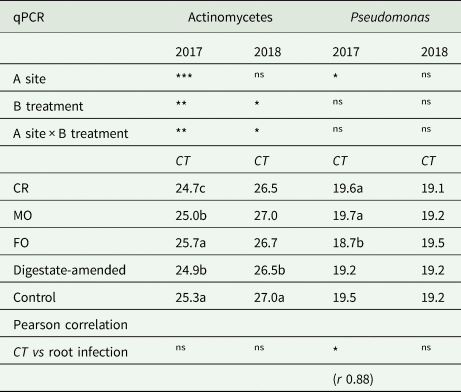
Data were expressed as CT values, which is the value inverse to the amount of target nucleic, therefore lower CT vales indicate higher amount.
***P < 0.001; **P < 0.01; *P < 0.05; nsnot significant.
Pseudomonas overall showed higher abundance in digestate-amended compared to unamended control soils; however, they significantly differed only for site in 2017. The FL site showed a higher occurrence of native Pseudomonas being the only in which digestate-amended soil showed higher abundance than control soil in both 2017 and 2018 (Supplementary material 1). Pseudomonas abundance was highly correlated with root infection (Table 4); as the CT value of qPCR is a value inverse to the amount of target nucleic, we can conclude that there was negative correlation between Pseudomonas abundance and root colonization by soil-borne pathogens. This finding indicates that this bacterial group was related to a highest natural soil ability to counteract root infection by fungal pathogens.
Biodiversity and difference in community composition
Actinomycetes and Pseudomonas composition highly differed for site and treatment in both the years showing also a high significant interaction site × treatment (P < 0.01) (Table 5). Shannon (H) and Simpson (1-Dominance) biodiversity indices of Pseudomonas and actinomycetes did not significantly differ between digestate-amended and unamended control soils neither within each site nor as a whole; conversely, they differ significantly between sites (P < 0.001) in both 2017 and 2018.
Table 5. Two-way NPMANOVA of species/abundance data of PCR-DGGE analysis of actinomycetes and Pseudomonas

****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; nsnot significant.
Discussion
This study highlighted benefits of anaerobic digestates for improving soil health in an intensively cultivated region. The main results of this study are the improvement of plant growth in periodically digestate-amended soils and a negative correlation between plant growth and root fungal infection. This suggested that periodical digestate amendments can increase the natural capacity of soils to counteract root colonization by soil-borne pathogens, that is soil suppressiveness. Furthermore, the higher abundance of actinomycetes and Pseudomonas in digestate-amended soils indicated that those bacterial groups could be involved in the observed plant growth improvement. Moreover, our findings provide evidence of high year-to-year variability of root fungal infection in response to precipitation patterns and inherent crop growth response.
The in-pot plant growth assay, which is one of the most common tests for soil suppressiveness (Neiendam Nielsen and Winding, Reference Neiendam Nielsen and Winding2002), associated with nitrogen fertilization, which was applied to reduce differences of N availability between the sampling sites of origin, permitted reducing a series of environmental variability factors thus allowing the identification of the main microbial components responsible of root development reduction as well as the mechanism of root defence. Indeed, though microorganisms are recognized as the major component of soil suppressiveness (Schlatter et al., Reference Schlatter, Kinkel, Thomashow, Weller and Paulitz2017), when plant growth reduction is caused by nonspecific root rot fungal agents such as in the case of this study, identifying major antagonists and biological control mechanisms is much more difficult.
Pseudomonas is a widespread bacterial genus, which includes a wide range of functional groups, such as plant growth promoters and antagonists toward many soil-borne pathogens, whose activity is mediated by releasing of a number of antibiotics and biologically active compounds (Patten and Glick, Reference Patten and Glick2002; Ganeshan and Kumar, Reference Ganeshan and Kumar2005; Mazurier et al., Reference Mazurier, Corberand, Lemanceau and Raaijmakers2009; Novik et al., Reference Novik, Savich and Kiselev2015; Chen et al., Reference Chen, Wu, Xiao, Wu, Wu, Qin, Wang, Wei, Khan, Lin and Lin2017). Although, quantitative analysis (qPCR) of Pseudomonas spp. showed an overall higher abundance in repeatedly digestate-amended soils compared to unamended ones, a clear correlation between Pseudomonas spp. abundance and soil suppressiveness was only observed in 2017, when they significantly correlated with root infection frequency thus suggesting Pseudomonas involvement in control action toward saprophytic soil-borne pathogens. This finding was in line with what was observed by Dignam et al. (Reference Dignam, O'Callaghan, Condron, Raaijmakers, Kowalchuk and Wakelin2019) who reported that long-term soil enrichment through plant residue incorporation into the soil increased activity of indigenous plant-beneficial bacterial taxa among which Pseudomonas was one of the most represented. As the V6/V7 region was amplified with primers targeting Pseudomonas genus (Chen et al., Reference Chen, Wu, Xiao, Wu, Wu, Qin, Wang, Wei, Khan, Lin and Lin2017) and qualitative (PCR-DGGE) analysis showed a large variability in the composition of Pseudomonas populations, no specific antagonistic mechanisms were identified.
Actinomycetes is a group of Gram-positive bacteria (order Actinomycetales) producing various bioactive compounds including antibiotics that are active in biological control of soil-borne pathogens (Crawford et al., Reference Crawford, Lynch, Whipps and Ousley1993; Mendes et al., Reference Mendes, Kruijt, De Bruijn, Dekkers, Van Der Voort, Schneider, Piceno, DeSantis, Andersen, Bakker and Raaijmakers2011; Latz et al., Reference Latz, Eisenhauer, Rall, Scheu and Jousset2016). Actinomycetes in this study did not directly correlate with plant growth or with root fungal infection frequency, but their abundance in digestate-amended soil was always higher than that in unamended ones. The latter finding suggested an overall beneficial action of actinomycetes, which is likely to have resulted from the sum of different functional groups that, according to the high variability of the community composition observed with the PCR-DGGE analysis, occurred in a very variable way across the experimental sites. Therefore, unlike what was observed for Pseudomonas, we cannot hypothesize a direct control effect for actinomycetes toward root pathogens, but we can hypothesize the involvement of a series of mechanisms capable of directly or indirectly promoting plant growth and health. Indeed, besides biocontrol (El-Tarabily and Sivasithamparam, Reference El-Tarabily and Sivasithamparam2006), actinomycetes can provide a series of other functions such as: (i) ability to make insoluble organic and inorganic phosphorus forms suitable for plant uptake, which was reported to vary widely according to climatic conditions (Ghorbani-Nasrabadi et al., Reference Ghorbani-Nasrabadi, Greiner, Alikhani, Hamedi and Yakhchali2013) and (ii) an active action in breaking down plant residues which contributes to maintaining the biotic equilibrium of soil by cooperating with nutrient cycling (Bhatti et al., Reference Bhatti, Haq and Bhat2017).
Communities of root fungal endophytes among which typical maize root pathogens such as H. turcicum was only occasionally recovered (Figs. 4 and 5), were rather composed of some secondary weak pathogens such as Fusarium spp. and M. phaseolina (Munkvold and White, Reference Munkvold and White2015) which however varied largely according to the sampling site and year. S. terrestris was the only soil-borne fungus occurring everywhere and present in both years of study. S. terrestris (past names Pyrenochaeta terrestris and Phoma terrestris), whose pathogenicity toward maize was confirmed by an ad hoc test in this study, is known for a certain specialization toward maize worldwide, but particularly in the USA where it has been reported to cause yield losses in continuous maize cropping systems either alone or associated with other soil fungi (Newby et al., Reference Newby, Carroll and Whittington1997; Koenning et al., Reference Koenning, Frye, Pataky, Gibbs and Cotton2007; Lević et al., Reference Lević, Petrović, Stanković, Krnjaja and Stanković2011). This soil-borne fungus, which has been already reported in Italy as the agent of pink root rot on canola (Yang et al., Reference Yang, Zuzak, Harding, Neilson, Feindel and Feng2017), seem to be selected by the return of maize on the same agricultural soils which is a well-known effect of monoculture (Shipton, Reference Shipton1977). Therefore, according to finding of this study, S. terrestris can be considered an additional indicator of maize monoculture effect in intensively cultivated areas such as the Po Valley.
Root fungal infection of maize at the end of in-pot testing, negatively correlated with plant biomass showing that root fungal endophytes impacted plant growth as pathogens thus resulting an indicator of agricultural soil health in this study. However, this correlation was high in 2017 and poor in 2018, when, respectively, a high and low average root infection frequency was recorded. A different plant response in soil samples taken from the same fields was in part explained by the different weather patterns in the previous months. Indeed, it was a favorable occurrence that the two subsequent years in which the soil was sampled were characterized by contrasting patterns of cumulated precipitation. We postulate that this had determined water stress in the early growth stages of maize in the first year (2017), but not in the second (2018), when optimal water availability was assured for maize in field up to six to ten leaves, the vegetative stage at which soil samples for in-pot testing were collected. As water stress can increase the release of root exudates into the soil in several plant species, including maize (Song et al., Reference Song, Han, Zhu and Herbert2012; Karst et al., Reference Karst, Gaster, Wiley and Landhäusser2017), this increase was hypothesized as being responsible for the increase of the inoculum of soil-borne pathogens of maize under water stress, by favoring the further root colonization, as observed in other plant species (Manici and Caputo, Reference Manici and Caputo2020). Therefore, different weather patterns during the early maize growing stages were probably responsible for a different soil inoculum of root pathogens at soil sampling time for the in-pot trials in 2017 and 2018.
As bacterial composition is deeply affected by the environment (Fierer and Jackson, Reference Fierer and Jackson2006; Manici et al., Reference Manici, Saccà, Caputo, Zanzotto, Gardiman and Fila2017), differences in actinomycetes and Pseudomonas composition between sites were expected. Indeed, such environmental variability overcomes differences in bacterial composition between digestate-amended and unamended fields across the three different agro-environments considered in this study. The latter observation was consistent with high resilience of the resident microbiota in response to digestate and compost amendment in microcosm experiments (Podmirseg et al., Reference Podmirseg, Waldhuber, Knapp, Insam and Goberna2019). Therefore, the overall increase of Pseudomonas and actinomycetes in digestate-amendment soils is likely to concern indigenous bacterial populations rather than allochthonous microorganisms introduced with digestate.
This study shows that repeated application of digestate to soils can improve its natural ability to control root pathogen. That ability appeared to be linked primarily to microbial factors implemented by repeated digestate amendment. Among these microbial factors we have not mentioned microbial biomass (Bongiorno et al., Reference Bongiorno, Postma, Bünemann, Brussaard, de Goede, Mäder, Tamm and Thuerig2019). However, the increase of basal respiration after digestate incorporation into the soil has been observed to totally decrease in 1–3 months after incorporation (Podmirseg et al., Reference Podmirseg, Waldhuber, Knapp, Insam and Goberna2019); whereas in this case, the last digestate amendment was applied to the soil more than 12 months earlier. Moreover, as soil fungi are the main component of microbial biomass in volume (Bailey et al., Reference Bailey, Smith and Bolton2002), no difference in total fungi DNA content between digestate-amended and unamended soils in each experimental site was found (data not shown).
In conclusion, this study based on multiple indicators for soil health indicates that further environmental and economic benefits may come from biogas production increased through the soil amendment with digestates. Although this type of waste-derived materials have well-known agronomic benefits (Tambone et al., Reference Tambone, Scaglia, D'Imporzano, Schievano, Orzi, Salati and Adani2010; Barzee et al., Reference Barzee, Edalati, El-Mashad, Wang, Scow and Zhang2019), in Italy they did not classified as by-products until 2016 and were, as such, considered for agronomic use according to the same rules as animal manure (MIPAAF, 2016). The improvement of soil suppressiveness is a so far disregarded effect that throws light on the additional benefits of soil amendment with digestates and indicates the advantages of their long-term application to agricultural soils. This improvement seems due to multiple and variable microbial functionalities; however, based on the findings of this study, it is possible to state that actinomycetes and Pseudomonas are two bacterial groups involved and they can be considered useful indicators of benefits from medium term soil amendment with biogas digestates. Finally, maize is commonly considered a crop that can be repeatedly cultivated in the same soil with no major disadvantages; nevertheless, findings of this study indicate that monocropping of maize for at least 15 yr reduces crop health. This result is consistent with those of Borrelli et al. (Reference Borrelli, Castelli, Ceotto, Cabassi and Tomasoni2014) who reported higher yield and yield stability for maize cultivated in a 3-yr rotation compared to maize monocrop in a 30-yr experiment conducted in the irrigated lowland of Lombardy (the Po Valley). This suggests that the current European rules, which discourages the use of maize as a feedstock for anaerobic digestion for biogas production, while encouraging the use of crop residues and no-food cover crops (European Parliament and the Council of the European Union, 2015), are likely to determine gains from the standpoints of biodiversity and soil suppressiveness and crop health.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1742170520000393
Acknowledgements
This study was financed by the Italian Ministry of Agriculture, Food and Forest Policy (MiPAAF) under the project AGROENER (D.D. no. 26329, 1 April 2016) (http://agroener.crea.gov.it/).
Author contributions
L.M. Manici: conceptualization; ideas, methodology, formal analysis and writing original draft. F. Caputo: investigation, resources and methodology. G.A. Cappelli: curation and analysis of weather data. E. Ceotto: supervision, writing, review and editing, support to conceptualization and formal analysis.














