Introduction
Schizophrenia (SZ) is a highly heritable disorder associated with disturbances in perception, cognition and affect, the biological basis of which is only partly understood. Successful identification of over 100 genetic risk loci to date has provided an important basis from which to begin to identify relevant biological mechanisms and their functional significance. Recently, a study of the major histocompatibility complex (MHC) region by Sekar et al. (Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016) identified one potential such mechanism involving a locus containing the complement component 4 (C4) gene isotypes C4A and C4B. In that study, C4 structural variation was associated with significantly altered C4 RNA expression (as measured in post-mortem brain tissue) such that copy number and structure of these genes could be used to predict C4A and C4B brain expression levels. Predicted C4A RNA expression was highly significantly associated with SZ risk (p = 3.6 × 10−24) in the Psychiatric Genomics Consortium (PGC) SZ GWAS data (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), driven by an allelic series of SZ risk levels that corresponded to each allele's relationship to C4A expression levels. The GWAS signal at the MHC region appeared to arise from at least three distinct genome-wide significant signals, one of which involves this collection of allelic influences on C4A expression. Finally, in a region of the mouse thalamus responsible for visual processing (an established model for experience-dependent synaptic refinement) C4 RNA was expressed in neurons during a period of peak synaptic pruning, and mediated synaptic refinement in this system (Sekar et al. Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016). Whether or how predicted C4 expression is associated with perceptual and cognitive function in humans is unknown.
The MHC region contains scores of genes with roles in the adaptive and innate immune systems and is the location of SZ's most significant genetic association (for common genetic variation) at a population level. Our group has previously reported a series of studies highlighting the cognitive and cortical effects of SZ-associated genetic risk loci in the MHC region and in non-MHC genes potentially related to complement regulation. We have shown that the SZ risk allele at rs10503253 within CSMD1, which encodes a regulator of C4, was associated with poorer general cognitive ability and episodic memory function in large independent samples of patients and healthy participants (Donohoe et al. Reference Donohoe, Walters, Hargreaves, Rose, Morris, Fahey, Bellini, Cummins, Giegling and Hartmann2013). We further showed that the same risk allele was associated with reduced cortical activation within the occipital cortex and cuneus during a spatial working memory task (Rose et al. Reference Rose, Morris, Hargreaves, Fahey, Greene, Garavan, Gill, Corvin and Donohoe2013). We have also shown that the SZ risk allele at rs6904071, a perfect proxy for the top MHC SZ risk SNP rs13194053 identified by both the International Schizophrenia Consortium (Purcell et al. Reference Purcell, Wray, Stone, Visscher, O'Donovan, Sullivan, Sklar, Ruderfer, McQuillin and Morris2009) and Molecular Genetics of Schizophrenia (Shi et al. Reference Shi, Levinson, Duan, Sanders, Zheng, Pe'Er, Dudbridge, Holmans, Whittemore and Mowry2009) studies, was associated with episodic memory performance in the same large datasets, and – in a third independent sample – with decreased hippocampal volume (Walters et al. Reference Walters, Rujescu, Franke, Giegling, Vásquez, Hargreaves, Russo, Morris, Hoogman and Da Costa2013). Given the demonstrated role for C4 in a model of experience-dependent synaptic pruning, we speculated that C4’s effects on synaptic pruning may also be apparent behaviourally and cortically during performance of perceptual and cognitive tasks. The findings from our previous CSMD1 and MHC studies, which have been supported by studies of other complement genetic variants (Athanasiu et al. Reference Athanasiu, Giddaluru, Fernandes, Christoforou, Reinvang, Lundervold, Nilsson, Kauppi, Adolfsson, Eriksson, Sundet, Djurovic, Espeseth, Nyberg, Steen, Andreassen and Le Hellard2017; Zhang et al. Reference Zhang, Lv, Fan, Tang and Yi2017), caused us to specifically hypothesize a role for C4 variation in memory function.
The purpose of the present study was to examine the relationship between predicted C4A RNA expression (based on structural variation in the C4 gene) and cognition in a large Irish sample of cases and healthy participants. In terms of the evidence and justification for the use of predicted C4A expression based on C4 structural variation, the following is noteworthy. In the Sekar et al. (Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016), based on eight panels of post-mortem human adult brain samples (674 samples from 245 distinct donors in three cohorts), RNA expression of C4A and C4B increased proportionally with copy number of C4A and C4B, respectively; the results of these expression analyses were consistent across all five brain regions analysed. Similarly, in serum, a previous study also reported that C4 gene dosage was positively correlated with serum C4 protein concentrations in vivo, mirroring the observations in the Sekar et al. post-mortem samples paper (Yang et al. Reference Yang, Chung, Zhou, Blanchong, Yu, Füst, Kovacs, Vatay, Szalai, Karadi and Varga2003). Sekar et al. (Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016) further measured C4A RNA expression levels in brain tissue samples from 35 SZ patients and 70 individuals without SZ. The median expression of C4A in brain tissues from SZ patients was 1.4-fold greater and was elevated in each of the five brain regions assayed. This was consistent with earlier reports that elevated the levels of complement proteins that were present in the serum of SZ patients (Rudduck et al. Reference Rudduck, Beckman, Franzen, Jacobsson and Lindström1985; Hakobyan et al. Reference Hakobyan, Boyajyan and Sim2005).
Based on this evidence above, and our previous studies, we hypothesised that increased predicted C4A RNA expression (which is associated with increased SZ risk) would be associated with poorer memory function in patients with SZ and in healthy participants. Given Sekar et al.’s report that C4 expression may influence visual development in an animal model, we also investigated, using functional MRI, whether predicted C4A expression would explain variation in cortical activity during a visual processing task in a healthy participant sample.
Methods
Participants
In total, 908 cases and 330 healthy participants completed a full neuropsychological assessment battery and had full genome-wide SNP data available on the basis of which predicted C4 expression levels could be calculated (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Cases consisted of n = 676 clinically stable patients with a diagnosis of SZ and schizoaffective disorder (SZA), and an additional n = 232 patients with ‘broad sense’ psychosis – diagnosed with either bipolar disorder with psychotic features, major depressive disorder with psychotic features, delusional disorder, or psychosis not otherwise specified. Patients were diagnosed by trained psychiatrists using the Structured Clinical Interview for DSM-IV Axis I Diagnosis (First, Reference First2005). These patients were recruited from five sites across Ireland. Inclusion criteria required participants to be clinically stable at the time of cognitive assessment, aged between 18–65 years, no history of co-morbid psychiatric disorder, no substance abuse in the preceding 6 months, no prior head injury with loss of consciousness, no history of seizures and with Irish ancestry (all four grandparents born in Ireland). Symptom severity was measured using the SAPS and SANS scores as previously described by us (Donohoe et al. Reference Donohoe, Hayden, McGLADE, O'GRÁDA, Burke, Barry, Behan, Dinan, O'Callaghan and Gill2009; Walters et al. Reference Walters, Corvin, Owen, Williams, Dragovic, Quinn, Judge, Smith, Norton and Giegling2010).
Healthy participants were recruited from the general population through local media advertisements. All were aged between 18 and 65 years and had Irish-born paternal and maternal grandparents, and satisfied, on the basis of clinical interview, the criteria of having no history of major mental health problems, intellectual disability or acquired brain injury, and no substance abuse in the preceding 6 months. Exclusion criteria also included having a first-degree relative with a history of psychosis. All assessments were conducted in accordance with the relevant ethics committees’ approval from each participating site, and all participants provided written informed consent. In this study, healthy participants did not represent a control group as no direct phenotypic comparison are made with patients; instead healthy participants are included both to establish whether comparable effects of predicted C4 expression levels were observed in both groups and, in a subset of these samples, to test for cortical effects using MRI.
Cognitive assessment
Memory recall was assessed using the Logical Memory subtest (immediate and delayed conditions) from the Wechsler Memory Scale, Third Edition (WMS III) (Wechsler, Reference Wechsler1997) and the Paired Associated Learning task (PAL; stages completed and total errors) from the Cambridge Automated Neuropsychological Test Battery (Robbins et al. Reference Robbins, James, Owen, Sahakian, McInnes and Rabbitt1994). Working memory was assessed using the Spatial Working Memory (SWM) subtest from the Cambridge Automated Neuropsychological Test Battery (Robbins et al. Reference Robbins, James, Owen, Sahakian, McInnes and Rabbitt1994) and Letter-Number Sequencing (LNS task) from the WMS III. Finally, measures of general cognitive ability (derived from the Wechsler Adult Intelligence Scale, Third Edition) (Wechsler, Reference Wechsler1997) and attentional control [Continuous Performance Task, Identical Pairs version; CPT-IP (Cornblatt et al. Reference Cornblatt, Risch, Faris, Friedman and Erlenmeyer-Kimling1988)] were also included as patients with SZ frequently show deficits in these areas of function. The published norms from the Wechsler test battery, the CANTAB test batteries, and the CPT-IP indicate a high level of test–retest validity, and, having been widely used in SZ research, have consistently showed a high sensitivity to cognitive deficits.
Functional MRI assessment
A subgroup of the healthy participants (n = 87) underwent functional imaging during a visual processing task as described by us previously (Grosbras & Paus, Reference Grosbras and Paus2006; Donohoe et al. Reference Donohoe, Morris, Robertson, Clarke, McGhee, Schwaiger, Nangle, Gill and Corvin2007; Rose et al. Reference Rose, Morris, Fahey, Robertson, Greene, O'Doherty, Newell, Garavan, McGrath, Bokde, Tropea, Gill, Corvin and Donohoe2012; Mothersill et al. Reference Mothersill, Morris, Kelly, Rose, Bokde, Reilly, Gill, Corvin and Donohoe2014a, Reference Mothersill, Morris, Kelly, Rose, Fahey, O'Brien, Lyne, Reilly, Gill and Corvinb). In this task, a face processing task developed by Grosbras & Paus (Reference Grosbras and Paus2006), participants watched a series of 2–5-s black-and-white videos of either contrasting circular images (expanding/contracting black-and-white concentric circles; ‘baseline’ condition), or faces which started from a neutral expression, and then turned into an angry expression or neutral expression. Overall, there were 28 blocks of 18-s duration each consisting of 4–7 video clips: nine blocks of concentric circles, five blocks of neutral face videos, five blocks of angry face videos. Attention to task was confirmed on the basis of a face recognition task following completion of the fMRI task and outside the scanner. Six of the 87 participants scored <4/5 on this task and were excluded from further analysis.
Imputation of C4 structural variation and genetically predicted C4A expression
Genotyping was conducted on DNA extracted from blood or saliva from patient and healthy participant participants. SNP data were obtained from two different sites; a GWAS using the Affymetrix SNP Array 6.0 platform, conducted as part of the Wellcome Trust Case Control Consortium 2 (Irish Schizophrenia Consortium & The Wellcome trust Case Control Consortium 2, 2012) and a collaborative GWAS with Cardiff University using an Illumina HumanCoreExome (+custom) SNP array. Direct genotypes for SNPs in the region of 23–35 Mb on chromosome 6 from the Affymetrix (n = 3657 SNPs) and Illumina (n = 3712) data were used to impute C4 structural alleles and predicted expression. This analysis of our data was undertaken by a member of the McCarroll group using the same methods described previously by them (Sekar et al. Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016). In brief, this involved imputation of C4 structural alleles in the study populations using a 222 haplotype integrated SNP and C4 reference panel. Imputed structural alleles were used to determine copy number of C4 structural elements (C4A, C4B, C4L and C4S and their co-occurrence) in each individual, and expected expression of C4A and C4B in the brain was inferred based on the previously determined relationship of copy number of C4 structural elements to gene expression in human brain samples. This resulted in a normally distributed range of predicted C4 expression scores of between 0 and 1.87 (mean 1.23, s.d. 0.45).
Statistical analysis – neuropsychological tests
To estimate the correlation between predicted C4A expression levels and performance of memory and other cognitive tasks, a series of correlational analysis was performed using Pearson's r, followed by multiple regression analysis for significant variables using IBM SPSS Statistics (IBM Corp, 2012). As this regression analysis focused on memory tasks known to be correlated with each other, and observed here to be correlated with predicted C4 expression levels, an unrotated principal components analysis was undertaken based on the four episodic memory test available to reduce the multiple testing burden. This resulted in one component which explained 72% of the variance in memory scores being extracted (with factor loadings of 0.881 for logical memory 1, 0.889 for logical memory 2, 0.766 for PAL stages and −0.813 for PAL total errors); participants scores on this factor were used as the dependent variable in the regression analysis. Age and gender were entered into the regression analysis as covariates of no interest. As cognitive profiles of patients with SZ and SZA are typically reported to differ from other kinds of psychosis (e.g. bipolar disorder), the analysis was undertaken both in the full group, and with psychosis patients with disorders other than SZ and SZA removed. Power calculations for these regression analyses indicated that sample sizes of n = 385 or greater would be required to observe small effects. This suggests that in the present study of 908 cases and 330 health participants (total sample N = 1238), we were adequately powered to detect small effects based on the full sample and the patient-only sample, but were somewhat underpowered to detect small effects in the healthy participant-only sample.
Imaging pre-processing and statistical analysis
Spatial pre-processing and statistical analysis of MRI data was performed using Statistical Parametric Mapping (SPM8, revision 4290, http://www.fil.ion.ucl.ac.uk/spm/software/spm8/) and MATLAB R2011b (v7.13; http://www.mathworks.co.uk/). Functional images were realigned to the mean functional image, normalised to Montreal Neurological Institute (MNI) space with a voxel size of 3 mm × 3 mm × 3 mm and smoothed using a 10 mm full width at half maximum (FWHM) isotropic Gaussian filter. After spatial pre-processing, graphical plots of the estimated time series of translations and rotations were inspected for excessive motion, which we defined as more than 3 mm translation and/or 3° rotation. One participant was excluded from further analysis due to movement, and six participants were excluded due to low-quality MRI data and/or significant artefacts, resulting in a final sample of 74 participants. For the face processing task, three task conditions (angry faces, neutral faces and baseline) and four contrasts consistent with our examination of neural activity associated with this task in SZ patients (Grosbras & Paus, Reference Grosbras and Paus2006; Mothersill et al. Reference Mothersill, Morris, Kelly, Rose, Bokde, Reilly, Gill, Corvin and Donohoe2014a): neutral faces v. baseline, angry faces v. baseline, all faces (angry and neutral) v. baseline and angry faces v. neutral faces. Participants’ contrast maps were entered into a second-level analysis to investigate effects of predicted C4 expression on neural activity. Results were examined at a p < 0.001 (uncorrected) level and clusters were considered statistically significant at a p < 0.05 level after family-wise error corrected for multiple comparisons across the whole brain at the cluster level. For each of these clusters, MNI coordinates of significant maxima were entered into the Anatomy toolbox in SPM 8 (Eickhoff et al. Reference Eickhoff, Stephan, Mohlberg, Grefkes, Fink, Amunts and Zilles2005, Reference Eickhoff, Heim, Zilles and Amunts2006, Reference Eickhoff, Paus, Caspers, Grosbras, Evans, Zilles and Amunts2007) and probable anatomical regions were identified using the AllAreas_v18_MPM atlas.
Results
C4 neuropsychological results
Demographic and clinical characteristics for patients and healthy participants appear in Table 1. Predicted C4A expression levels were not associated with age, gender or years of education. In terms of clinical symptom severity, no association was observed between predicted C4A RNA expression levels and either positive, negative or disorganized symptom factor scores [based on a principal components analysis of SAPS and SANS scores previously described by us (Donohoe et al. Reference Donohoe, Hayden, McGLADE, O'GRÁDA, Burke, Barry, Behan, Dinan, O'Callaghan and Gill2009)]. Similarly, no association between predicted C4A expression and medication dosage, measured in terms of chlorpromazine equivalents was observed.
Table 1. Mean demographic, clinical and predicted C4A expression levels for participants included in the neuropsychological analysis

Based on a correlational analysis, increased predicted C4A RNA expression levels were associated with poorer performance on all indexes of both verbal and non-verbal episodic memory performance (see Table 2). Given the correlation between these measures, to estimate the amount of variance in memory function explained by predicted C4A expression levels, these four memory scores were combined using an unrotated principal components analysis, the first extracted component of which explained 72% of variance on these measures. Participant's scores on this memory factor were then used as the dependent variable in the regression analysis. After the effects of age and gender were accounted for (as covariates of no interest), predicted C4A expression continued to significantly predict variation in memory performance (F change = 8.07; df = 1653; p = 0.005), explaining 1.2% of variation in memory factor scores (see Table 3). On the basis of a Bonferroni correction for the four cognitive constructs included in this study, this finding survives correction for multiple testing [corrected p value (0.005 × 4) = 0.02]. Re-running the analysis to account for diagnosis (entered as a covariate on the step prior to entering predicted C4 expression level), the results were unchanged (F change = 9.3; df1639; p = 0.002; r 2 change = 1.1%). Similarly, results remained significant when only patients and not healthy participants were included in the analysis (F change = 4.71; df1499; p = 0.030; r 2 change = 0.8%), or when only narrow psychosis and healthy participants were included and not non-SZ psychotic cases (F change = 8.2; d = 1513; p = 0.004; r 2 change = 1.3%). Finally, in an analysis of the healthy participant group only (which was less than half the size of the patient sample), predicted C4 expression showed the same direction of association as in patients but was not statistically significant.
Table 2. Correlation coefficients for predicted C4A expression and cognitive function in psychosis cases and healthy controls

Table 3. Regression analysis of C4 expression levels and episodic memory scores in patients and controls

− Analysis without adding covariates.
+ Analysis including covariates of age and gender in regression block 1.
++ Analysis including covariates of age, gender and diagnosis as other covariate.
This relationship between predicted C4A expression and episodic memory was observed in the absence of any correlation with working memory. Similarly, predicted C4A expression was not observed to correlate with either general cognitive ability or attentional control (see Table 2).
Two other variants within the MHC region were each associated with the risk in the Sekar et al. study, independently of C4 and of each other. For one of these, rs210133, we did not find any association with memory (r 2 change = .001, n.s.). The other SNP, rs13194504, was not available in our dataset. Instead we use a linkage disequilibrium (LD) proxy SNP rs148082388 (r 2 = 0.87) 82.5 kb away to investigate whether the same memory effects were associated with this SNP; a comparable association with poorer memory function was observed (r 2 change 0.6%; F change = 4.46; p = 0.035). This SNP is also in moderately high LD (r 2 = 0.67) with the MHC risk variant rs115329265 reported on by the PGC (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), for which we observed a similar association with poorer memory function (r 2 change 0.5%; F change = 5.23; p = 0.022). Finally, to relate our C4 predicted expression findings to our earlier cognitive findings with MHC SNP rs6904071 (Walters et al. Reference Walters, Rujescu, Franke, Giegling, Vásquez, Hargreaves, Russo, Morris, Hoogman and Da Costa2013), a Pearson's r correlation was carried out, based on which a statistically significant positive correlation was observed (r = 0.32, df = 610, p = 7.56 × 10–16).
C4 fMRI analysis in healthy participants
In the subset of participants for whom fMRI data were available, differences in predicted C4A expression were not observed to associate with either age or gender (p > 0.05; see Table 4). A nominally significant (positive) correlation with years of education was observed (p = 0.04). We therefore examined the effects of education on neural activity across our sample for all experimental conditions examined but no significant effects of education were observed, so education was not considered further.
Table 4. MRI participant demographics

a Mean ± standard deviation reported.
Neural activity during face processing task
Based on a whole brain analysis, increasing levels of genetically predicted C4A expression significantly correlated with decreased activity in a cluster incorporating the middle temporal gyrus during neutral face processing compared to baseline [t (74) = 5.49; corrected p < 0.05; see Table 5 and Fig. 1]. This relationship was also observed during angry face processing v. baseline and all faces v. baseline, but only at trend levels (uncorrected p < 0.001). To check for outlier effects, each participant's mean parameter estimates for all voxels were calculated for the temporal cluster showing a significant correlation with predicted C4A expression. These parameter estimates were then inputted into SPSS to check for outlier values, which were defined as any value more than 1.5 times the interquartile range of the values. No outliers were detected.
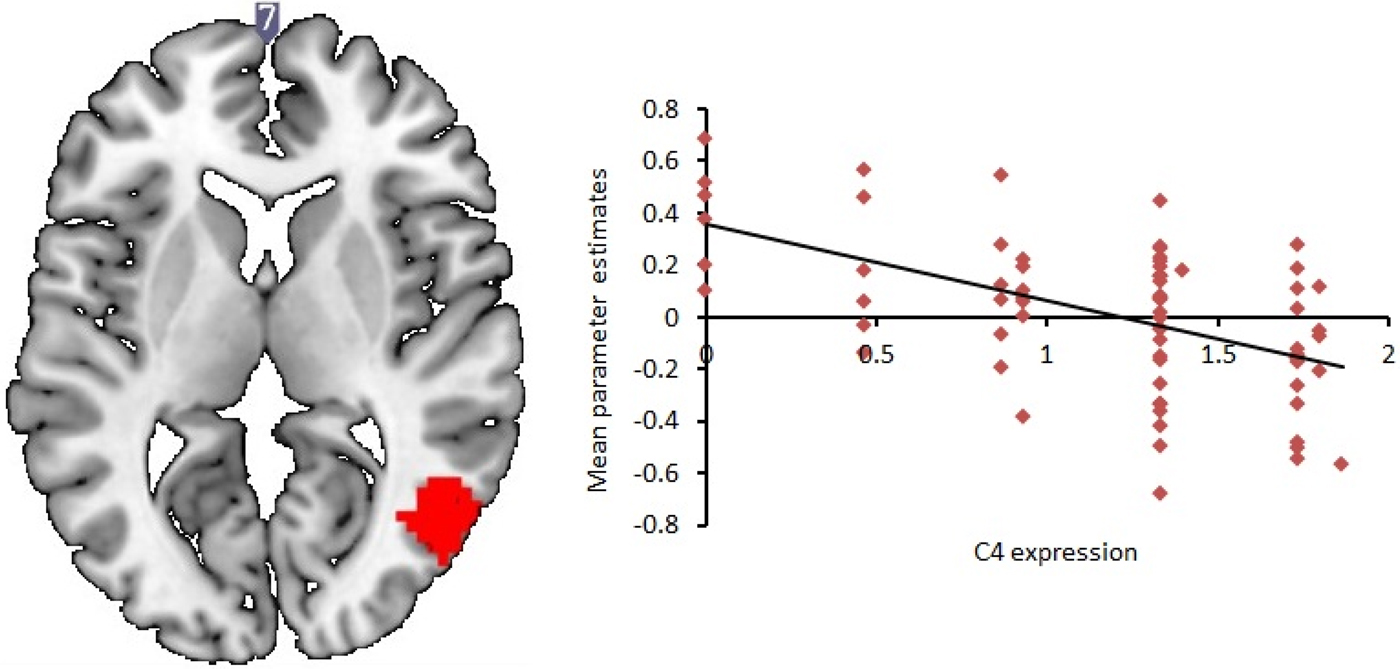
Fig. 1. Increased C4A expression is associated with decreased right middle temporal response during neutral face processing. Red: cluster showing decreased neural response with increasing C4A predicted expression levels [N = 74; multiple regression with C4A expression as covariate of interest; significance set at p < 0.05, family-wise error (FWE) corrected for multiple comparisons across the whole brain at the cluster level; df = 71]. The two-dimensional axial slice is labelled with an MNI coordinate. Cluster is rendered on the ‘ch256’ brain template using MRIcroGL (http://www.mccauslandcenter.sc.edu/mricrogl/). Parameter estimates displayed in arbitrary units.
Table 5. Cluster showing significantly decreased activity with increasing predicted C4A expression during neutral face processing relative to baseline, corrected for multiple comparisons at the cluster level

Discussion
This study examined the effects of genetically predicted C4A RNA expression on neuropsychological function in a large dataset of psychosis cases and healthy participants, and on task-dependent cortical activation during a visual task in a subset of healthy samples. Based on recent evidence of an association between predicted C4A RNA expression and increased SZ risk in humans, and between C4 deficiency and altered synaptic pruning in mice (Sekar et al. Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016), and our previous neurocognitive studies of variants at this locus, we hypothesised that variation in predicted C4A RNA expression would be associated with reduced memory function and altered neural activity. In testing this hypothesis, we observed that increased predicted C4A RNA expression was significantly correlated with, and predictive of, poorer performance on measures of episodic memory in both patients and healthy participants. Furthermore, based on an analysis carried out in a subset of our healthy participants, we found that increased predicted C4A RNA expression was associated with a pattern of reduced cortical activity in the middle temporal gyrus during a measure of visual processing.
Among the cognitive deficits associated with SZ, deficits in memory function are among the largest observed (Heinrichs & Zakzanis, Reference Heinrichs and Zakzanis1998). The association between predicted C4A RNA expression and poorer episodic memory observed in this study are highly consistent with our previous studies of other genetic risk variants either at this locus or known to directly interact with C4. C4 was selected for a study by Sekar et al. (Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016) on the basis of the MHC signal previously reported both in the PGC GWAS and by previous GWAS (Ripke et al. Reference Ripke, O'Dushlaine, Chambert, Moran, Kähler, Akterin, Bergen, Collins, Crowley and Fromer2013). On the basis of our analysis of the MHC risk allele at rs6904071, we previously reported an association with poorer episodic memory and, in an independent cohort, with decreased hippocampal volume. Even though the correlation between rs6904071 and predicted C4 expression moderate (r 2 estimate of shared variance ~10.2%), the patterns of cognitive results here are highly consistent with both the specific phenotype and direction of those previous findings. At present, other cognitive datasets in which predicted C4 expression levels have been calculated are not available; although supportive of our earlier MHC findings, independent replication of these results will be required to confirm C4’s effects on cognition. Finally, the association with memory performance observed here is unlikely to be solely attributable to inattentiveness, as these associations were observed in the absence of an association with attentional performance as measured by the CPT-IP.
Sekar et al. reported two other variants within the MHC region which were each associated with risk, independently of C4 and of each other. Based on an analysis of an LD proxy for one of these – rs148082388, a comparable association with poorer memory function was observed. As noted, this SNP is in moderately high LD with the MHC risk variant rs115329265 reported on by the PGC (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), and for which we observed a similar association with poorer memory function. While it is highly unlikely that all SZ-associated variants within the MHC locus would show the same phenotypic effects, the consistency of these genetic effects on memory function is interesting. Returning to C4 in particular, the basis for this study reported here, it is interesting to note that Sekar et al. found that of the five brain regions assessed, cells expressing C4 were most abundant in the hippocampus, the subcortical region most strongly associated with memory recall.
A key observation of the Sekar et al. (Reference Sekar, Bialas, de Rivera, Davis, Hammond, Kamitaki, Tooley, Presumey, Baum and Van Doren2016) C4 study was the observation of reduced levels of synaptic refinement in mice that lacked C4. In an experimental model of synaptic pruning in the visual system, Sekar et al. reported that C4-deficient mice showed decreased C4 expression in the lateral geniculate nucleus (LGN) of the visual thalamus, and that this was associated with defects in experience-dependent synaptic remodelling. In linking these findings to our cortical activation findings, in which we observed predicted C4 expression-related difference in the middle temporal gyrus and not the thalamic regions, the following points are noteworthy: (1) the functional specialization of C4 into C4A and C4B in humans does not have an analogy in mice, and (2) the mice findings related to developmental (rather than cross-sectional) differences in synaptic pruning) in the thalamic dLGN region; furthermore, (3) our study employed a visual processing task designed to index face processing – an aspect of visual information processing involving the ventral stream that is consistently shown to be impaired in patients with SZ (Mothersill et al. Reference Mothersill, Morris, Kelly, Rose, Bokde, Reilly, Gill, Corvin and Donohoe2014a, Reference Mothersill, Morris, Kelly, Rose, Fahey, O'Brien, Lyne, Reilly, Gill and Corvinb). Given that this task is unlikely to specifically highlight regions serving basic visual processing, it is therefore unsurprising that the between-group differences in thalamic activation are not observed; (4) in genetic terms, using the same task, Dickie et al. (Dickie et al. Reference Dickie, Tahmasebi, French, Kovacevic, Banaschewski, Barker, Bokde, Büchel, Conrod and Flor2014) found that task-related BOLD response within a cluster incorporating the middle temporal cortex was strongly genetically influenced. Consistent with these findings, our study highlights the role of C4 in the activity of the right middle temporal gyrus during task performance. Given that this effect was significant for the neutral faces v. baseline contrast but not others (e.g. association between predicted C4 expression and activation during angry faces v. baseline, all faces v. baseline, did not survive correction), confirmation of these results in further samples will be important.
The right middle temporal gyrus plays an important role in facial recognition (Carvajal et al. Reference Carvajal, Rubio, Serrano, Ríos-Lago, Alvarez-Linera, Pacheco and Martín2013), and is activated by both neutral and angry facial expressions (Fusar-Poli et al. Reference Fusar-Poli, Placentino, Carletti, Landi, Allen, Surguladze, Benedetti, Abbamonte, Gasparotti and Barale2009; Dickie et al. Reference Dickie, Tahmasebi, French, Kovacevic, Banaschewski, Barker, Bokde, Büchel, Conrod and Flor2014), consistent with the view that healthy participants respond similarly to both neutral and angry faces at both a behavioural and neural level (Lee et al. Reference Lee, Kang, Park, Kim and An2008; Ille et al. Reference Ille, Holl, Kapfhammer, Reisinger, Schäfer and Schienle2011). Nevertheless, participants may interpret neutral faces differently, not only due to the fact that no overt anger is being displayed, but also due to the presentation context – for example, neutral faces are sometimes interpreted more positively if immediately following negative faces and more negatively if following happy faces (Lee et al. Reference Lee, Kang, Park, Kim and An2008). In this study, we found that C4A expression affected right middle temporal activity during both neutral and angry face processing, but this effect was only significant at a corrected level during neutral face processing. Future imaging genetics studies based on face processing will be needed to examine why neural response to neutral faces might be more sensitive to C4A genetic variation compared to angry faces.
Finally, in the absence of a memory component to this visual fMRI task, whether these cortical abnormalities are related to, and account for, the behavioural memory impairments observed on neuropsychological testing is unknown. Similarly, as there was not a behavioural component to this task, it was not possible to correlate task performance with memory task performance. Whether these findings implicate the pleiotropic effects of predicted C4 expression differences, or the behavioural and cortical effects of a common pathway, therefore, remains to be elucidated. From a translational perspective, this will be important for determining the extent to which any pharmacological attempt to target the deleterious cortical effects of C4 variation should be specific to, or broader than, memory function alone.
The finding of comparable cognitive effects of predicted C4 expression in patients and healthy participants is consistent with our general expectation that while risk-associated biological processes will, by definition, occur at higher frequency in cases than controls, the phenotypic effects will be comparable in healthy participants who carry that risk factor. Comparable phenotypic effects in cases and healthy participants have previously been reported for other SZ risk variants (e.g. MIR137; Mothersill et al. Reference Mothersill, Morris, Kelly, Rose, Fahey, O'Brien, Lyne, Reilly, Gill and Corvin2014b), although for some cases this expectation has not been met (e.g. Walters et al. Reference Walters, Corvin, Owen, Williams, Dragovic, Quinn, Judge, Smith, Norton and Giegling2010). The cortical effects of predicted C4 expression reported here are based on the analysis of healthy participants only, an approach previously used in psychiatric genetics studies given the challenges of imaging sufficiently large samples of cases. Whether the same cortical effects of C4, based on one contrast (neutral faces v. baseline) but not others (angry faces v. either neutral faces or baseline), will be observed in patients is currently unknown, and further imaging studies of patients will be required to establish how C4 expression effects visual processing in this group.
Conclusion
The recent association of SZ risk with increased predicted C4 expression is a major step towards understanding the aetiology of SZ. Based on the hypothesis that C4’s effect would be most pronounced in cortical regions whose development is highly experience-dependent, we hypothesised and then observed that increased predicted C4A RNA expression was predictive of poorer memory performance and reduced cortical activity in middle temporal cortex during a measure of visual processing. Doing so further elucidates the pathway between genetically mediated altered development and illness-related disability.
Acknowledgements
The authors have no conflict of interest to declare. The authors wish to thank all patients and their support staff, and all healthy volunteers for participating in the data collection on which this manuscript is based. Recruitment, genotyping and analysis were supported by the European Research Council (grant 677467) Science Foundation Ireland (grants 12/IP/1670, 12/IP/1359 and 08/IN.1/B1916) and the Wellcome Trust Case Control Consortium 2 project (grants 085475/B/08/Z and 085475/Z/08/Z) and the Wellcome Trust (grants 072894/Z/03/Z, 090532/Z/09/Z and 075491/Z/04/B). The authors thank Dr Avery Davis and Dr Steven A. McCarroll at Harvard Medical School for generating the predicted C4A RNA expression data from chromosome 6 SNP data for all samples. The authors also thank Lucinda Hopkins and Lesley Bates for assistance in generating genotype data at Cardiff University. They also thank Dr Davis and Dr McCarroll for their helpful comments on the manuscript. The Wellcome Trust Case Control Consortium 2 investigators include: Peter Donnelly, Lesley Bates, Ines Barroso, Jenefer M. Blackwell, Elvira Bramon, Matthew A. Brown, Juan P. Casas, Aiden Corvin, Panos Deloukas, Audrey Duncanson, Janusz Jankowski, Hugh S. Markus, Christopher G. Mathew, Colin N. A. Palmer, Robert Plomin, Anna Rautanen, Stephen J. Sawcer, Richard C. Trembath, Ananth C. Viswanathan, Nicholas W. Wood, Chris C. A. Spencer, Gavin Band, Céline Bellenguez, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, Lucinda Hopkins, Matti Pirinen, Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, Cordelia Langford, Sarah E. Hunt, Sarah Edkins, Rhian Gwilliam, Hannah Blackburn, Suzannah J. Bumpstead, Serge Dronov, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T. McCann, Jennifer Liddle, Simon C. Potter, Radhi Ravindrarajah, Michelle Ricketts, Matthew Waller, Paul Weston, Sara Widaa and Pamela Whittaker.








