Introduction
Depression and anxiety disorders (AD) are among the most common psychiatric disorders. According to the latest report by the World Health Organization, depression affects more than 300 million people worldwide and is the leading cause of disability (World Health Organization, 2017). AD affect more than 260 million people and is the sixth leading cause of disability (World Health Organization, 2017). The two disorders are highly comorbid and might share common pathophysiologies (Frances et al., Reference Frances, Manning, Marin, Kocsis, Mckinney, Hall and Kline1992; Goodwin, Reference Goodwin2015). Nevertheless, pharmacological treatment for major depressive disorder (MDD) or AD has not seen much advance in the last two decades or so, with a lack of therapies having novel mechanisms of action. In addition, only about one-third of MDD patients achieve complete remission after a single antidepressant trial (Trivedi et al., Reference Trivedi, Rush, Wisniewski, Nierenberg, Warden, Ritz, Norquist, Howland, Lebowitz, Mcgrath, Shores-Wilson, Biggs, Balasubramani, Fava and Team2006) and around 10–30% of patients are treatment-resistant (Al-Harbi, Reference Al-Harbi2012).
On the other hand, with the advent of high-throughput technologies such as genome-wide association studies (GWAS) in the last decade, we have gained a much better understanding of the genetic bases of many complex diseases. It is hoped that human genomics data will accelerate drug development for psychiatric disorders, especially due to the difficulties for animal models to fully mimic human psychiatric conditions such as depression (Papassotiropoulos and de Quervain, Reference Papassotiropoulos and De Quervain2015).
We hypothesize that gene-sets associated with antidepressants or anxiolytics, or more generally with psychiatric medications, will be enriched among the GWAS results of depression and anxiety phenotypes. If the gene-set analysis (GSA) approach is able to ‘re-discover’ known treatments, it might also be able to reveal new therapies for depression and AD. Practically, if the pathway targeted by a particular drug (or chemical) is significantly enriched in GWAS but the drug is not a known treatment for the disease, it may serve as a good candidate for repositioning.
With regards to GWAS-based drug discovery, the current focus is mainly on identification of new drug targets from the top GWAS hits. In an earlier study, Sanseau et al. (Reference Sanseau, Agarwal, Barnes, Pastinen, Richards, Cardon and Mooser2012) identified the most significant GWAS hits from a range of diseases and compared them against known drug targets to find ‘mismatches’ (i.e. drug indication different from the studied disorder) as candidates for repurposing. While it is intuitive and legitimate to focus on the most significant SNPs, for many complex traits, the genetic architecture may be highly polygenic and variants of weaker effects may be ‘hidden’. Moreover, given the complex and multifactorial etiologies of many complex diseases, the development of multi-target drugs with wide-ranging biological activities (known as ‘polypharmacology’) is gaining increased attention [please refer to, e.g. Anighoro et al. (Reference Anighoro, Bajorath and Rastelli2014) for a review]. It is argued that multi-target drugs may have improved efficacy over highly selective pharmacological agents, as they tackle multiple pathogenic pathways in the system. A gene-set or pathway-based approach to drug repositioning follows this multi-target paradigm.
GSA is an established approach to gain biological insight into expression microarrays, GWAS, or other high-throughput ‘omics’ studies (Maciejewski, Reference Maciejewski2014; de Leeuw et al., Reference De Leeuw, Neale, Heskes and Posthuma2016). De Jong et al. made use of GSA to identify repurposing opportunities for schizophrenia (de Jong et al., Reference De Jong, Vidler, Mokrab, Collier and Breen2016) and several top results were supported by the literature. In another very recent study, Gaspar and Breen (Reference Gaspar and Breen2017) performed further analyses of GSA results, and reported that GWAS signals of schizophrenia are enriched for neuropsychiatric medications as sample size increases. Another related study on schizophrenia was conducted by Ruderfer et al. (Reference Ruderfer, Charney, Readhead, Kidd, Kahler, Kenny, Keiser, Moran, Hultman, Scott, Sullivan, Purcell, Dudley and Sklar2016), who collected genome-wide significant GWAS variants and exome sequencing results and compared the identified genes against known or predicted drug targets. Significant enrichment was observed for antipsychotics.
In this study, we take a different focus on depression and AD, which are highly prevalent and disabling disorders. Besides considering individual GWAS studies of phenotypes related to depression and anxiety, we also performed analyses on combined GWAS summary statistics to improve the power.
Methods
Overview of analytic approach
Our analyses can be broadly divided into two steps. Firstly, we extracted gene-sets associated with a variety of drugs (or chemicals), and tested whether the gene-set associated with each individual drug is enriched among the GWAS results. The drugs ranked among the top (i.e. those with lower p values) were considered potential candidates for repositioning. In the second step, we performed analyses on the drugs. We evaluated the prioritized drugs and tested which drug classes were enriched. As we have hypothesized above, we would specifically test whether antidepressants/anxiolytics and other psychiatric medications were enriched among the repositioning results. Figure 1 describes an overview of our analytic approach.

Fig. 1. A schematic diagram showing our analysis workflow. We first extracted gene-sets associated with a variety of drugs (and chemicals), and examined whether the gene-set associated with each drug is enriched among the GWAS results. The drugs ranked among the top were considered potential candidates for repositioning, and a systemic literature search was performed for the top 20 repositioning hits of each psychiatric trait and meta-analyzed results. We also evaluated which drug classes were enriched. We specifically tested whether antidepressants/anxiolytics and other psychiatric medications were enriched, and then also examined all ATC (level 3) drug classes for enrichment in a hypothesis-free manner (ATC, Anatomical Therapeutic Chemical Classification System).
GWAS data
In the main analysis, we considered five GWAS datasets that are associated with depression and anxiety. Two are studies of MDD, namely MDD-2018 (Wray and Sullivan, Reference Wray, Ripke, Mattheisen, Trzaskowski, Byrne, Abdellaoui, Adams, Agerbo, Air, Andlauer, Bacanu, Baekvad-Hansen, Beekman, Bigdeli, Binder, Blackwood, Bryois, Buttenschon, Bybjerg-Grauholm, Cai, Castelao, Christensen, Clarke, Coleman, Colodro-Conde, Couvy-Duchesne, Craddock, Crawford, Crowley, Dashti, Davies, Deary, Degenhardt, Derks, Direk, Dolan, Dunn, Eley, Eriksson, Escott-Price, Kiadeh, Finucane, Forstner, Frank, Gaspar, Gill, Giusti-Rodriguez, Goes, Gordon, Grove, Hall, Hannon, Hansen, Hansen, Herms, Hickie, Hoffmann, Homuth, Horn, Hottenga, Hougaard, Hu, Hyde, Ising, Jansen, Jin, Jorgenson, Knowles, Kohane, Kraft, Kretzschmar, Krogh, Kutalik, Lane, Li, Li, Lind, Liu, Lu, MacIntyre, MacKinnon, Maier, Maier, Marchini, Mbarek, McGrath, McGuffin, Medland, Mehta, Middeldorp, Mihailov, Milaneschi, Milani, Mill, Mondimore, Montgomery, Mostafavi, Mullins, Nauck, Ng, Nivard, Nyholt, O'Reilly, Oskarsson, Owen, Painter, Pedersen, Pedersen, Peterson, Pettersson, Peyrot, Pistis, Posthuma, Purcell, Quiroz, Qvist, Rice, Riley, Rivera, Saeed Mirza, Saxena, Schoevers, Schulte, Shen, Shi, Shyn, Sigurdsson, Sinnamon, Smit, Smith, Stefansson, Steinberg, Stockmeier, Streit, Strohmaier, Tansey, Teismann, Teumer, Thompson, Thomson, Thorgeirsson, Tian, Traylor, Treutlein, Trubetskoy, Uitterlinden, Umbricht, Van der Auwera, van Hemert, Viktorin, Visscher, Wang, Webb, Weinsheimer, Wellmann, Willemsen, Witt, Wu, Xi, Yang, Zhang, Arolt, Baune, Berger, Boomsma, Cichon, Dannlowski, de Geus, DePaulo, Domenici, Domschke, Esko, Grabe, Hamilton, Hayward, Heath, Hinds, Kendler, Kloiber, Lewis, Li, Lucae, Madden, Magnusson, Martin, McIntosh, Metspalu, Mors, Mortensen, Muller-Myhsok, Nordentoft, Nothen, O'Donovan, Paciga, Pedersen, Penninx, Perlis, Porteous, Potash, Preisig, Rietschel, Schaefer, Schulze, Smoller, Stefansson, Tiemeier, Uher, Volzke, Weissman, Werge, Winslow, Lewis, Levinson, Breen, Borglum and Sullivan2017) and MDD-CONVERGE (Cai et al., Reference Cai, Bigdeli, Kretzschmar, Li, Liang, Song, Hu, Li, Jin, Hu, Wang, Wang, Qian, Liu, Jiang, Lu, Zhang, Yin, Li, Xu, Gao, Reimers, Webb, Riley, Bacanu, Peterson, Chen, Zhong, Liu, Wang, Sun, Sang, Jiang, Zhou, Li, Li, Zhang, Wang, Fang, Pan, Miao, Zhang, Hu, Yu, Du, Sang, Li, Chen, Cai, Yang, Yang, Ha, Hong, Deng, Li, Li, Song, Gao, Zhang, Gan, Meng, Pan, Gao, Zhang, Sun, Li, Niu, Zhang, Liu, Hu, Zhang, Lv, Dong, Wang, Tao, Wang, Xia, Rong, He, Liu, Huang, Mei, Shen, Liu, Shen, Tian, Liu, Wu, Gu, Fu, Shi, Chen, Gan, Liu, Wang, Yang, Cong, Marchini, Yang and Wang2015). However, the samples included in these studies are different. MDD-CONVERGE (N = 10 640) is a cohort of Chinese women, and this sample mainly consists of hospital-ascertained cases affected by severe depression, of whom ~85% had melancholic symptoms (Cai et al., Reference Cai, Bigdeli, Kretzschmar, Li, Liang, Song, Hu, Li, Jin, Hu, Wang, Wang, Qian, Liu, Jiang, Lu, Zhang, Yin, Li, Xu, Gao, Reimers, Webb, Riley, Bacanu, Peterson, Chen, Zhong, Liu, Wang, Sun, Sang, Jiang, Zhou, Li, Li, Zhang, Wang, Fang, Pan, Miao, Zhang, Hu, Yu, Du, Sang, Li, Chen, Cai, Yang, Yang, Ha, Hong, Deng, Li, Li, Song, Gao, Zhang, Gan, Meng, Pan, Gao, Zhang, Sun, Li, Niu, Zhang, Liu, Hu, Zhang, Lv, Dong, Wang, Tao, Wang, Xia, Rong, He, Liu, Huang, Mei, Shen, Liu, Shen, Tian, Liu, Wu, Gu, Fu, Shi, Chen, Gan, Liu, Wang, Yang, Cong, Marchini, Yang and Wang2015). The MDD-2018 (N = 173 005, for which full summary statistics are available) samples are composed of Caucasians of both sexes, and they are more heterogeneous and not specifically enriched for any subtypes of depression (Wray et al., Reference Ripke, Wray, Lewis, Hamilton, Weissman, Breen, Byrne, Blackwood, Boomsma, Cichon, Heath, Holsboer, Lucae, Madden, Martin, McGuffin, Muglia, Noethen, Penninx, Pergadia, Potash, Rietschel, Lin, Müller-Myhsok, Shi, Steinberg, Grabe, Lichtenstein, Magnusson, Perlis, Preisig, Smoller, Stefansson, Uher, Kutalik, Tansey, Teumer, Viktorin, Barnes, Bettecken, Binder, Breuer, Castro, Churchill, Coryell, Craddock, Craig, Czamara, De Geus, Degenhardt, Farmer, Fava, Frank, Gainer, Gallagher, Gordon, Goryachev, Gross, Guipponi, Henders, Herms, Hickie, Hoefels, Hoogendijk, Hottenga, Iosifescu, Ising, Jones, Jones, Jung-Ying, Knowles, Kohane, Kohli, Korszun, Landen, Lawson, Lewis, Macintyre, Maier, Mattheisen, McGrath, McIntosh, McLean, Middeldorp, Middleton, Montgomery, Murphy, Nauck, Nolen, Nyholt, O'Donovan, Oskarsson, Pedersen, Scheftner, Schulz, Schulze, Shyn, Sigurdsson, Slager, Smit, Stefansson, Steffens, Thorgeirsson, Tozzi, Treutlein, Uhr, van den Oord, Van Grootheest, Völzke, Weilburg, Willemsen, Zitman, Neale, Daly, Levinson and Sullivan2018). The MDD-2018 study represents the largest and most recent GWAS meta-analysis on MDD, and includes all subjects from an earlier GWAS, i.e. the MDD-PGC-2012 study by Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al. (Reference Ripke, Wray, Lewis, Hamilton, Weissman, Breen, Byrne, Blackwood, Boomsma, Cichon, Heath, Holsboer, Lucae, Madden, Martin, McGuffin, Muglia, Noethen, Penninx, Pergadia, Potash, Rietschel, Lin, Müller-Myhsok, Shi, Steinberg, Grabe, Lichtenstein, Magnusson, Perlis, Preisig, Smoller, Stefansson, Uher, Kutalik, Tansey, Teumer, Viktorin, Barnes, Bettecken, Binder, Breuer, Castro, Churchill, Coryell, Craddock, Craig, Czamara, De Geus, Degenhardt, Farmer, Fava, Frank, Gainer, Gallagher, Gordon, Goryachev, Gross, Guipponi, Henders, Herms, Hickie, Hoefels, Hoogendijk, Hottenga, Iosifescu, Ising, Jones, Jones, Jung-Ying, Knowles, Kohane, Kohli, Korszun, Landen, Lawson, Lewis, Macintyre, Maier, Mattheisen, McGrath, McIntosh, McLean, Middeldorp, Middleton, Montgomery, Murphy, Nauck, Nolen, Nyholt, O'Donovan, Oskarsson, Pedersen, Scheftner, Schulz, Schulze, Shyn, Sigurdsson, Slager, Smit, Stefansson, Steffens, Thorgeirsson, Tozzi, Treutlein, Uhr, van den Oord, Van Grootheest, Völzke, Weilburg, Willemsen, Zitman, Neale, Daly, Levinson and Sullivan2013) (N = 18 759). Note that for MDD-2018, full summary statistics (on which the current analysis is based) are only publicly available for samples excluding 23andMe subjects.
Another two GWAS studies were meta-analyses on depressive symptoms and neuroticism conducted by the Social Science Genetics Association Consortium (SSGAC) (Okbay et al., Reference Okbay, Baselmans, De Neve, Turley, Nivard, Fontana, Meddens, Linner, Rietveld, Derringer, Gratten, Lee, Liu, de Vlaming, Ahluwalia, Buchwald, Cavadino, Frazier-Wood, Furlotte, Garfield, Geisel, Gonzalez, Haitjema, Karlsson, van der Laan, Ladwig, Lahti, van der Lee, Lind, Liu, Matteson, Mihailov, Miller, Minica, Nolte, Mook-Kanamori, van der Most, Oldmeadow, Qian, Raitakari, Rawal, Realo, Rueedi, Schmidt, Smith, Stergiakouli, Tanaka, Taylor, Thorleifsson, Wedenoja, Wellmann, Westra, Willems, Zhao, LifeLines Cohort, Amin, Bakshi, Bergmann, Bjornsdottir, Boyle, Cherney, Cox, Davies, Davis, Ding, Direk, Eibich, Emeny, Fatemifar, Faul, Ferrucci, Forstner, Gieger, Gupta, Harris, Harris, Holliday, Hottenga, De Jager, Kaakinen, Kajantie, Karhunen, Kolcic, Kumari, Launer, Franke, Li-Gao, Liewald, Koini, Loukola, Marques-Vidal, Montgomery, Mosing, Paternoster, Pattie, Petrovic, Pulkki-Raback, Quaye, Raikkonen, Rudan, Scott, Smith, Sutin, Trzaskowski, Vinkhuyzen, Yu, Zabaneh, Attia, Bennett, Berger, Bertram, Boomsma, Snieder, Chang, Cucca, Deary, van Duijn, Eriksson, Bultmann, de Geus, Groenen, Gudnason, Hansen, Hartman, Haworth, Hayward, Heath, Hinds, Hypponen, Iacono, Jarvelin, Jockel, Kaprio, Kardia, Keltikangas-Jarvinen, Kraft, Kubzansky, Lehtimaki, Magnusson, Martin, McGue, Metspalu, Mills, de Mutsert, Oldehinkel, Pasterkamp, Pedersen, Plomin, Polasek, Power, Rich, Rosendaal, den Ruijter, Schlessinger, Schmidt, Svento, Schmidt, Alizadeh, Sorensen, Spector, Starr, Stefansson, Steptoe, Terracciano, Thorsteinsdottir, Thurik, Timpson, Tiemeier, Uitterlinden, Vollenweider, Wagner, Weir, Yang, Conley, Smith, Hofman, Johannesson, Laibson, Medland, Meyer, Pickrell, Esko, Krueger, Beauchamp, Koellinger, Benjamin, Bartels and Cesarini2016). The meta-analysis on depressive symptoms (SSGAC-DS) included the MDD-PGC-2012 study (N = 18 759) and a case–control sample from the Genetic Epidemiology Research on Aging (GERA) Cohort (N = 56 368), and it also comprised a UK BioBank sample made up of general population (N = 105 739). Depressive symptoms were measured by a self-reported questionnaire. We also included another study on neuroticism (SSGAC-NEU) (N = 170 906), as this personality trait is known to be closely associated with depression and AD (Lahey, Reference Lahey2009). In addition, antidepressants may affect personality traits, including a reduction in neuroticism, independent of their effects on depressive symptoms (Tang et al., Reference Tang, Derubeis, Hollon, Amsterdam, Shelton and Schalet2009). The aim of including SSGAC-DS and SSGAC-NEU is to investigate whether the study of depression/anxiety-related phenotypes in the general population, other than clinical depression or AD, may affect the potential of guiding drug discovery. An analogous example in cardiovascular medicine is that some genetic variants associated with raised fasting glucose may not be linked to diabetes (Florez et al., Reference Florez, Jablonski, Mcateer, Franks, Mason, Mather, Horton, Goldberg, Dabelea, Kahn, Arakaki, Shuldiner and Knowler2012; Merino et al., Reference Merino, Leong, Posner, Porneala, Masana, Dupuis and Florez2017); hence the results from our gene-set and drug repositioning analysis might also differ when different phenotypes are studied.
The last dataset is a GWAS meta-analysis of AD, including generalized AD, panic disorder, social phobia, agoraphobia, and specific phobias (Otowa et al., Reference Otowa, Hek, Lee, Byrne, Mirza, Nivard, Bigdeli, Aggen, Adkins, Wolen, Fanous, Keller, Castelao, Kutalik, Der Auwera, Homuth, Nauck, Teumer, Milaneschi, Hottenga, Direk, Hofman, Uitterlinden, Mulder, Henders, Medland, Gordon, Heath, Madden, Pergadia, Van Der Most, Nolte, Van Oort, Hartman, Oldehinkel, Preisig, Grabe, Middeldorp, Penninx, Boomsma, Martin, Montgomery, Maher, Van Den Oord, Wray, Tiemeier and Hettema2016b). We extracted the GWAS results of the case–control analyses (N = 17 310).
GWAS summary results were downloaded from https://www.med.unc.edu/pgc/results-and-downloads and https://www.thessgac.org/data.
Extracting gene-sets associated with each drug
We made use of the DSigDB database (Yoo et al., Reference Yoo, Shin, Kim, Ryall, Lee, Lee, Jeon, Kang and Tan2015) to extract gene-sets related to each drug. DSigDB holds gene-sets for a total of 17 839 unique compounds. The gene-sets were compiled according to multiple sources: (1) bioassay results from PubChem (Kim et al., Reference Kim, Thiessen, Bolton, Chen, Fu, Gindulyte, Han, He, He, Shoemaker, Wang, Yu, Zhang and Bryant2016) and ChEMBL (Gaulton et al., Reference Gaulton, Bellis, Bento, Chambers, Davies, Hersey, Light, Mcglinchey, Michalovich, Al-Lazikani and Overington2012); (2) kinase profiling assay from the literature and two kinase databases (Medical Research Council Kinase Inhibitor database and Harvard Medical School Library of Integrated Network-based Cellular Signatures database); (3) differentially expressed genes after drug treatment (with >2 fold-change compared with controls), as derived from the Connectivity Map (Lamb et al., Reference Lamb, Crawford, Peck, Modell, Blat, Wrobel, Lerner, Brunet, Subramanian, Ross, Reich, Hieronymus, Wei, Armstrong, Haggarty, Clemons, Wei, Carr, Lander and Golub2006); and (4) manually curated and text mined drug targets from the Therapeutics Targets Database (Li et al., Reference Li, Yu, Li, Zhang, Tang, Yang, Fu, Zhang, Cui, Tu, Zhang, Li, Yang, Sun, Qin, Zeng, Chen, Chen and Zhu2018) and the Comparative Toxicogenomics Database (Mattingly et al., Reference Mattingly, Colby, Forrest and Boyer2003). We downloaded the entire database from http://tanlab.ucdenver.edu/DSigDB. The above sources captured different aspects of drug properties, for example, differentially expressed genes from perturbation experiments might be different from drug target genes defined in bioassay studies. Besides performing analyses based on the whole database which incorporates a broad definition of drug-related genes, we also performed a separate enrichment analysis for genes derived from PubChem and ChEMBL only, as they represent conventionally defined ‘drug targets’ that are more well-studied and perhaps more directly associated with drug actions.
It should be noted that although the focus is on drug ‘repositioning’, the analytic framework is general and can apply to any drugs with some known associated genes. Indeed DSigDB contains a substantial number of drugs which do not have an approved indication yet, which were still included in our analyses.
GSA approach
We first converted the SNP-based test results to gene-based test results. We employed fastBAT (Bakshi et al., Reference Bakshi, Zhu, Vinkhuyzen, Hill, Mcrae, Visscher and Yang2016) (included in the software package GCTA) for gene-based analyses. FastBAT computes the sum of χ2 statistics over all SNPs within a gene and uses an analytic approach to compute the p value. Gene size and linkage disequilibrium patterns are taken into account when computing the p values. The same statistical approach for gene-based tests is also used by two other popular programs, VEGAS (Liu et al., Reference Liu, Mcrae, Nyholt, Medland, Wray, Brown, Hayward, Montgomery, Visscher, Martin, Macgregor and Investigators2010) and PLINK (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, De Bakker, Daly and Sham2007), although they computed p values by simulations or permutations. FastBAT has been shown to be equivalent to VEGAS and PLINK at higher p values (>1E-06) and more accurate than these two programs for smaller p (Bakshi et al., Reference Bakshi, Zhu, Vinkhuyzen, Hill, Mcrae, Visscher and Yang2016). We ran fastBAT with the default settings and used the 1000 Genomes genotype data as the reference panel.
We then performed a standard GSA by comparing gene-based test statistics within and outside the gene-set. We adopted the same approach as implemented in MAGMA (de Leeuw et al., Reference De Leeuw, Mooij, Heskes and Posthuma2015), which is also reviewed in de Leeuw et al. (Reference De Leeuw, Neale, Heskes and Posthuma2016). Briefly, gene-based p values are first converted to z-statistics by z = Φ−1(p), where Φ−1 is the probit function (more negative z-values represent stronger statistical associations). We then employed a single-sided two-sample t test to see if the mean z-statistics of genes within the gene-set is lower than that outside the gene-set. To avoid results driven by only a few genes, we only considered drugs with at least five genes in their gene-sets. A total of 5232 drugs were included for final analyses.
Combining p values across datasets
Besides analyzing each GWAS dataset in turn for repositioning opportunities, we also considered the aggregate contribution of all datasets, as depression, anxiety, and neuroticism are closely connected to each other. Depression and AD are highly clinically comorbid (Lamers et al., Reference Lamers, Van Oppen, Comijs, Smit, Spinhoven, Van Balkom, Nolen, Zitman, Beekman and Penninx2011; Kessler et al., Reference Kessler, Sampson, Berglund, Gruber, Al-Hamzawi, Andrade, Bunting, Demyttenaere, Florescu, De Girolamo, Gureje, He, Hu, Huang, Karam, Kovess-Masfety, Lee, Levinson, Medina Mora, Moskalewicz, Nakamura, Navarro-Mateu, Browne, Piazza, Posada-Villa, Slade, Ten Have, Torres, Vilagut, Xavier, Zarkov, Shahly and Wilcox2015), demonstrate significant genetic correlations (Otowa et al., Reference Otowa, Hek, Lee, Byrne, Mirza, Nivard, Bigdeli, Aggen, Adkins, Wolen, Fanous, Keller, Castelao, Kutalik, Der Auwera, Homuth, Nauck, Teumer, Milaneschi, Hottenga, Direk, Hofman, Uitterlinden, Mulder, Henders, Medland, Gordon, Heath, Madden, Pergadia, Van Der Most, Nolte, Van Oort, Hartman, Oldehinkel, Preisig, Grabe, Middeldorp, Penninx, Boomsma, Martin, Montgomery, Maher, Van Den Oord, Wray, Tiemeier and Hettema2016a), and share similar pharmacological treatments (Ballenger, Reference Ballenger2000). As mentioned above, neuroticism is associated with both depression and anxiety, and levels of neuroticism may be affected by antidepressant treatment. Studies also suggest a shared genetic basis between neuroticism and depression or AD (Jardine et al., Reference Jardine, Martin, Henderson and Rao1984; Gale et al., Reference Gale, Hagenaars, Davies, Hill, Liewald, Cullen, Penninx, Boomsma, Pell, Mcintosh, Smith, Deary, Harris and Gwas2016).
The combined analysis is complementary to the study of individual phenotypes. Based on previous studies and as discussed earlier, depression, anxiety, and neuroticism may have some common biological basis, although most likely their etiologies do not completely overlap. A combined analysis may improve the power to detect drugs which target the shared pathophysiological processes, by increasing the total sample size. On the other hand, study of individual phenotypes may reveal drugs or drug classes that are specifically useful for particular disease symptoms or subgroups, for example, melancholic depression.
We performed meta-analysis of p values based on two methods, the Simes’ method (Simes, Reference Simes1986) and the Brown's approach (Brown, Reference Brown1975; Poole et al., Reference Poole, Gibbs, Shmulevich, Bernard and Knijnenburg2016). The Simes’ method is valid under positive regression dependencies (Sarkar and Chang, Reference Sarkar and Chang1997). Brown's method is similar to Fisher's method but also accounts for dependencies in p values. Briefly, assuming k p values, Fisher showed that the statistic ![]() $T = \sum {-2\log P_i} $ should follow a χ2 distribution with 2k degrees of freedom if the p values are independent. Brown's method is an extension of Fisher's approach by estimating the statistic T with a re-scaled χ2 distribution
$T = \sum {-2\log P_i} $ should follow a χ2 distribution with 2k degrees of freedom if the p values are independent. Brown's method is an extension of Fisher's approach by estimating the statistic T with a re-scaled χ2 distribution ![]() $T\sim c{\rm \chi} _{2f}^2 $ where
$T\sim c{\rm \chi} _{2f}^2 $ where
The expectation of the statistic T can be estimated by E(T) = 2k and the variance by ![]() ${\mathop{\rm var}} (T) = 4k + 2\sum\limits_{i \lt j} {{\mathop{\rm cov}} (-2\log P_i,-2\log P_j)} $. We estimated the covariance empirically from the observed p value vectors of different phenotypes. The Simes’ method is more powerful when there are few false null hypotheses (H0) (i.e. few true associations) whereas Fisher's or Brown's method are more powerful when there are multiple false H0.
${\mathop{\rm var}} (T) = 4k + 2\sum\limits_{i \lt j} {{\mathop{\rm cov}} (-2\log P_i,-2\log P_j)} $. We estimated the covariance empirically from the observed p value vectors of different phenotypes. The Simes’ method is more powerful when there are few false null hypotheses (H0) (i.e. few true associations) whereas Fisher's or Brown's method are more powerful when there are multiple false H0.
The MDD-CONVERGE sample includes only Chinese subjects and does not overlap with other datasets, but we accounted for overlapping samples (with Brown's or Simes’ method) in the remaining GWAS studies.
Testing for enrichment of psychiatric and other drug classes
We considered three sources when defining psychiatric drug-sets in our analyses. The first set came from drugs listed in the Anatomical Therapeutic Classification (ATC) system downloaded from KEGG. We extracted three groups of drugs: (1) all psychiatric drugs (coded ‘N05’ or ‘N06’); (2) antipsychotics (coded ‘N05A’); (3) antidepressants and anxiolytics (coded ‘N05B’ or ‘N06A’). The second source was from MEDication Indication resource (MEDI) (Wei et al., Reference Wei, Cronin, Xu, Lasko, Bastarache and Denny2013) derived from four public medication resources, namely RxNorm, Side Effect Resource 2 (SIDER2), Wikipedia, and MedlinePlus. A random subset of the extracted indications was checked by physicians. The MEDI high-precision subset (MEDI-HPS), with an estimated curation precision of 92%, was used in our analyses (Wei et al., Reference Wei, Cronin, Xu, Lasko, Bastarache and Denny2013). Since only known drug indications are included in ATC or MEDI-HPS, we also included an expanded set of drugs that are considered for clinical trials (as listed on https://clinicaltrials.gov). These drugs are usually promising candidates supported by preclinical or clinical studies. A precompiled list of these drugs was obtained from https://doi.org/10.15363/thinklab.d212. We also examined enrichment for closely related disorders in combinations, including schizophrenia with bipolar disorder (BD), as well as depression with anxiety.
The above represents a hypothesis-driven analysis of psychiatric drug classes. To explore whether other drug groups may be repositioned for disease treatment, we also performed a comprehensive enrichment analysis of all ATC level 3 drug classes. To avoid the results being driven by too few drugs in a class, we limited the analyses to drug classes with at least five members. Note that in the previous step we have found individual drugs as repositioning candidates, but we also hope to find out which drug classes may be promising for repositioning, which can also provide insights into potentially new mechanisms of actions in future development. A total of 129 and 79 ATC drug classes (for gene-sets derived from the entire DSigDB and PubChem/ChEMBL, respectively) were included in the final analyses.
We performed enrichment tests of repositioning hits for known drug classes, in a manner similar to the GSA described above. The p values are first converted to z-statistics, and the mean z-score within each drug class is compared against the theoretical null of zero (self-contained test) and against other drugs outside the designated drug class (competitive test) with one-sided tests.
In the context of GSA, the self-contained test examines the null hypothesis that none of the genes in the gene-set are associated with the phenotype, while the competitive test examines the null hypothesis that genes in the set are no more strongly associated with the phenotype than those outside the set. If only a small number of genes contribute to the phenotype, the difference between the two tests is small. However, if the trait is highly polygenic (which may be the case for many complex traits), the competitive test is more appropriate and usually preferred for revealing the biological processes involved. Using clinical trials as an analogy, self-contained analysis is akin to a treatment-only design, whereas the competitive analysis is akin to a treatment versus control design.
Here we are testing for enrichment of sets of drugs, and there is one complication to be noted. It is reasonable to believe that the current antidepressants or anxiolytics are not the only drugs that have therapeutic effects; in other words, a certain proportion of drugs in the ‘competing set’ might also have therapeutic potential against depression or anxiety. Therefore, results of the competitive tests should be interpreted with this potential limitation in mind. In this paper, we presented the drug-set enrichment results of both self-contained and competitive tests. Ideally, the results will be significant regardless of the type of analysis (i.e. large ‘treatment’ effects as well as an edge over the ‘control’ group, or in our case, significant enrichment for each psychiatric drug gene-set as well as stronger enrichment over non-psychiatric drug gene-sets).
Literature support of results
We extracted the top 20 repositioning hits of each psychiatric trait and meta-analyzed results, with drug-related gene-sets derived from either the entire database or drug targets defined by PubChem or ChEMBL. We performed a systematic search in PubMed and Google scholar using the following terms: Drug_name AND (depression OR depressive OR antidepressant OR anxiety OR panic OR phobia OR anxiolytic). References therein were looked up as necessary. The search was performed in June to August 2017 and March 2018 (for MDD-2018 results).
A limitation of manual search is that assessment of all drugs is almost impossible due to the time involved. As will be discussed later, we revealed literature support for the top repositioning candidates, but it could be argued that evidence of support may also be found for lower ranking drugs. One may also wish to evaluate whether drugs with less (or no) support by the literature are associated with weaker significance (i.e. higher p values) in our analysis. These drugs are roughly analogous to ‘negative controls’ in an experiment.
As a complementary strategy to manual literature search of the top results, we conducted an ‘automated’ literature search on all drugs in PubMed in an unbiased manner. We extracted the number of research articles supporting each drug's association with depression or AD using a Python script. To enhance specificity of the search, we employed MeSH terms for the search using the following terms: [‘drug_name’(Title/Abstract)] AND [anxiety(MeSH Terms) OR anxiety disorders(MeSH Terms) OR depressive disorder major(MeSH Terms) OR depressive disorder(MeSH Terms)] AND [therapeutics(MeSH Terms) OR antidepressive agents(MeSH Terms) OR anti-anxiety agents(MeSH Terms)]. We then examined the correlation between the number of research articles of support and p value for each drug. We hypothesize that drugs with lower p values (i.e. stronger candidates from our analysis) will correspond to a greater number of articles retrieved from PubMed, leading to negative correlations. Similar approaches for validating repositioning candidates based on literature mining have also been used in other studies (e.g. Huang et al., Reference Huang, Li, Sheng, Xia, Ma, Zhan and Wong2014). As the number of articles is typically skewed and not normally distributed, we employed Spearman and Kendall correlation measures, which are based on ranks of observations. In addition, we compared the p values of drugs with no article support versus those with at least one article support, and hypothesized that the former group would have higher p values. We used the Wilcoxon rank-sum test for such comparison. Statistical tests were carried out in R and tests were one-tailed.
We note that the number of research articles is only a crude proxy of the level of literature support. This is considered a secondary analysis and the results should be interpreted with these limitations in mind.
Correction for multiple testing
We employed the false discovery rate (FDR) approach to account for multiple testing (Benjamini and Hochberg, Reference Benjamini and Hochberg1995). The FDR approach controls the expected proportion of false positives among those declared to be significant (with consideration of the multiple testing performed). It has been widely adopted in genomic studies (So and Sham, Reference So and Sham2011). As a simple example, if there are 100 hypotheses with ‘FDR-adjusted p value’ ⩽0.05, we would expect that among these 100 hypotheses, there are ⩽5% (i.e. five) false positives. ‘FDR-adjusted p values’ (also known as ‘q-values’) were computed by the R function p.adjust with the Benjamini–Hochberg procedure (Benjamini and Hochberg, Reference Benjamini and Hochberg1995). The primary threshold for FDR-adjusted p values was set at 0.05, while results with FDR-adjusted p between 0.05 and 0.1 were regarded as suggestive associations.
Results
Enrichment of psychiatric drug classes among the drugs repositioned from GSA
Tables 1–3 show the FDR-adjusted p values for enrichment of major psychiatric drug classes among the drugs repositioned from GSA. Gene-sets were derived from the entire DSigDB. We observed that the drugs repositioned from most GWAS of anxiety and depressive traits are generally enriched for known psychiatric medications.
Table 1. Enrichment of repositioning hits derived from GWAS of major depressive disorder (MDD) and depressive symptoms for psychiatric drug categories (FDR-adjusted p values presented; gene-sets derived from entire DSigDB)
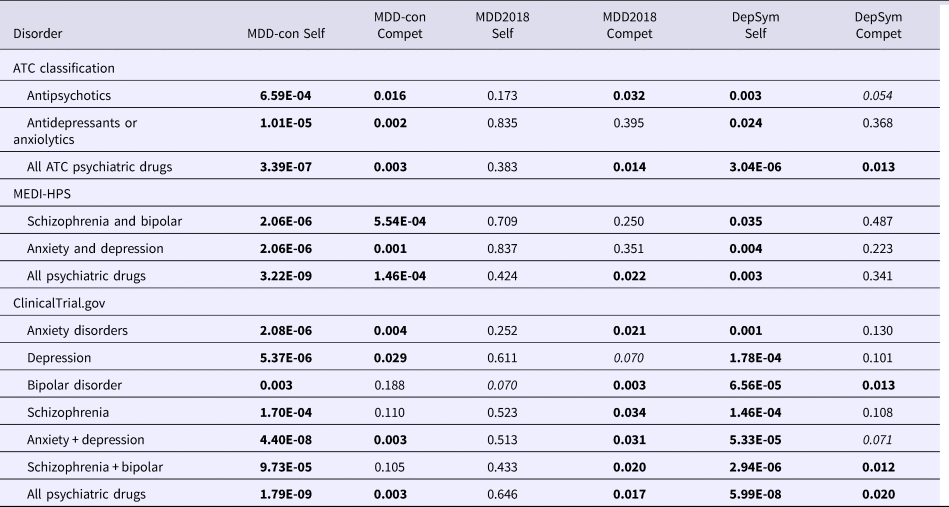
Note that we present the FDR-adjusted p values, which are already corrected for multiple testing. Gene-sets were derived from the entire DSigDB. We used three databases to define psychiatric drug categories. ATC and MEDI-HPS records known psychiatric drugs while clinicalTrial.gov records the drugs that were tested in clinical trials. Test results with FDR < 0.05 are in bold. Results with FDR between 0.05 and 0.1 are in italics.
Self: self-contained test; Compet, competitive test. In the context of gene-set analysis, the self-contained test examines the null hypothesis that none of the genes in the gene-set are associated with the phenotype, while the competitive test examines the null hypothesis that genes in the set are no more strongly associated with the phenotype than those outside the set. MDD-CONVERGE, MDD with GWAS data from the CONVERGE Consortium; MDD with GWAS data from the Psychiatric Genomics Consortium; DepSym, GWAS of depressive symptoms from the Social Science Genetics Association Consortium (SSGAC).
Table 2. Enrichment of repositioning hits derived from GWAS of anxiety disorders and neuroticism for psychiatric drug categories (FDR-adjusted p values presented; gene-sets derived from entire DSigDB)
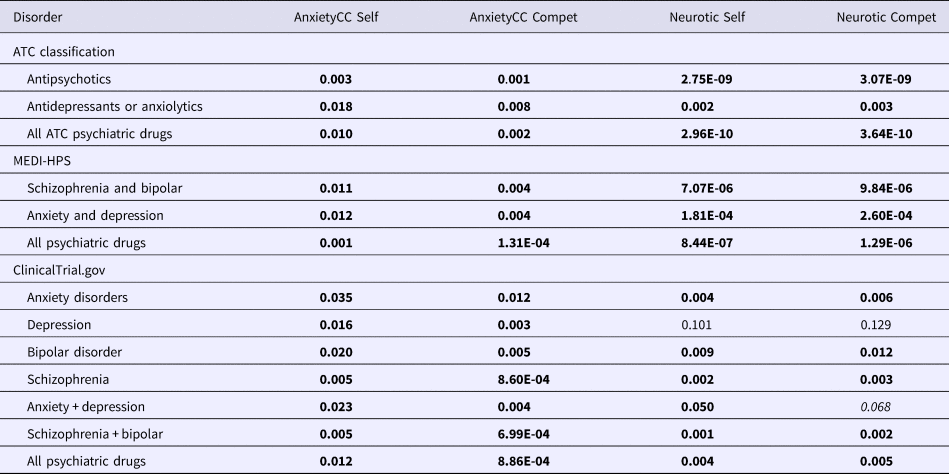
The FDR-adjusted p values are presented, which are already corrected for multiple testing. Gene-sets were derived from the entire DSigDB.
Test results with FDR < 0.05 are in bold. Results with FDR between 0.05 and 0.1 are in italics. Self: self-contained test; Compet, competitive test. Anxiety CC, GWAS of anxiety disorders case–control sample; neurotic, GWAS of neuroticism in general population.
Table 3. Enrichment of repositioning hits derived from meta-analyzed GWAS p values for psychiatric drug categories (FDR-adjusted p values presented; gene-sets derived from entire DSigDB)
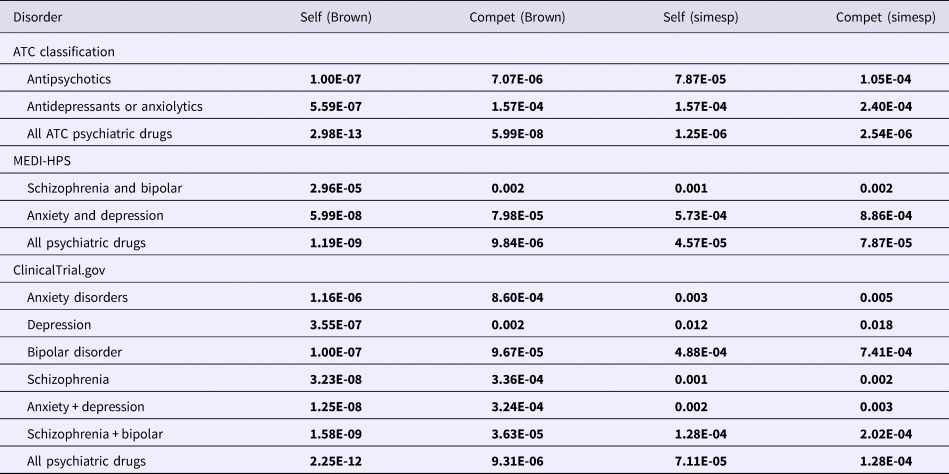
Self, self-contained test; Compet, competitive test.
The presented Brown and Simes p values are derived from meta-analyzing all five datasets, i.e. MDD-CONVERGE, MDD2018, DepSym, AnxietyCC, and neuroticism.
The FDR-adjusted p values are presented, which are already corrected for multiple testing. Gene-sets were derived from the entire DSigDB.
Test results with FDR < 0.05 are in bold. Results with FDR between 0.05 and 0.1 are in italics.
First we consider the datasets (MDD-CONVERGE, MDD2018, SSGAC-DS) which focus on depression traits (Table 1). We observed significant enrichment for various psychiatric drug classes in all the above datasets. The MDD-CONVERGE sample showed significant enrichment in both self-contained and competitive analyses. Significant results were seen for antipsychotics and antidepressants/anxiolytics within ATC and MEDI-HPS categories. For SSGAC-DS, we observed enrichment of drugs for schizophrenia and BD, and suggestive associations with medications for anxiety and depression listed in clinicalTrial.gov. For the MDD2018 sample, we also observed enrichment for antipsychotics and most psychiatric drug groups listed in clinicalTrial.gov. As a subsidiary analysis, we also examined whether there is any enrichment for psychiatric drugs based on the much smaller MDD-PGC-2012 sample, but no significant results were observed (online Supplementary Table S4)
As for GWAS studies on neuroticism (SSGAC-NEU) and AD (Table 2), there was evidence of enrichment for most psychiatric drug classes under study. Interestingly, for neuroticism, the strongest enrichment was for antipsychotics (lowest FDR-adjusted p = 2.28E-09) instead of antidepressants.
For analyses involving meta-analyzed GWAS data across all datasets (Table 3), enrichment was observed for all psychiatric drug classes, with generally stronger or at least comparable statistical associations when compared with enrichment tests of individual GWAS. The results of Brown's and Simes’ tests were largely consistent with each other.
We also performed an analysis based on drug target genes derived from PubChem or ChEBML only. The results (FDR-adjusted p values) were given in online Supplementary Tables S1–S3 and were largely similar to the above findings.
Table 4 and online Supplementary Table S5 show the results of enrichment tests across all ATC level 3 drug classes based on gene-sets derived from the whole DSigDB database. Table 5 and online Supplementary Table S6 show the findings from the same analysis but with gene-sets derived from PubChem or ChEBML only. We consider results from both of the analyses here. (Note that we mainly focus on the top 5 ATC drug classes, in contrast to top 20 repositioning candidates in the subsequent section, as the number of drug classes considered was smaller than the number of drugs.) For MDD-CONVERGE, drug classes pertaining to corticosteroids were ranked highly. Notably, antidepressants and antipsychotics were ranked within the top 5 in our two sets of analyses. For MDD-PGC, anti-infective and anti-inflammatory agents were significantly enriched. Interestingly, hypnotics/sedatives and anxiolytics, which were commonly used as short-term therapies especially at the acute phase of illness (Kanba, Reference Kanba2004), are also ranked among the top. Antidepressants or other psychiatric drugs however were not enriched. For MDD2018, interestingly, antiepileptics was ranked the highest. Other top-ranked drug classes that may be clinically relevant include antimigraine preparations, hypnotics, and dopaminergic agents.
Table 4. Top 5 enriched ATC drug classes (gene-sets derived from the whole DSigDB database)

The FDR-adjusted p values are presented, which are already corrected for multiple testing. Test results with FDR < 0.05 are in bold. Results with FDR between 0.05 and 0.1 are in italics. Self, self-contained test; Compet, competitive test. The definitions of self-contained and competitive tests are very similar to those in Tables 1–3. In the context, the above ‘drug-group’ analysis, the self-contained test examines the null hypothesis that none of the drugs in the group are associated with repositioning potential, while the competitive test examines the null hypothesis that drugs inside the group are no more significantly associated with repositioning potential than those outside the group.
Table 5. Top 5 enriched ATC drug classes with gene-sets derived from PubChem and ChEMBL only

The FDR-adjusted p values are presented, which are already corrected for multiple testing. Test results with FDR <0.05 are in bold. Results with FDR between 0.05 and 0.1 are in italics. Self, self-contained test; Compet, competitive test. The definitions of self-contained and competitive tests are very similar to those in Tables 1–3. In the context the above ‘drug-group’ analysis, the self-contained test examines the null hypothesis that none of the drugs in the group are associated with repositioning potential, while the competitive test examines the null hypothesis that drugs inside the group are no more significantly associated with repositioning potential than those outside the group.
For AD, antipsychotics and antidepressants were the two most strongly enriched medication classes. The β-blocking agents were ranked among the top for AD. The β-adrenergic blockers, especially propranolol, have been clinically used in AD for a long time, although the efficacy still remains uncertain (Steenen et al., Reference Steenen, Van Wijk, Van Der Heijden, Van Westrhenen, De Lange and De Jongh2016). As for depressive symptoms, dopaminergic and antiepileptic agents were listed among the top, and interestingly lipid-lowering agent was also on the top list. As for neuroticism, antipsychotics were the most strongly enriched drug class, and antidepressants were also ranked within the top 5. Other drug classes listed in top 5 also included hypnotics/sedatives, anxiolytics, and dopaminergic agents. The full results are shown in online Supplementary Tables S5 and S6.
We also examined the presence and extent of overlap between the top-listed ATC drug categories. The results are presented in online Supplementary Table S7. In general, there is a little overlap across most categories and almost zero overlap for categories starting with N (neuropsychiatric drugs). Overlap was concentrated in categories of anti-inflammatory agents, anti-infective agents, and corticosteroids.
Correlation of level of drug p values with degree of literature support
Online Supplementary Table S8 shows the correlation between p value of each drug (derived from GSA) with the number of research articles retrieved (gene-sets derived from the whole DSigDB database). All results were statistically significant and passed FDR correction for multiple testing (FDR<0.05), suggesting that drugs with lower p values from our GSA may have stronger literature support, and those with higher p values have weaker support. The strongest correlations were observed for combined analysis using Simes’ or Brown's method. When restricted to gene-sets from PubChem and ChEMBL (online Supplementary Table S9), the correlations were weaker, with significant results (FDR<0.05) observed for depressive symptoms, MDD2018, and combined p values using Brown's method.
However, we observed that the levels of correlation were only modest (<0.1). This analysis alone may not be sufficient to provide evidence for the validity of repositioning candidates; however, it may be considered a complementary validation strategy for our repositioning approach as a whole (as compared with manual literature review of the top candidates), as this analysis is more quantitative, unbiased, and comprehensive (covering all drugs).
Top repositioning hits and literature support from previous studies
Some of the top hits are mentioned below. However, we emphasize that these findings are tentative and require validations in further experimental and clinical studies. In addition, although some of the drugs received support from the literature, a certain degree of publication bias is likely present. Also, our literature search is not meant to be a systematic review due to the number of drugs involved.
Based on gene-sets derived from the whole DSigDB database
We found several interesting repositioning hits that were supported by previous studies (Table 6; online Supplementary Table S10 gives a fully annotated list of the top 20 drugs). The top-ranked repositioning hit identified in the meta-analysis was fendiline (Brown's p = 1.06E-11, q = 5.55E-8), a non-selective calcium channel blocker (CCB). Fendiline was recently shown to exert antidepressant-like effects in a mouse model by inhibition of acid sphingomyelinase activity and reduction of ceramide concentrations in the hippocampus (Gulbins et al., Reference Gulbins, Palmada, Reichel, Luth, Bohmer, Amato, Muller, Tischbirek, Groemer, Tabatabai, Becker, Tripal, Staedtler, Ackermann, Van Brederode, Alzheimer, Weller, Lang, Kleuser, Grassme and Kornhuber2013). A drop in ceramide concentrations might lead to increased neurogenesis and improved neuronal maturation and survival (Gulbins et al., Reference Gulbins, Palmada, Reichel, Luth, Bohmer, Amato, Muller, Tischbirek, Groemer, Tabatabai, Becker, Tripal, Staedtler, Ackermann, Van Brederode, Alzheimer, Weller, Lang, Kleuser, Grassme and Kornhuber2013). The drug was ranked among the top for MDD-PGC-2012, MDD2018, SSGAC-DS and neuroticism.
Table 6. Selected repositioning hits (ranked within top 20 for a phenotype) with literature support
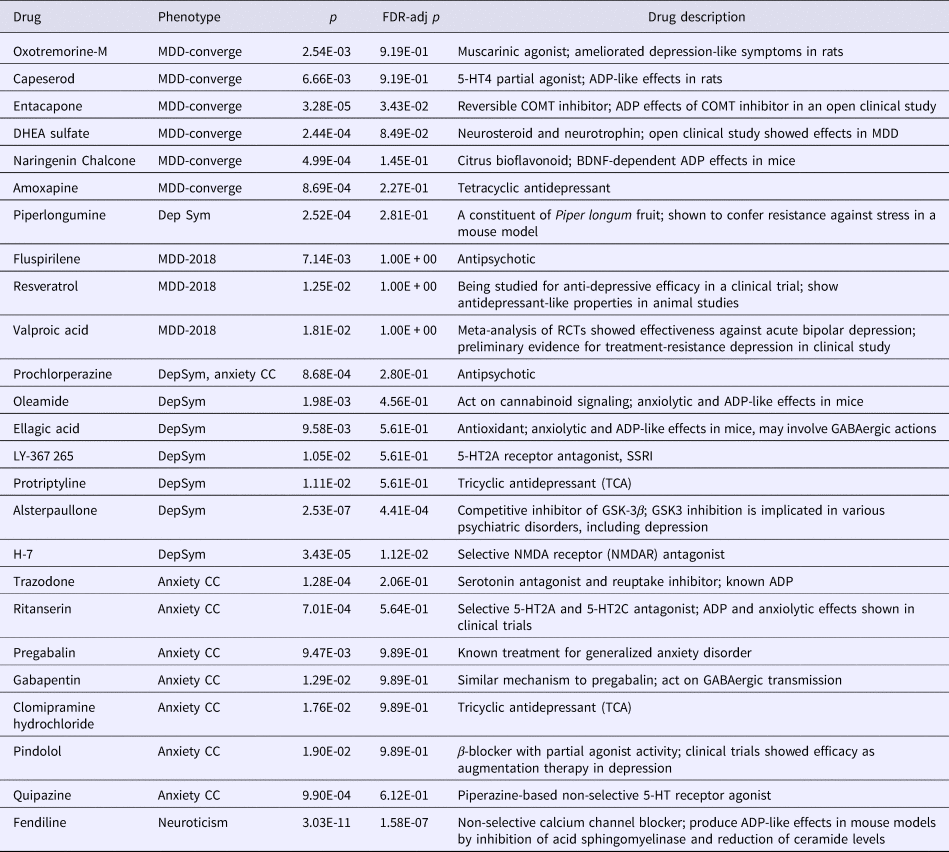
FDR-adj p, FDR-adjusted p values.
Here the repositioning hits are listed regardless of whether the underlying gene-sets were derived from the entire DsigDB or from PubChem/ChEBML only. Results from meta-analysis (Simes’ or Brown's method) are not specifically listed as they are included in the results of individual phenotypes.
Please also refer to the legends of Table 1 and the main text for the references and more detailed discussions.
As for individual phenotypes, some interesting candidates for MDD-CONVERGE include DHEA, a neurosteroid with evidence of antidepressive effects in a double-blind randomized controlled trial (RCT); naringenin chalcone, a citrus bioflavonoid shown to be effective in mouse models (Yi et al., Reference Yi, Liu, Li, Luo, Liu, Geng, Tang, Xia and Wu2014); amoxapine, a tetracyclic antidepressant (Jue et al., Reference Jue, Dawson and Brogden1982). Other candidates listed for MDD-PGC or MDD2018 include ibuprofen, an NSAID shown to be associated with lower depressive symptoms in osteoarthritis patients; piperlongumine, a constituent of the fruit of Piper longum, which was shown to confer resistance against stress in a mouse model (Yadav et al., Reference Yadav, Chatterjee, Majeed and Kumar2015); and sanguinarine, a selective mitogen-activated protein kinase phosphatase-1 (Mkp-1) inhibitor which produced antidepressant-like effect in rats (Chen et al., Reference Chen, Wang, Zhang, Wang, Peng, Sun and Tan2012).
For AD, candidates included a known antidepressant trazodone and a serotonin agonist quipazine that may increase brain serotonin levels (Fuller et al., Reference Fuller, Snoddy, Perry, Roush, Molloy, Bymaster and Wong1976); for depressive symptoms, alsterpaullone, a glycogen synthase kinase-3β (GSK-3β) inhibitor (Leost et al., Reference Leost, Schultz, Link, Wu, Biernat, Mandelkow, Bibb, Snyder, Greengard, Zaharevitz, Gussio, Senderowicz, Sausville, Kunick and Meijer2000). Increased activation of GSK-3β was also associated with depression-like behavior in mouse models, which could be alleviated by GSK-3β inhibitors (Beaulieu et al., Reference Beaulieu, Zhang, Rodriguiz, Sotnikova, Cools, Wetsel, Gainetdinov and Caron2008). In addition, inhibition of GSK3 has been postulated as a major mechanism of action by the mood stabilizer lithium (Freland and Beaulieu, Reference Freland and Beaulieu2012). A NMDA receptor antagonist H-7 was also top-listed (Amador and Dani, Reference Amador and Dani1991).
Among the top repositioning hits from the individual and meta-analysis results, a number of them are CCB. These include fendiline, perhexiline, prenylamine, and felodipine [prenylamine was withdrawn from the market due to risk of QT prolongation and torsades de pointes (Fung, Reference Fung2001)]. Although with the exception of fendiline, no direct experimental or clinical studies have shown antidepressant or anxiolytic properties of the above drugs, CCB as a whole has been proposed as treatment for various psychiatric disorders. CCB has been mostly studied for the treatment of mania, recently reviewed in Cipriani et al. (Reference Cipriani, Saunders, Attenburrow, Stefaniak, Panchal, Stockton, Lane, Tunbridge, Geddes and Harrison2016). However, the number of quality double-blind RCTs was small, and there is yet no sufficient evidence to suggest the use of CCB in treating manic symptoms. As for depression, a recent pilot (patient-only) study of isradipine on bipolar depression showed positive results (Ostacher et al., Reference Ostacher, Iosifescu, Hay, Blumenthal, Sklar and Perlis2014). Another CCB, nicardipine, was reported to enhance the antidepressant action of electroconvulsive therapy (Dubovsky et al., Reference Dubovsky, Buzan, Thomas, Kassner and Cullum2001). Notwithstanding the mixed evidence, CCB are probably still worthy of further investigation for depression and AD, given the biological relevance of calcium signaling, preliminary support from clinical studies, and the wide availability as well as known safety profiles of this drug class.
Based on gene-sets derived from PubChem and ChEMBL
A manually annotated list of the top 20 prioritized drugs is given in online Supplementary Table S11. Some highlighted candidates are also shown in Table 6. Some repositioning hits for MDD-CONVERGE included oxotremorine-M, a muscarinic acetylcholine receptor agonist that may ameliorate depressive symptoms and restore hippocampal neurogenesis in an animal model (Veena et al., Reference Veena, Srikumar, Mahati, Raju and Shankaranarayana Rao2011) and capeserod, a 5-HT4 receptor agonist shown effective in a mouse model (Tamburella et al., Reference Tamburella, Micale, Navarria and Drago2009). For MDD-PGC-2012, top-listed drugs include three NSAIDs, namely ibuprofen, meclofenamate, and flufenamic acid, which possess anti-inflammatory actions that may also be beneficial for depression (Iyengar et al., Reference Iyengar, Gandhi, Aneja, Thorpe, Razzouk, Greenberg, Mosovich and Farkouh2013; Zaminelli et al., Reference Zaminelli, Gradowski, Bassani, Barbiero, Santiago, Maria-Ferreira, Baggio and Vital2014). Remarkably, our analysis recovered a known augmentation therapy for depression, namely Cytomel [or synthetic triiodothyronine (T3)] (Touma et al., Reference Touma, Zoucha and Scarff2017). Another candidate was guanidinonaltrindole di-trifluoroacetate, a κ-opioid receptor inhibitor with antidepressant-like and anxiolytic-like efficacy in rat models (Peters et al., Reference Peters, Zacco, Gordon, Maciag, Litwin, Thompson, Schroeder, Sygowski, Piser and Brugel2011). For MDD2018, resveratrol was reported to have antidepressant-like effects (de Oliveira et al., Reference De Oliveira, Chenet, Duarte, Scaini and Quevedo2017) and is currently under study in an RCT (https://clinicaltrials.gov/ct2/show/NCT03384329); valproate was also on the list which is effective against bipolar depression (Smith et al., Reference Smith, Cornelius, Azorin, Perugi, Vieta, Young and Bowden2010). Preliminary evidence from a small clinical study showed possible benefits for treatment-resistant depression (Ghabrash et al., Reference Ghabrash, Comai, Tabaka, Saint-Laurent, Booij and Gobbi2016).
For depressive symptoms, the top-listed candidates include prochlorperazine and raclopride, which are antipsychotics; protriptyline, a tricyclic antidepressant; oleamide, a cannabinoid receptor type 1 (CB1) receptor agonist of potential antidepressant effects in animal models (Fedorova et al., Reference Fedorova, Hashimoto, Fecik, Hedrick, Hanus, Boger, Rice and Basile2001; Wei et al., Reference Wei, Yang, Dong and Wu2007); ellagic acid and chebulinic acid (an ellagitannin), natural phenol antioxidants with some evidence of improving depressive traits again in animal models (Dhingra and Chhillar, Reference Dhingra and Chhillar2012; Girish et al., Reference Girish, Raj, Arya and Balakrishnan2013; Onasanwo et al., Reference Onasanwo, Faborode, Agrawal, Ijiwola, Jaiyesimi and Narender2014). Also of note is the nicotinic agonist DMPP, as nicotine has been shown to have antidepressant properties in pre-clinical and clinical studies (Philip et al., Reference Philip, Carpenter, Tyrka and Price2012); the antidepressant effects may be due to initial activation of the nicotinic receptor followed by desensitization leading to long-term antagonism (Philip et al., Reference Philip, Carpenter, Tyrka and Price2012). For neuroticism, again quite a number of top hits are CCB, and the CCB fendiline was the top-ranked candidate. The repositioning hits derived from meta-analyzed p values were covered above.
There were a number of noteworthy candidates from the analysis results of AD. Notably, the top-ranked drugs contain pregabalin and gabapentin, two drugs with similar mechanisms which have been shown anxiolytic effects in clinical trials (especially pregabalin) (Pande et al., Reference Pande, Crockatt, Feltner, Janney, Smith, Weisler, Londborg, Bielski, Zimbroff, Davidson and Liu-Dumaw2003; Rickels et al., Reference Rickels, Pollack, Feltner, Lydiard, Zimbroff, Bielski, Tobias, Brock, Zornberg and Pande2005); clomipramine, a known tricyclic antidepressant; and trazodone, another known antidepressant belonging to the serotonin antagonist and reuptake inhibitor class. Some other noteworthy candidates include ritanserin, a selective 5-HT2A and 5-HT2C receptor antagonist (Ceulemans et al., Reference Ceulemans, Hoppenbrouwers, Gelders and Reyntjens1985; Nappi et al., Reference Nappi, Sandrini, Granella, Ruiz, Cerutti, Facchinetti, Blandini and Manzoni1990; Strauss and Klieser, Reference Strauss and Klieser1991); pindolol, a β-blocker with partial β-adrenergic receptor agonist activity and a possible augmentation therapy for depression (Blier and Bergeron, Reference Blier and Bergeron1995; Artigas et al., Reference Artigas, Celada, Laruelle and Adell2001); atomoxetine, a norepinephrine reuptake inhibitor.
Discussion
In this study, we leveraged large-scale GWAS summary data and analyzed gene-sets associated with drugs to uncover repositioning opportunities for depression and AD. It is encouraging that we observed significant enrichment for known psychiatric medications or drugs considered in clinical trials. Remarkably, antipsychotics and antidepressants were the two most significantly enriched drug classes in our combined analysis (with gene-sets derived from the whole DSigDB). Our findings provide support for the validity of GSA in drug repurposing. In addition, we revealed a number of interesting candidates for repurposing, some of which were supported by prior studies. Although relatively few susceptibility variants of genome-wide significance have been found for depression and AD, our findings suggest that leveraging variants with weaker associations, for example, by GSA, might still contribute valuable information to the prioritization of new drug candidates.
While we have included several datasets (SSGAC-DS, MDD2018, MDD-CONVERGE) directly related to depression, the enrichment results are not entirely the same. MDD2018 and MDD-CONVERGE are case–control GWAS studies on MDD; however, the drugs and drug classes enriched were quite different. Another noteworthy finding is that despite similar sample sizes, MDD-CONVERGE demonstrated much stronger enrichment for psychiatric drugs than MDD-PGC-2012, suggesting its better value in guiding drug discoveries. The discrepancy in results between MDD-CONVERGE and other samples might be due to different characteristics of the recruited subjects. The MDD-CONVERGE sample differs from other MDD GWAS samples by gender, ethnicity, and other characteristics of the case and control subjects (Flint et al., Reference Flint, Chen, Shi, Kendler and Consortium2012; Cai et al., Reference Cai, Bigdeli, Kretzschmar, Li, Liang, Song, Hu, Li, Jin, Hu, Wang, Wang, Qian, Liu, Jiang, Lu, Zhang, Yin, Li, Xu, Gao, Reimers, Webb, Riley, Bacanu, Peterson, Chen, Zhong, Liu, Wang, Sun, Sang, Jiang, Zhou, Li, Li, Zhang, Wang, Fang, Pan, Miao, Zhang, Hu, Yu, Du, Sang, Li, Chen, Cai, Yang, Yang, Ha, Hong, Deng, Li, Li, Song, Gao, Zhang, Gan, Meng, Pan, Gao, Zhang, Sun, Li, Niu, Zhang, Liu, Hu, Zhang, Lv, Dong, Wang, Tao, Wang, Xia, Rong, He, Liu, Huang, Mei, Shen, Liu, Shen, Tian, Liu, Wu, Gu, Fu, Shi, Chen, Gan, Liu, Wang, Yang, Cong, Marchini, Yang and Wang2015). The CONVERGE sample only included women, and cases were required to have two or more depressive episodes. The proportion of patients with melancholia is ~85%, which is higher than the prevalence in a general MDD sample (~23.5%) (McGrath et al., Reference Mcgrath, Khan, Trivedi, Stewart, Morris, Wisniewski, Miyahara, Nierenberg, Fava and Rush2008). To reduce the probability that some patients initially diagnosed with MDD may go on to develop BD, cases were required to be at least 30 years of age. Regarding control subjects, all controls had been screened by personal interview to exclude history of depressive episode(s). There was also an age limit for controls (>40), to reduce the chance that some of them may develop MDD at later ages. Finally, all four grandparents of cases and controls were required to be of Chinese ancestry, in order to maximize genetic homogeneity.
It is widely accepted that MDD is a heterogeneous disorder, with a variety of clinical presentations and possibly divergent pathophysiologies (Goldberg, Reference Goldberg2011). By recruiting a more homogeneous group of patients, the CONVERGE study might have better power in detecting susceptibility genes despite a lower sample size. Indeed, MDD-CONVERGE revealed two genome-wide significant loci while none was found in the MDD-PGC-2012 study although the sample sizes were similar. It is also worth mentioning that previous meta-analyses showed that the response to antidepressant depends on the baseline severity of depression (Kirsch et al., Reference Kirsch, Deacon, Huedo-Medina, Scoboria, Moore and Johnson2008; Fournier et al., Reference Fournier, Derubeis, Hollon, Dimidjian, Amsterdam, Shelton and Fawcett2010). The studies reported that effects of antidepressants were largest for the most severely depressed group, but smaller for mild-to-moderate depression. Our findings, although based on a different study paradigm, are broadly in line with this clinical observation.
Given that the prioritized candidates from MDD-CONVERGE and other samples of European ancestry are different, an interesting question is whether there may exist ‘ethnicity-specific drugs’, or at least drug efficacy differing by ethnicities. This is an interesting direction for future work, but our present understanding of MDD and the data at hand do not allow a concrete conclusion to be drawn yet. While the CONVERGE sample differs from other samples in ethnicity (Chinese versus European ancestry), there are also marked differences in patient characteristics and other ascertainment criteria as detailed above. It is therefore difficult to tease out whether the differences in results are due to ethnic-specific factors alone. Our current understanding of MDD in different ethnic groups is still relatively limited. Also, few studies have explored differences in genetic basis of MDD across ethnicities. A recent work reported statistically significant but only partial genetic correlations between MDD in Chinese and European populations (0.33 for lifetime MDD; 0.40 and 0.41 for female-only and recurrent MDD, respectively; the Chinese sample was based on MDD-CONVERGE) (Bigdeli et al., Reference Bigdeli, Ripke, Peterson, Trzaskowski, Bacanu, Abdellaoui, Andlauer, Beekman, Berger, Blackwood, Boomsma, Breen, Buttenschon, Byrne, Cichon, Clarke, Couvy-Duchesne, Craddock, De Geus, Degenhardt, Dunn, Edwards, Fanous, Forstner, Frank, Gill, Gordon, Grabe, Hamilton, Hardiman, Hayward, Heath, Henders, Herms, Hickie, Hoffmann, Homuth, Hottenga, Ising, Jansen, Kloiber, Knowles, Lang, Li, Lucae, Macintyre, Madden, Martin, Mcgrath, Mcguffin, Mcintosh, Medland, Mehta, Middeldorp, Milaneschi, Montgomery, Mors, Muller-Myhsok, Nauck, Nyholt, Nothen, Owen, Penninx, Pergadia, Perlis, Peyrot, Porteous, Potash, Rice, Rietschel, Riley, Rivera, Schoevers, Schulze, Shi, Shyn, Smit, Smoller, Streit, Strohmaier, Teumer, Treutlein, Van Der Auwera, Van Grootheest, Van Hemert, Volzke, Webb, Weissman, Wellmann, Willemsen, Witt, Levinson, Lewis, Wray, Flint, Sullivan and Kendler2017). With regards to drug response, Murphy et al. (Reference Murphy, Hou, Maher, Woldehawariat, Kassem, Akula, Laje and Mcmahon2013) reported that patients of African ancestry had poorer response to treatment based on the STAR*D trial; genetic ancestry explained a significant proportion of residual variance even after accounting for socioeconomic and baseline clinical differences. However, some studies reported no racial group differences in treatment response (Lesser et al., Reference Lesser, Myers, Lin, Bingham Mira, Joseph, Olmos, Schettino and Poland2010; Lesser et al., Reference Lesser, Zisook, Gaynes, Wisniewski, Luther, Fava, Khan, Mcgrath, Warden, Rush and Trivedi2011). However, to our knowledge, there are no similar studies comparing Chinese with other ethnicities. Overall, we believe that further studies are required to elucidate the differences and similarities in clinical/genetic determinants of depression and drug response across ethnic groups.
It is noteworthy that the repositioning hits are not only enriched for antidepressants or anxiolytics but also antipsychotics. A meta-analysis by Spielmans et al. revealed that atypical antipsychotics are effective as adjunctive treatment for treatment-resistant depression (Spielmans et al., Reference Spielmans, Berman, Linardatos, Rosenlicht, Perry and Tsai2013). Zhou et al. also reached a similar conclusion in a recent network meta-analysis (Zhou et al., Reference Zhou, Keitner, Qin, Ravindran, Bauer, Del Giovane, Zhao, Liu, Fang, Zhang and Xie2015). Atypical antipsychotics may also be useful for AD and symptoms (Gao et al., Reference Gao, Muzina, Gajwani and Calabrese2006; LaLonde and Van Lieshout, Reference Lalonde and Van Lieshout2011; Samuel et al., Reference Samuel, Zimovetz, Gabriel and Beard2011), although further studies are required and that the benefits need to be balanced against the side-effects. Furthermore, a shared genetic basis between schizophrenia and depression is well-established (Cross-Disorder Group of the Psychiatric Genomics et al., Reference Lee, Ripke, Neale, Faraone, Purcell, Perlis, Mowry, Thapar, Goddard, Witte, Absher, Agartz, Akil, Amin, Andreassen, Anjorin, Anney, Anttila, Arking, Asherson, Azevedo, Backlund, Badner, Bailey, Banaschewski, Barchas, Barnes, Barrett, Bass, Battaglia, Bauer, Bayes, Bellivier, Bergen, Berrettini, Betancur, Bettecken, Biederman, Binder, Black, Blackwood, Bloss, Boehnke, Boomsma, Breen, Breuer, Bruggeman, Cormican, Buccola, Buitelaar, Bunney, Buxbaum, Byerley, Byrne, Caesar, Cahn, Cantor, Casas, Chakravarti, Chambert, Choudhury, Cichon, Cloninger, Collier, Cook, Coon, Cormand, Corvin, Coryell, Craig, Craig, Crosbie, Cuccaro, Curtis, Czamara, Datta, Dawson, Day, De Geus, Degenhardt, Djurovic, Donohoe, Doyle, Duan, Dudbridge, Duketis, Ebstein, Edenberg, Elia, Ennis, Etain, Fanous, Farmer, Ferrier, Flickinger, Fombonne, Foroud, Frank and Franke2013), and a recent study also found significant genetic correlation between neuroticism and schizophrenia (Smith et al., Reference Smith, Escott-Price, Davies, Bailey, Colodro-Conde, Ward, Vedernikov, Marioni, Cullen, Lyall, Hagenaars, Liewald, Luciano, Gale, Ritchie, Hayward, Nicholl, Bulik-Sullivan, Adams, Couvy-Duchesne, Graham, Mackay, Evans, Smith, Porteous, Medland, Martin, Holmans, Mcintosh, Pell, Deary and O'Donovan2016). Epidemiology studies also demonstrated associations of neuroticism with schizophrenia (Malouff et al., Reference Malouff, Thorsteinsson and Schutte2005).
Antidepressants and antipsychotics were the most consistently associated categories based on our analysis. Here we also briefly discuss a few other drug categories listed among the top. However, we caution that the other drug categories are less well-studied for depression and anxiety, with considerably weaker support of therapeutic effect from the literature. They are also less consistently implicated in our analysis. While discussed below, we stress that our results are tentative and further validation studies are necessary to verify the findings.
For MDD-CONVERGE, which is composed of mainly severe melancholic depressive patients, we found that corticosteroids were highly ranked. A number of studies (Gold and Chrousos, Reference Gold and Chrousos2002; Stetler and Miller, Reference Stetler and Miller2011; Lamers et al., Reference Lamers, Vogelzangs, Merikangas, De Jonge, Beekman and Penninx2013) have shown that hyperactivity of the hypothalamic–pituitary axis is an important feature of melancholic depression. Nevertheless, as with other kinds of GSA, the current approach of GSA does not delineate the exact direction of drug effects. It may be concluded that corticosteroids or drugs targeting relevant pathways might be of clinical significance to melancholic depression; however, whether corticosteroids themselves or drugs blocking their actions would be useful remains to be investigated. It is also worth noting that currently there is no reliable approach to subtype depression yet. While some researchers propose melancholic features may be used to define subtypes of depression (Parker et al., Reference Parker, Fink, Shorter, Taylor, Akiskal, Berrios, Bolwig, Brown, Carroll, Healy, Klein, Koukopoulos, Michels, Paris, Rubin, Spitzer and Swartz2010), the biological and clinical validity of such classification remains to be established (Melartin et al., Reference Melartin, Leskela, Rytsala, Sokero, Lestela-Mielonen and Isometsa2004; Harald and Gordon, Reference Harald and Gordon2012).
We briefly highlight a few other drug classes here. Dopaminergic agents were ranked among the top for depressive symptoms; stimulants including dopaminergic agents have been tested in clinical trials for unipolar and bipolar depression (Nierenberg et al., Reference Nierenberg, Dougherty and Rosenbaum1998; Dell'Osso et al., Reference Dell'Osso, Ketter, Cremaschi, Spagnolin and Altamura2013; Corp et al., Reference Corp, Gitlin and Altshuler2014; Szmulewicz et al., Reference Szmulewicz, Angriman, Samame, Ferraris, Vigo and Strejilevich2017). However, further randomized controlled trials of large sample sizes are required to confirm the efficacy and clarify possible side-effects of such treatment. We also observed that anti-migraine medications were highly ranked in the list. An increased rate of depression among migraine patients is well-established (Antonaci et al., Reference Antonaci, Nappi, Galli, Manzoni, Calabresi and Costa2011), and antidepressants (mainly tricyclics) have been used for migraine prophylaxis (Koch and Jurgens, Reference Koch and Jurgens2009). The effects of anti-migraine medications on depressive symptoms however remain to be elucidated. Lipid-lowering agents was top-listed for depressive symptoms (based on all genes in DSigDB) and AD (based on drug target genes from PubChem and ChEMBL), and studies have shown potential benefits of statins in depression (Salagre et al., Reference Salagre, Fernandes, Dodd, Brownstein and Berk2016), and when used with concomitant SSRI (Kohler et al., Reference Kohler, Gasse, Petersen, Ingstrup, Nierenberg, Mors and Ostergaard2016). However, some studies have also reported increased depressive symptoms with statins (You et al., Reference You, Lu, Zhao, Hu and Zhang2013), hence the exact relationship may be complex and may differ by patient characteristics. Whether other types of lipid-lowering drugs may be beneficial is another topic worthy of further investigations.
In this study, we employed the GSA approach to drug repositioning. The current study is complementary to our recent repositioning attempt using a new framework in which the drug-induced transcriptome is compared against GWAS-imputed expression profiles (So et al., Reference So, Chau, Chiu, Ho, Lo, Yim and Sham2017). Each of these two methods has their own advantages and disadvantages. The methodology of finding reversed expression patterns [as detailed in So et al. (Reference So, Chau, Chiu, Ho, Lo, Yim and Sham2017)] has a unique advantage of accounting for the directions of associations. It also takes into account the functional impact of variants on expression and is intuitive from a biological point of view. While differentially expressed genes can be included in gene-sets, the actual (quantitative) expression changes are not considered which results in a loss of information. GSA also does not delineate the directions of effects. Nevertheless, GSA can directly make use of knowledge concerning known drug targets and other information on drug-related genes, for which more databases are available. Moreover, in the ‘reversed transcriptome’ approach (So et al., Reference So, Chau, Chiu, Ho, Lo, Yim and Sham2017), one only considers functional effects of the SNPs on expression, but genetic variants may also contribute to disease pathogenesis via other mechanisms, such as splicing or changes in protein function. The gene-based test in the current analysis is based on combining statistical evidence of individual SNPs in a general manner, and may capture a wider range of effects on disease risk. Also, the transcriptome comparison approach involves ‘imputing’ expression levels; since the major reference transcriptome dataset (GTEx) is mainly composed of Caucasians (84.6%) with greater proportion of males (65.6%) (https://www.gtexportal.org/home/tissueSummaryPage, accessed 7 September 2017), the quality of imputation for other ethnicities and females might be less reliable, for example, when applied to the MDD-CONVERGE dataset.
Just as medications acting on different pathways might have synergistic therapeutic effects, we believe that it is beneficial to have different approaches to drug repositioning to complement each other. Of course, computational methods leveraging human genomic data are not the only means to drug discovery. We believe that a combination of a variety of approaches, including experimental and computational ones, is required to speed up drug repurposing and discoveries.
Our enrichment analyses support the application of GSA in drug repositioning in depression and anxiety. However, we stress that our repositioning results should be validated in further pre-clinical and clinical studies before translation to practice. All the findings should be regarded tentative until further validations. In addition, GSA analyses do not provide information on the direction of effects, as discussed previously. Measures of statistical significance also do not provide definitive evidence for the actual therapeutic effects of the repositioned drugs.
The current study provides a proof-of-concept of how human genomic data may assist drug discovery or repositioning in depression/AD. However, each candidate has to be individually and carefully assessed (e.g. for their side-effects, efficacy, interaction with other agents, ability to cross blood–brain barrier, etc.) before being tested in clinical trials or applied in the clinic. For the above reasons, we do not believe every top ‘hit’ will be useful clinically, although in the future one may develop a more ‘intelligent’ repositioning framework to factor in the above elements. Our described methodology may be considered as a general way to prioritize promising candidates for further research. Given the huge cost [up to ~US$2.6 billion (105)] in developing a new drug (the cost is lower but still considerable for repositioning), even a minute reduction in failure rate will represent very substantial savings in absolute terms.
In summary, we have performed a drug repositioning analyses on depression and AD, using a GSA approach considering five related GWAS studies. We showed that the repositioned drugs are in general enriched for known psychiatric medications or those considered in clinical trials. Remarkably, antipsychotics and antidepressants were ranked among the top even if we considered all level 3 ATC drug classes. Our findings lend further support to the usefulness of human genomic data in guiding drug development in psychiatry.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0033291718003641.
Author ORCIDs
Hon-Cheong So 0000-0002-7102-833X.
Acknowledgements
This work is partially supported by the Lo Kwee Seong Biomedical Research Fund and a Direct Grant from the Chinese University of Hong Kong. We also thank Professor Stephen K.W. Tsui and the Hong Kong Bioinformatics Centre for computing support. We would also like to acknowledge the Psychiatric Genomics Consortium, the CONVERGE Consortium, the Social Science Genetics Association Consortium and Otawa et al. for providing open access to full GWAS summary results.
Author contributions
H.-C.S. conceived and designed the study. H.-C.S. performed data analyses with input from C.K.L.C and K.Z. H.-C.S. interpreted the data. A.L. and S-Y.W. performed drug annotations. H.-C.S. wrote the manuscript and supervised the study.
Conflict of interest
None.









