INTRODUCTION
Human remains dating to the late Middle Paleolithic and early Upper Paleolithic remain rare. Recent research has demonstrated how important direct dating and DNA analyses are for investigating biological and cultural developments: for such small datasets, errors in the remains assumed age and bio-cultural context rapidly impact our ability to adequately reconstruct human evolution (e.g., Conard et al. Reference Conard, Grootes and Smith2004; Davies et al. Reference Davies, White, Lewis and Stringer2015; Devièse et al. Reference Devièse, Karavanić, Comeskey, Kubiak, Korlević, Hajdinjak, Radović, Procopio, Buckley, Pääbo and Higham2017; Street et al. Reference Street, Terberger and Orschiedt2006; Talamo et al. Reference Talamo, Hajdinjak, Mannino, Fasani, Welker, Martini, Romagnoli, Zorzin, Meyer and Hublin2016; Trinkaus Reference Trinkaus2005). This holds especially true for specimens found without secure archaeological context or in secondary deposition. In some of these cases, direct dating was able to recontextualize the fossils, thus contributing tremendously to our understanding of possible Neanderthal/Modern-Human interactions and human evolution more generally (e.g. Fu et al. Reference Fu, Hajdinjak, Moldovan, Constantin, Mallick, Skoglund, Patterson, Rohland, Lazaridis, Nickel, Viola, Prüfer, Meyer, Kelso, Reich and Pääbo2015, Reference Fu, Li, Moorjani, Jay, Slepchenko, Bondarev, Johnson, Aximu-Petri, Prüfer, de Filippo, Meyer, Zwyns, Salazar-García, Kuzmin, Keates, Kosintsev, Razhev, Richards, Peristov, Lachmann, Douka, Higham, Slatkin, Hublin, Reich, Kelso, Viola and Pääbo2014). Therefore, this research set out to directly date the remains of two Neanderthal individuals that were found in secondary deposition in a river ravine near Šal’a, Slovakia (Figure 1). It is thought that the original fossil accumulation was covered with loess that began eroding since the Holocene, resulting in material being transported downstream along the river Váh (Kaminská Reference Kaminská2014: 75). As a result, previous age estimations relied on biostratigraphic information of its secondary find context (stratigraphy, paleoenvironmental and geological context), cranial morphology, bone preservation and patina, as well as faunal fragments that may have originated from the same primary deposition as the Neanderthal remains.

Figure 1 Map showing location of Šal’a (star) and sites with Neanderthal remains referenced in this paper: 1 – Les Rochers-de-Villeneuve, 2 – Ferassie (France), 3 – Spy (Belgium), 4 –Kleine Feldhöfergrotte (Germany), 5 – Vindija (Croatia), 6 – Gánovce (Slovakia), 7 – Mezmaiskaya (Russia). The site of Okladnikov is in the Altai Mountains (Russia) and not shown on the map due to its location further east.
Šal’a I
Šal’a I was originally found in 1961 (Vlček Reference Vlček1968, Reference Vlček1969). The nearly complete frontal bone (with squamous, orbital and nasal portions preserved) belongs to an adult classified as a Late Pleistocene Central European Neanderthal (Sládek et al. Reference Sládek, Trinkaus, Šefčáková and Halouzka2002), possibly a woman between 20–39 years of age (Vlček Reference Vlček1968, Reference Vlček1969). Based on bone preservation, indirect association with interstadial faunal remains (Vlček Reference Vlček1968; Ďurišová Reference Ďurišová1989, Reference Ďurišová1993, Reference Ďurišová1994) and the stratigraphy of the secondary deposit, the cranial fragment was previously estimated to be 100-80k years old (Šefčáková et al. Reference Šefčáková, Halouzka and Thurzo2005, with references).
The bone is heavily mineralized, resulting in a dark brown-black color. The almost fully intact teeth of the coronal suture (sutura coronalis) indicate that the remains were only briefly exposed to flowing water. However, the peripheral parts of the bone show some smoothing, which may have been caused by sand or water. An elliptical depression is visible in the area of the right supraorbital plane — a scar measuring about 10x12 mm. It penetrates the entire thickness of the external lamina and creates a shallow defect in the diploë (Sládek et al. Reference Sládek, Trinkaus, Šefčáková and Halouzka2002; Vlček Reference Vlček1968). The walls of the disruption show newly formed bone, while the area around the scar is highly atrophied and the torus supraorbitalis is locally thinned. It may be a healed wound from a penetrating trauma caused by a sharp object (e.g., stone, tooth). On the upper edge of the right orbit, just below the injury, there are three ‘notches’ of 4.2–7.4 mm length and with a V-shaped cross-section. Furthermore, there are several shallow linear scratches. They are located above the root of the nose and above the left orbit, and are of similar color as the rest of the bone. They are likely the result of exposure to moving gravel in the river. By contrast, the interior of the skull fragment opposite the eye sockets shows 12 small holes (1.5 mm in diameter) that have been filled with wax and painted to match the color of the bone patina (Figure 2). These are sampling traces from the fluorine analysis conducted to confirm the degree of fossilization (Vlček Reference Vlček1968), and none of them seem to penetrate the outer bone surface.
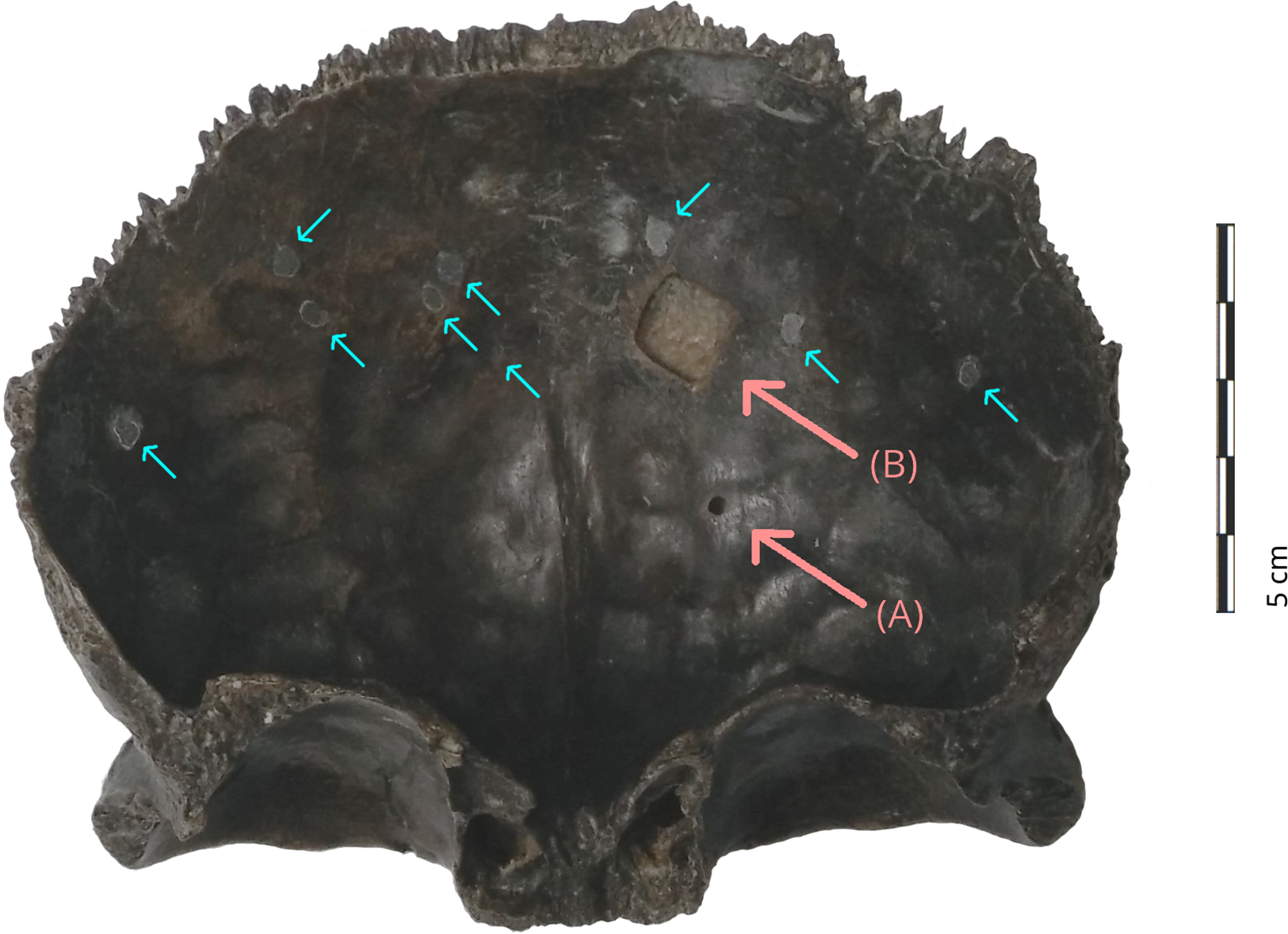
Figure 2 Interior surface of Šal’a I (frontal bone) showing sampling area for (A) %N and aDNA analyses as collected in April 2016, and for (B) radiocarbon dating as collected in June 2016. The circular, filled holes (small arrows) are the result of previous sampling for fluorine analysis (photo: R. Hopkins).
Before being transferred to the ‘Slovak National Museum – Natural History Museum’ in Bratislava, the Šal’a I remains were originally part of the collections of the ‘Institute of Archaeology of the Slovak Academy of Sciences’ (IA SAS) in Nitra. As a result, Šal’a I has two archival identification codes: ‘A 6915 P 12/93’ and ‘AP no. 18258’, respectively.
Šal’a II
Šal’a II consists of two cranial fragments that were found independently and are presently stored at the Institute of Archaeology of the Slovak Academy of Sciences (IA SAS) in Nitra (Slovakia). The left parietal bone was discovered in 1993, followed by the left half of a frontal bone in 1995. Both fragments refitted along a fresh fracture line, thus establishing that they belonged to the same individual; an adult male estimated to be 40–59 years old (Jakab Reference Jakab1996, Reference Jakab1998, Reference Jakab2005). The frontal bone of Šal’a II was split along the sutura frontale and the frontal tuber shows some erosion. The parietal bone showed two flat areas, one between the parietal eminence and the sagittal suture, the other between the parietal eminence and the lambdoid suture. They could have been caused by injury or taphonomic processes. As the scratches do not differ in color, a healed wound is more likely. While the inside of the cranial fragment is intact, there is a shallow impression which may stem from an epidural hematoma (Jakab Reference Jakab2005). Overall, Šal’a II shows notable modern conservation and restoration work (Figure 3), which has to be addressed when applying radiocarbon dating. In 1997, Prof M. Schultz cut an L-shaped sample (1.5 g) from the frontal bone for histological and DNA analyses at the Zentrum für Anatomie der Georg-August-Universität Göttingen (Jakab Reference Jakab2005), but no results have been reported (pers. comm. L’. Kaminská, August 2015). The missing area has since been filled, and the two bone fragments rejoined using glue of unknown composition. In addition, modifications were painted over to match the surface patina.

Figure 3 Interior surface of Šal’a II showing sampling area for (A) %N and aDNA analyses from August 2015, and for (B) radiocarbon dating from March 2016. The frontal bone was previously sampled for histological and aDNA analyses in 1997, when an L-shaped fragment (42x18 cm) was cut close to the glued fracture line (small arrow). The original analyses did not lead to published results (photo: R. Hopkins).
METHODS
As the remains of both Neanderthal individuals were found without a secure and datable context, methods to contextualize the specimens were necessary. Considering the possibility that these were late Neanderthals, the main aim was to establish whether these individuals date to the Middle to Upper Paleolithic biocultural shift. This was approximated by whether they dated within the measurable radiocarbon dating timescale. Additionally, ancient DNA analysis may provide further information to biologically contextualize the individuals. As there was no information on why the dating from 1997 failed, we took small test samples for percent nitrogen (%N) (proxy for collagen preservation) and ancient DNA analyses, before collecting sufficient material for single-amino-acid radiocarbon dating. Sampling and radiocarbon dating were conducted by R. Hopkins in Slovakia and at the Oxford Radiocarbon Accelerator Unit (ORAU), University of Oxford, respectively, while the aDNA analyses were run by M. Hajdinjak at the Max Planck Institute for Evolutionary Anthropology in Leipzig.
Sampling
Using the key-hole drilling technique, Šal’a I was sampled for %N (5 mg) and aDNA (85 mg) analyses in April 2016 (Figure 2, A). Contamination was minimized by placing the sampling location towards the center of the bone (away from the restoration work). Surface material was discarded. Anatomical features of the interior surface were avoided, and the exterior surface left intact. In June, 2016, a further 940 mg of bone powder was drilled from an area of ca. 1 cm2 for single-amino-acid radiocarbon dating (Figure 2, B).
Šal’a II was initially sampled for %N (5 mg) and aDNA (70 mg) analyses in August 2015 via key-hole drilling (Figure 3, A). In an effort to restrict sampling to one of the two cranial fragments, drilling was located to the inside of the frontal bone and away from the previous L-shaped sample location. Unfortunately, maneuverability in this area proved insufficient for a larger sample and radiocarbon sampling (930 mg) had to be moved to an area just under 1 cm2 on the left parietal bone (Figure 3, B). Surface material was discarded.
Radiocarbon Dating
The radiocarbon samples were first pretreated according to Brock et al. (Reference Brock, Higham, Ditchfield and Bronk Ramsey2010). The bone powder (Šal’a I: 367.90 mg, Šal’a II: 494.88 mg) was demineralized with three 0.5 M hydrochloric acid (HCl) treatments (2h,2h,18h, RT), and decontaminated with 0.1 M sodium hydroxide (NaOH) (30 min., RT), and with 0.5 M HCl (15 min., RT). After each treatment step, the samples were rinsed three times with ultrapure water, and the supernatant discarded. The resulting collagen was gelatinized (75° C, 20h, pH=3), filtered with Ezee-filters, and subsequently freeze-dried for 48h.
To avoid possible museum contamination, we targeted the amino acid hydroxyproline for dating (instead of bulk collagen). For this, we followed the protocol developed by Devièse et al. (Reference Devièse, Comeskey, McCullagh, Bronk Ramsey and Higham2018). The extracted collagen (Šal’a I: 15.55 mg, Šal’a II: 17.76 mg) was treated with 2mL of 6 M HCl in a nitrogen atmosphere (24h, 110°C). The solution was evaporated to dryness in a vacuum evaporator. The samples were then redissolved in 700 μL of 0.1 NaOH, and filtered into a 1 mL HPLC total recovery vial using a 2 mL BP Plastipak syringe with a 0.2 μm PTFE syringe filter to remove insoluble matter. In order to avoid losing any amino acid residue, 300 μL of ultrapure water was added and filtered. Amino acids were separated via HPLC (1mL sample injection size) with a mixed-mode chromatographic column (Primesep A), MilliQ water as eluent A, and 0.3v% phosphoric acid diluted with MilliQ water as eluent B. The oven was set to 30°C, and the flow rate to 18 mL/min. The hydroxyproline fraction was collected between 15.5–17.5 min. (total of 36 mL), and concentrated in a vacuum evaporator. The amount of hydroxyproline collected (Table 1) was calculated via a six-point calibration curve which was developed based on known injection sizes (Devièse et al. Reference Devièse, Comeskey, McCullagh, Bronk Ramsey and Higham2018). To extract the carbon dioxide (CO2) for combustion, the hydroxyproline was resolubilised in 25 μL of MilliQ water and transferred onto Chromasorb in a combustion tin capsule. Combustion, graphitization and AMS measurement followed standard ORAU procedures (Bronk Ramsey et al. Reference Bronk Ramsey, Higham and Leach2004; Dee and Bronk Ramsey Reference Dee and Bronk Ramsey2000; Hedges et al. Reference Hedges, Humm, Foreman, van Klinken and Bronk Ramsey1992).
Table 1 Radiocarbon dating results for both Šal’a I and Šal’a II, as well as the two internal bone background standards (Bison bone from Ash Bend, Canada). Both samples and standards followed the same pretreatment. %N = nitrogen weight content before chemical pretreatment. Weight = bone powder pretreated in mg. Collagen = collagen yield after gelatinization and freeze-drying, in mg (wt.%). HYP = hydroxyproline yield (in mg) calculated using a six-point calibration curve (Devièse et al. Reference Devièse, Comeskey, McCullagh, Bronk Ramsey and Higham2018). mgC = amount of carbon in HYP combusted for graphitization (used for HPLC background correction). δ15N and δ13C = stable isotope measurements on combustion. %C = carbon yield on combustion. C/N = carbon to nitrogen atomic ratio of combusted sample. F14C (AMS) = fraction modern measurement as obtained by AMS measurement (used for HPLC background correction). F14C (corr.) = fraction modern value after HPLC background correction (used for calculating corrected radiocarbon age). 14C (BP) = HPLC background corrected radiocarbon age in years Before Present.

At the time of processing, the single-amino-acid protocol was was still being tested at ORAU. Consequently, any measurements were given a ‘OxA-X’ lab prefix, and manual HPLC background corrections were necessary (Devièse et al. Reference Devièse, Comeskey, McCullagh, Bronk Ramsey and Higham2018). For the latter, the ORAU internal background standard P19651 was used (Table 1). This is a bison bone (left Calcaneum) from Ash Bend (Canada) that was found below the Sheep Creek-Klondike tephra. Two bone powder fractions were prepared (P19651.165 and P19651.167), which underwent the same pretreatment as the Šal’a samples. To avoid overloading the HPLC column, a reduced amount of collagen (29.0 mg) extracted from the P19651.167 background standard was run for hydroxyproline collection. By contrast, the entire collagen yield was used for hydroxyproline extraction from the background standard P19651.165 and both Šala samples.
The routine corrections for carbon contamination picked up during standard procedures (wet-chemistry, combustion, graphitization) is automatically applied to all dates produced by ORAU, and is in this paper referred to as resulting in the HPLC uncorrected radiocarbon measurement. To account for exogenous carbon that was added during the hydroxyproline separation, the following equations were used.
Equation 1 calculates the corrected F14C for a sample’s single amino acid age (
![]() ${F^{14}}{C_{Hyp}}$
), where AMS is the measured F14C of the sample,
${F^{14}}{C_{Hyp}}$
), where AMS is the measured F14C of the sample,
![]() $M{F_{Mod}}$
the mass fraction of modern contamination (Equation 3); and
$M{F_{Mod}}$
the mass fraction of modern contamination (Equation 3); and
![]() $M{F_{Hyp}}$
the mass fraction of a sample’s hydroxyproline (Equation 4). Exogenous dead carbon contribution was deemed insignificant based on two factors: there is substantial amount of dead carbon needed to affect a date, and our samples are from the European Palaeolithic, therefore already close to or at radiocarbon background.
$M{F_{Hyp}}$
the mass fraction of a sample’s hydroxyproline (Equation 4). Exogenous dead carbon contribution was deemed insignificant based on two factors: there is substantial amount of dead carbon needed to affect a date, and our samples are from the European Palaeolithic, therefore already close to or at radiocarbon background.
 $$\sigma {F^{14}}{C_{Hyp}} = \sqrt {\left( {{{\left( {{1 \over {M{F_{Hyp}}}}*\Delta AMS} \right)}^2} + {{\left( {{1 \over {M{F_{Hyp}}}}*\Delta M{F_{Mod}}} \right)}^2} + {{\left( {{{\left( {M{F_{Mod}} - AMS} \right)} \over {{{\left( {M{F_{Hyp}}} \right)}^2}}}*\Delta M{F_{Hyp}}} \right)}^2}} \right)} $$
$$\sigma {F^{14}}{C_{Hyp}} = \sqrt {\left( {{{\left( {{1 \over {M{F_{Hyp}}}}*\Delta AMS} \right)}^2} + {{\left( {{1 \over {M{F_{Hyp}}}}*\Delta M{F_{Mod}}} \right)}^2} + {{\left( {{{\left( {M{F_{Mod}} - AMS} \right)} \over {{{\left( {M{F_{Hyp}}} \right)}^2}}}*\Delta M{F_{Hyp}}} \right)}^2}} \right)} $$
And Equation 2 calculated the new dating uncertainties that accompany a sample’s single amino acid age.
Equation 3 approximates
![]() $M{F_{Mod}}$
by using the measured F14C of the background standards (
$M{F_{Mod}}$
by using the measured F14C of the background standards (
![]() $AM{S_{Std}}$
), the mass of carbon found within the background standards (
$AM{S_{Std}}$
), the mass of carbon found within the background standards (
![]() ${C_{Std}}$
), and the mass of carbon found within the sample (
${C_{Std}}$
), and the mass of carbon found within the sample (
![]() ${C_T}$
). The latter two values are both obtained on the mass spectrometer.
${C_T}$
). The latter two values are both obtained on the mass spectrometer.
Equation 4 approximates
![]() $M{F_{Hyp}}$
by assuming its true value to be that minus the mass fraction of modern contamination (
$M{F_{Hyp}}$
by assuming its true value to be that minus the mass fraction of modern contamination (
![]() $M{F_{Mod}}$
).
$M{F_{Mod}}$
).
Ancient DNA
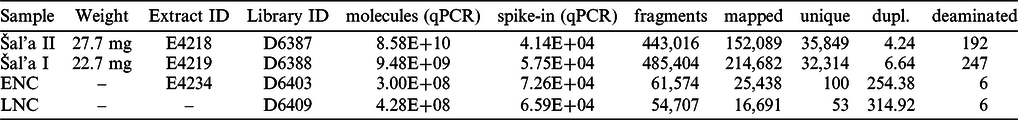
DNA was extracted from 27.7 mg and 22.7 mg of bone powder of Šala II and Šala I, respectively, using a silica based method (Dabney et al. Reference Dabney, Knapp, Glocke, Gansauge, Weihmann, Nickel, Valdiosera, Garcia, Paabo, Arsuaga and Meyer2013) with modifications (Korlevic et al. Reference Korlevic, Gerber, Gansauge, Hajdinjak, Nagel, Aximu-Petri and Meyer2015). Ten µL (20%) of each extract was converted into a single stranded DNA library using an automated protocol (Gansauge et al. Reference Gansauge, Aximu-Petri, Nagel and Meyer2020). These libraries, along with the DNA extraction and library negative controls, were quantified, amplified and double indexed (Kircher et al. Reference Kircher, Sawyer and Meyer2012) using automated protocols as detailed in Gansauge et al. (Reference Gansauge, Aximu-Petri, Nagel and Meyer2020). An aliquot of each library was further enriched for human mitochondrial DNA (mtDNA) (Fu et al. Reference Fu, Mittnik, Johnson, Bos, Lari, Bollongino, Sun, Giemsch, Schmitz, Burger, Ronchitelli, Martini, Cremonesi, Svoboda, Bauer, Caramelli, Castellano, Reich, Paabo and Krause2013; Maricic et al. Reference Maricic, Whitten and Paabo2010; Slon et al. Reference Slon, Hopfe, Weiß, Mafessoni, de la Rasilla, Lalueza-Fox, Rosas, Soressi, Knul and Miller2017) by hybridization capture and the enriched libraries were paired-end sequenced on an Illumina MiSeq (2 x 76 cycles) (Kircher et al. Reference Kircher, Sawyer and Meyer2012).
The adapters were trimmed and sequencing reads overlap-merged using leeHom (Renaud et al. Reference Renaud, Stenzel and Kelso2014). The Burrows-Wheeler Aligner (Li and Durbin Reference Li and Durbin2010) with ancient DNA parameters (“-n 0.01 –o 2 –l 16500”) (Meyer et al. Reference Meyer, Fu, Aximu-Petri, Glocke, Nickel, Arsuaga, Martinez, Gracia, de Castro, Carbonell and Paabo2014) was used to align the sequences to the revised Cambridge Reference Sequence (rCRS) (Andrews et al. Reference Andrews, Kubacka, Chinnery, Lightowlers, Turnbull and Howell1999). We restricted all downstream analyses to DNA fragments with a perfect match to the expected index combinations. We used SAMtools (version: 1.3.1) (Li et al. Reference Li, Handsaker, Wysoker, Fennell, Ruan, Homer, Marth, Abecasis, Durbin and Genome Project Data2009) to filter for DNA fragments of at least 35 base pairs (bp) with a mapping quality of 25 or more (MQ >= 25), and bam-rmdup (version: 0.6.3; https://github.com/mpieva/biohazard-tools) to remove PCR duplicates. Table 2 summarizes the number of DNA fragments retained after filtering steps. To determine if some of the recovered DNA fragments are of ancient origin, we evaluated the frequency at which cytosines (C) in the reference genome are substituted by thymines (T) at their ends, as elevated frequencies of C-to-T substitutions are expected to occur in genuine ancient DNA (Briggs et al. Reference Briggs, Stenzel, Johnson, Green, Kelso, Prufer, Meyer, Krause, Ronan, Lachmann and Paabo2007) and are largely absent in recent contamination (Krause et al. Reference Krause, Briggs, Kircher, Maricic, Zwyns, Derevianko and Paabo2010a; Sawyer et al. Reference Sawyer, Krause, Guschanski, Savolainen and Paabo2012).
Table 2 General characteristics of DNA libraries from Šal’a II and Šal’a I enriched for mitochondrial DNA. “molecules” = number of molecules (qPCR); “spike-in (qPCR)” = number of control oligonucleotide (qPCR); “fragments” = number of fragments sequenced; “mapped” = number of mapped fragments ≥ 35bp, MQ≥25; “unique” = number of unique fragments ≥ 35bp, MQ≥25; “dupl.” = Average number of duplicates; “deaminated” = number of fragments with C→T substitution ≥ 35bp, MQ≥25. qPCR – quantitative polymerase chain reaction, ENC – extraction negative control, LNC – library negative control, mg – milligrams, bp – base pairs, MQ – mapping quality.
RESULTS
The %N results for Šal’a I and Šal’a II showed promisingly high values for expected old samples (Table 1). Therefore, further bone powder was collected for radiocarbon dating. To minimize complete sample loss, only half of the bone powder for each sample was initially pretreated for radiocarbon dating. Again, collagen yields suggested low but sufficient preservation for dating: 4.83 wt.% for Šal’a I, and 3.14 wt.% for Šal’a II. However, the freeze-dried collagen was extremely brittle and retained an ashy color. Nonetheless, the carbon extracted from the hydroxyproline was sufficient (0.64 mg and 0.54 mg). The C/N atomic ratio for hydroxyproline is
![]() $ {\approx} $
5.0 (Devièse et al. Reference Devièse, Karavanić, Comeskey, Kubiak, Korlević, Hajdinjak, Radović, Procopio, Buckley, Pääbo and Higham2017). However, the mass-spectrometer used showed a reproducible shift in C/N values for small mass samples, as revealed by tests conducted on standards. As a result, the values obtained for Šal’a I and Šal’a II of 5.4 and 5.2, respectively, lie within acceptable range. Even before HPLC background correction, both Neanderthal specimens gave F14C values of 0.00178 ± 0.00162 and 0.00127 ± 0.00192, dating them likely beyond the radiocarbon time scale. The corrected radiocarbon dates are >44,800 BP (OxA-X-2731-16) for Šal’a I and >45,100 BP (OxA-X-2731.15) for Šal’a II
Footnote 1
. The radiocarbon activities obtained for the standards were 0.00116 ± 0.00099 (19651.165) and 0.00143 ± 0.00113 (19651.167), and was used to obtain the samples’ corrected single amino acid ages (Table 1).
$ {\approx} $
5.0 (Devièse et al. Reference Devièse, Karavanić, Comeskey, Kubiak, Korlević, Hajdinjak, Radović, Procopio, Buckley, Pääbo and Higham2017). However, the mass-spectrometer used showed a reproducible shift in C/N values for small mass samples, as revealed by tests conducted on standards. As a result, the values obtained for Šal’a I and Šal’a II of 5.4 and 5.2, respectively, lie within acceptable range. Even before HPLC background correction, both Neanderthal specimens gave F14C values of 0.00178 ± 0.00162 and 0.00127 ± 0.00192, dating them likely beyond the radiocarbon time scale. The corrected radiocarbon dates are >44,800 BP (OxA-X-2731-16) for Šal’a I and >45,100 BP (OxA-X-2731.15) for Šal’a II
Footnote 1
. The radiocarbon activities obtained for the standards were 0.00116 ± 0.00099 (19651.165) and 0.00143 ± 0.00113 (19651.167), and was used to obtain the samples’ corrected single amino acid ages (Table 1).
The DNA analysis recovered 35,849 and 32,314 unique mtDNA fragments from Šala II and Šala I libraries, respectively (Table 2). Each unique DNA fragment was observed between 4.6 and 6.6 times on average, indicating that deeper sequencing of these libraries would not substantially increase the number of unique mtDNA fragments. However, the frequencies of C-to-T substitutions in Šala II and Šala I ranged between 0.6% and 1.1% on the 5’-ends and between 0.4% and 0.5% on the 3’-ends of DNA fragments, respectively (Figure 4). These frequencies are much lower than what is observed in other specimens of similar age or even younger (Benazzi et al. Reference Benazzi, Slon, Talamo, Negrino, Peresani, Bailey, Sawyer, Panetta, Vicino, Starnini, Mannino, Salvadori, Meyer, Paabo and Hublin2015; Fu et al. Reference Fu, Hajdinjak, Moldovan, Constantin, Mallick, Skoglund, Patterson, Rohland, Lazaridis, Nickel, Viola, Prüfer, Meyer, Kelso, Reich and Pääbo2015, Reference Fu, Mittnik, Johnson, Bos, Lari, Bollongino, Sun, Giemsch, Schmitz, Burger, Ronchitelli, Martini, Cremonesi, Svoboda, Bauer, Caramelli, Castellano, Reich, Paabo and Krause2013, Reference Fu, Posth, Hajdinjak, Petr, Mallick, Fernandes, Furtwangler, Haak, Meyer, Mittnik, Nickel, Peltzer, Rohland, Slon, Talamo, Lazaridis, Lipson, Mathieson, Schiffels, Skoglund, Derevianko, Drozdov, Slavinsky, Tsybankov, Cremonesi, Mallegni, Gely, Vacca, Morales, Straus, Neugebauer-Maresch, Teschler-Nicola, Constantin, Moldovan, Benazzi, Peresani, Coppola, Lari, Ricci, Ronchitelli, Valentin, Thevenet, Wehrberger, Grigorescu, Rougier, Crevecoeur, Flas, Semal, Mannino, Cupillard, Bocherens, Conard, Harvati, Moiseyev, Drucker, Svoboda, Richards, Caramelli, Pinhasi, Kelso, Patterson, Krause, Paabo and Reich2016; Hajdinjak et al. Reference Hajdinjak, Fu, Hubner, Petr, Mafessoni, Grote, Skoglund, Narasimham, Rougier, Crevecoeur, Semal, Soressi, Talamo, Hublin, Gusic, Kucan, Rudan, Golovanova, Doronichev, Posth, Krause, Korlevic, Nagel, Nickel, Slatkin, Patterson, Reich, Prufer, Meyer, Paabo and Kelso2018; Krause et al. Reference Krause, Briggs, Kircher, Maricic, Zwyns, Derevianko and Paabo2010a,b; Meyer et al. Reference Meyer, Kircher, Gansauge, Li, Racimo, Mallick, Schraiber, Jay, Prufer, de Filippo, Sudmant, Alkan, Fu, Do, Rohland, Tandon, Siebauer, Green, Bryc, Briggs, Stenzel, Dabney, Shendure, Kitzman, Hammer, Shunkov, Derevianko, Patterson, Andres, Eichler, Slatkin, Reich, Kelso and Paabo2012), indicating that the recovered DNA from the two specimens is not of ancient origin (Krause et al. Reference Krause, Briggs, Kircher, Maricic, Zwyns, Derevianko and Paabo2010a; Sawyer et al. Reference Sawyer, Krause, Guschanski, Savolainen and Paabo2012) and rather stems from recent contamination. These frequencies dropped to 0% (Figure 4) when filtering for fragments with a C-to-T substitution at the opposite end (‘conditional’ substitutions) (Meyer et al. Reference Meyer, Fu, Aximu-Petri, Glocke, Nickel, Arsuaga, Martinez, Gracia, de Castro, Carbonell and Paabo2014), further supporting there is no evidence for ancient DNA preservation in Šala II and Šala I to the limits of our resolution.
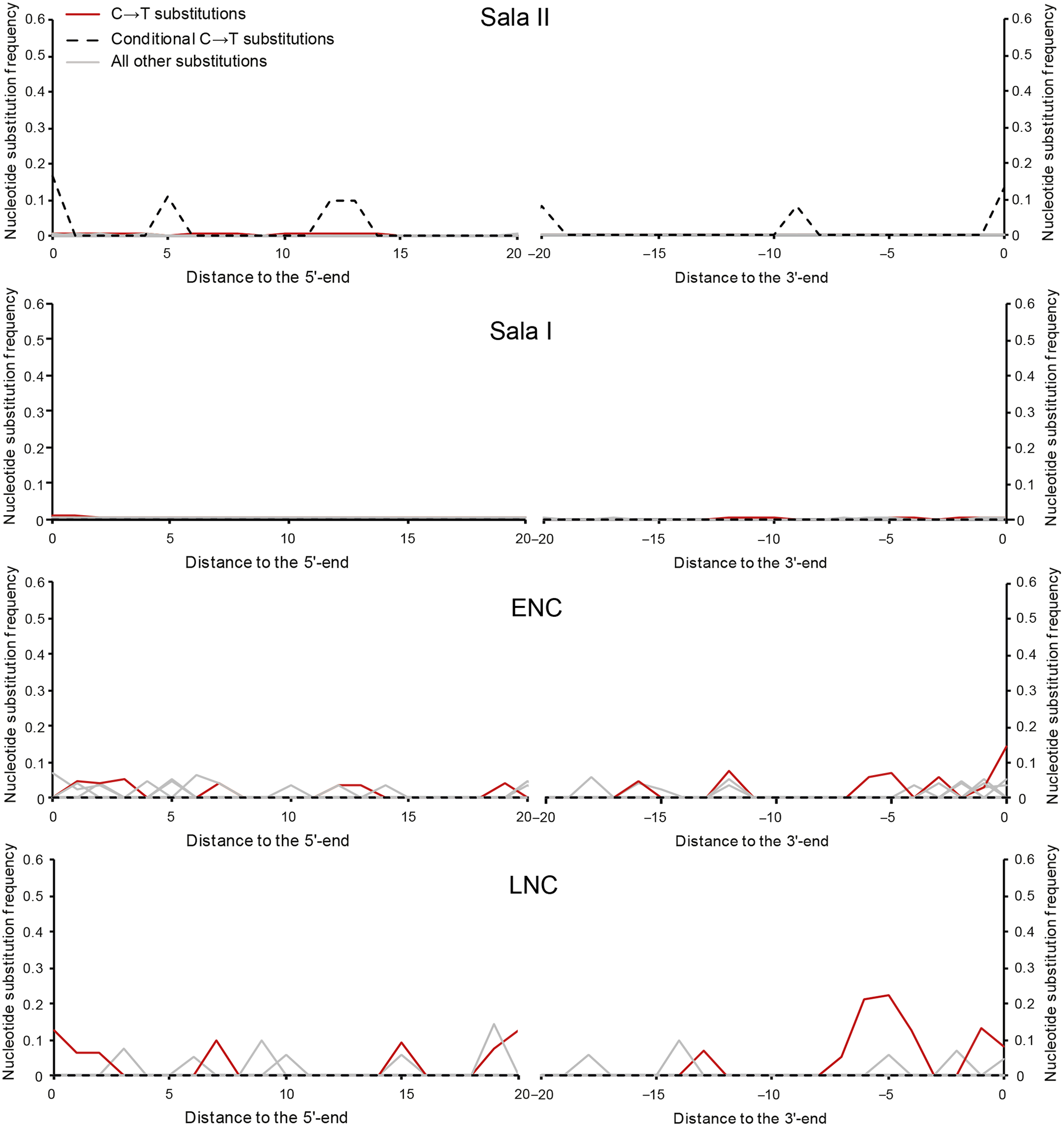
Figure 4 C-to-T substitution frequencies at the beginning and the end of mtDNA alignments of Šal’a II and Šal’a I libraries. ENC — extraction negative control, LNC — library negative control.
DISCUSSION
Despite poor preservation, we were able to successfully single-amino-acid radiocarbon date the two rare Neanderthal individuals from Šal’a (Slovakia). By targeting hydroxyproline, potential contamination sources sometimes associated with old museum specimens and poorly preserved bones were avoided. Concerns that modern bone glue might have been used on Šal’a I were not validated, as such treatments would have led to substantially younger radiocarbon dates.
The resulting radiocarbon ages for both Šal’a I and Šal’a II firmly place the specimens in the European Middle Paleolithic, possibly predating the very young individuals from Spy (Crevecoeur et al. Reference Crevecoeur, Bayle, Rougier, Maureille, Higham, van der Plicht, De Clerck and Semal2010; Semal et al. Reference Semal, Rougier, Crevecoeur, Jungels, Flas, Hauzeur, Maureille, Germonpré, Bocherens, Pirson, Cammaert, De Clerck, Hambucken, Higham, Toussaint and van der Plicht2009; Toussaint and Pirson Reference Toussaint and Pirson2006), Ferassie (Higham et al. Reference Higham, Douka, Wood, Bronk Ramsey, Brock, Basell, Camps, Arrizabalaga, Baena, Barroso-Ruíz, Bergman, Boitard, Boscato, Caparrós, Conard, Draily, Froment, Galván, Gambassini, Garcia-Moreno, Grimaldi, Haesaerts, Holt, Iriarte-Chiapusso, Jelinek, Jordá Pardo, Maíllo-Fernández, Marom, Maroto, Menéndez, Metz, Morin, Moroni, Negrino, Panagopoulou, Peresani, Pirson, de la Rasilla, Riel-Salvatore, Ronchitelli, Santamaria, Semal, Slimak, Soler, Soler, Villaluenga, Pinhasi and Jacobi2014, SI), and Kleine Feldhöfergrotte (Schmitz et al. Reference Schmitz, Serre, Bonani, Feine, Hillgruber, Krainitzki, Paabo and Smith2002). However, the formerly late Neanderthals from Vindija (SP3563 and SP3562) turned out to be 42,700 ± 1600 BP (OxA-X-2689-09) and 43,900 ± 2000 BP (OxA-X-2689-10), respectively, when dated using the single-amino-acid method; and not 28–32,000 BP as initial dating with less rigorous pretreatment protocols suggested (Devièse et al. Reference Devièse, Karavanić, Comeskey, Kubiak, Korlević, Hajdinjak, Radović, Procopio, Buckley, Pääbo and Higham2017). More rigorous dating may therefore find those other ‘young’ Neanderthals to be close to or older than 50,000 BP. Similarly, the two Neanderthals from Mezmaiskaya (Higham et al. Reference Higham, Douka, Wood, Bronk Ramsey, Brock, Basell, Camps, Arrizabalaga, Baena, Barroso-Ruíz, Bergman, Boitard, Boscato, Caparrós, Conard, Draily, Froment, Galván, Gambassini, Garcia-Moreno, Grimaldi, Haesaerts, Holt, Iriarte-Chiapusso, Jelinek, Jordá Pardo, Maíllo-Fernández, Marom, Maroto, Menéndez, Metz, Morin, Moroni, Negrino, Panagopoulou, Peresani, Pirson, de la Rasilla, Riel-Salvatore, Ronchitelli, Santamaria, Semal, Slimak, Soler, Soler, Villaluenga, Pinhasi and Jacobi2014, SI) and Okladnikov (Krause et al. Reference Krause, Orlando, Serre, Viola, Prüfer, Richards, Hublin, Hänni, Derevianko and Pääbo2007) — currently dated to 39,000 ± 1100 BP (OxA-21839) and 37,800 ± 450 BP (OxA-15481) — may yet yield older dates. Whether Šal’a I and Šal’a II are contemporaneous with Individual I from Les Rochers-de-Villeneuve (Beauval et al. Reference Beauval, Lacrampe-Cuyaubère, Maureille and Trinkaus2006) is difficult to assess, as there remains some, but limited, overlap of the age probability distributions.
This research also concludes that the presently known Slovakian Neanderthals predate any archaeological or skeletal evidence of Modern Humans in Europe (Benazzi et al. Reference Benazzi, Douka, Fornai, Bauer, Kullmer, Svoboda, Pap, Mallegni, Bayle, Coquerelle, Condemi, Ronchitelli, Harvati and Weber2011; Hublin et al. Reference Hublin, Sirakov, Aldeias, Bailey, Bard, Delvigne, Endarova, Fagault, Fewlass, Hajdinjak, Kromer, Krumov, Marreiros, Martisius, Paskulin, Sinet-Mathiot, Meyer, Pääbo, Popov, Rezek, Sirakova, Skinner, Smith, Spasov, Talamo, Tuna, Wacker, Welker, Wilcke, Zahariev, McPherron and Tsanova2020). Apart from Šal’a I and Šal’a II, the only other evidence of Neanderthal remains from Slovakia are an endocranial cast and two long bone imprints from Gánovce (Eisová et al. Reference Eisová, Velemínský and Bruner2019; Vlček Reference Vlček1953, Reference Vlček1955, Reference Vlček1969). The endocast is presently dated to 105ka (+10.2 ka, −9.4 ka) via uranium-thorium dating of the travertine in which it was found and is associated with artifacts belonging to a Taubachian microlithic industry (Eisová et al. Reference Eisová, Velemínský and Bruner2019; Jäger Reference Jäger1989; Vlček Reference Vlček1995). While it will be challenging to obtain a more precise age for the Šal’a individuals, morphological features and possibly associated fauna may be seen as tangential evidence for inferring an age younger than the Neanderthals from Gánovce, and potentially coeval with later Micoquian and/or Mousterian Neanderthal sites. It remains to be seen whether further improvements in ancient DNA analysis will eventually lead to the identification of (highly degraded) endogenous DNA, thereby further contributing to the temporal and evolutionary placement of those precious finds from Šal’a.
CONCLUSIONS
We successfully radiocarbon dated Šal’a I to >44,800 BP (OxA-X-2731-16) and Šal’a II to >45,100 BP (OxA-X-2731-15). Nonetheless, the successful collagen and hydroxyproline extraction did not translate to sufficiently preserved DNA for analysis. Therefore, we conclude that both Neanderthal individuals firmly belong to the Middle Paleolithic, and predate late Neanderthals (e.g., from Vindija and possibly Spy) as well as early AMHs in the region.
ACKNOWLEDGMENTS
This research has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 324139 PalaeoChron awarded to Professor Tom Higham. The publication process was facilitated by the Hunt Fellowship granted to Rachel J.A. Hopkins by the Wenner Gren Foundation (Gr. 9881), and some of the travel expenses were covered by Wolfson College, Oxford. We would further like to thank Sarah Nagel and Birgit Nickel for their help in ancient DNA laboratory. The ancient DNA work was funded by the Max Planck Society and the European Research Council (grant agreement no. 694707 to Svante Pääbo). Additionally, none of this research would have been possible without the support of Mgr. Ján Kautman (Director of the Slovak National Museum – Natural History Museum, Bratislava) and Dr. Matej Ruttkay (Director of the Institute of Archaeology, Slovak Academy of Sciences, Nitra), who gave us permission to sample Šal’a I and Šal’a II, respectively. We would also like to thank museum donors and museum staff, who preserve these important finds for both the public and future research.









