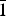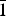I. INTRODUCTION
Clotrimazole (an imidazole) and fluconazole (a triazole) are common antifungal agents which belong to the azole family of pharmaceutical compounds (see Figure 1). Both active pharmaceutical ingredients (APIs) can be administered systemically and topically. The azoles exhibit the same wide antifungal spectrum and mechanism of action. They stop fungi from producing ergosterol, an essential component of fungal cell membranes. If ergosterol synthesis is completely or partially inhibited, the fungal cell is unable to construct an intact cell membrane, and dies. Most of the azole formulations are available as over-the-counter medications in various forms: tablets, lozenges, creams, ear drops, etc. Fungal infections are one of the most common causes of skin disease. They are usually treated with topical and oral imidazoles and triazoles among other types of antifungal agents. In particular, tinea pedis (athlete's foot) and tinea cruris (jock itch) are commonly treated topically with azoles. The latest developments in systemic antifungals are based on triazoles (Bennet, Reference Bennet, Brunton, Lazo and Parker2006).

Figure 1. Chemical structure of (a) clotrimazole and (b) fluconazole.
A search of the PDF-4/Organics 2013 database lead to three reports related to clotrimazole: PDF 00-035-1915, 00-051-2002, and 02-071-0743 (ICDD, Reference Kabekkodu2013). The first two contain low-quality experimental patterns (Hoogerheide and Wyka, Reference Hoogerheide, Wyka and Florey1982 and Mayer, Reference Mayer1999, respectively). The third (Song and Shin, Reference Song and Shin1998) corresponds to a calculated pattern using the crystallographic information contained in the Cambridge Structural Database (CSD, V 5.33) (Allen, Reference Allen2002). This is the only report included in the CSD for this important pharmaceutical compound (Refcode PUVRIH).
For fluconazole, there are eight patterns reported in the PDF-4/Organics 2013 database. Five of them are unindexed experimental patterns (PDF 00-057-1444, 00-058-1926, 00-059-1308, 00-062-1568, and 00-062-1569). The other three are calculated using the crystal structure data reported by Caira et al. (Reference Caira, Khouloud and Rana2004) and contained in the CSD. The first one (PDF 02-085-0756) corresponds to a phase containing one-quarter of an ethyl acetate molecule (Refcode IVUQEV), another (PDF 02-085-0757) to a monohydrated fluconazole (Refcode IVUQIZ), and the third (PDF 02-085-0758) to anhydrous fluconazole (Refcode IVUQOF).
As part of the work being carried out in our laboratory to explore the possible formation of polymorphs of important APIs under different crystallization conditions, high-quality powder diffraction data for anhydrous clotrimazole and monohydrated fluconazole were recorded and analyzed. The chemical nature of the materials under study was examined by FT-IR spectroscopy and thermogravimetric analysis/differential thermal analysis (TGA/DTA).
II. EXPERIMENTAL
A. Crystallization experiments
Clotrimazole and fluconazole, provided by Laboratorios CAM, C.A., were recrystallized by slow evaporation from different solvents. Several experiments were carried out in acetone, methanol, and methanol/H2O (with a 4:1 volume ratio) at room temperature. Recrystallization of clotrimazole was also tried in glacial acetic acid. The materials obtained after each crystallization experiment were analyzed by IR spectroscopy, thermal analysis and X-ray powder diffraction.
B. IR spectroscopy and thermal analysis
FT-IR spectra were recorded in KBr pellets, using a Perkin Elmer RX1 spectrophotometer with Spectrum software. Thermogravimetric and derivative thermogravimetric analysis (TGA/DTG) and DTA were carried out in a SDT Q600V3 thermal analyzer using samples of 7–8 mg, heated up to 600 °C at a rate of 10 °C min−1, under a dynamic nitrogen atmosphere at 100 ml min−1.
C. X-ray powder diffraction data collection
Powder diffraction patterns were recorded at room temperature on a BRUKER D8 ADVANCE diffractometer working in the Bragg–Brentano geometry using CuKa radiation (Λ = 1.54184 Å), operating at 40 kV and 30 mA. The patterns were recorded in steps of 0.015 26° (2Θ), from 5 to 70° at 1.5 s step−1. The diffractometer was equipped with primary and secondary Soller slits of 2.5°, divergence slit of 0.2 mm, Ni filter of 0.02 mm, and a LynxEye detector. The profile fit of each pattern was carried out with the FULLPROF software (Rodriguez-Carvajal, Reference Rodriguez-Carvajal1990). After the peak positions were established, the indexing of the patterns was performed with the program DICVOL06 (Boultif and Louër, Reference Boultif and Loüer2004).
III. RESULTS AND DISCUSSION
The FT-IR spectra of clotrimazole recrystallized in all the solvents used were very similar and contain the expected vibrations associated with this molecule. The stretching vibration of the imidazole C–H appears at 3111.49 cm−1, as expected. Aromatic C–H stretching vibrations appear at 3083.41, 3062.12, 3027.05, and 3005.49 cm−1. The C = C stretchings in the aromatic rings are observed at 1617.81, 1586.78, 1565.56, and 1490.81 cm−1. The absorption band of the C = N in the imidazole ring appears at 1347.38 cm−1 and the C–Cl stretch at 765.38 cm−1. No absorption bands associated with presence of water or other solvent were observed. Similarly, the spectra registered for all of the recrystallized fluconazole samples were the same. The absorption observed at 3156.02 cm−1 is consistent with the presence of water. The absorption at 3107.85 cm−1 is associated with the C–H stretch of the triazole rings. The C–H stretches of the 2,4-difluorobenzyl group appear at 3062.46 and 3019.94 cm−1 and the C = C stretches of this group appear at 1619.65 and 1592.44 cm−1. The absorptions at 1369.94 and 1248.99 cm−1 correspond to the C = N stretches of the triazole rings. The C–F and C–OH stretches appear at 1276.24 and 1018.70 cm−1, respectively. The results obtained from the fluconazole IR spectra are consistent with those reported by Caira et al. (Reference Caira, Khouloud and Rana2004).
The TGA–DTG curves for raw and all recrystallized clotrimazole show the absence of solvent and no weight loss up to approximately 200 °C. Above this temperature, a series of decomposition processes take place in at least four steps up to 600 °C. The corresponding DTA trace of the methanol recrystallized material shows an endotherm at 147.71 °C, which corresponds to the melting point and agrees with the reported value of 147–149 °C, in the Merck Index (O'Neil et al., Reference O'Neil, Heckelman, Merck Koch and Roman2006).
For the recrystallized fluconazole samples, the TGA–DTG traces exhibit a weight loss of about 5.18% indicating the presence of a water molecule. This water content may come from the hydrated raw material and/or from the presence of water in the methanol used. In the sample recrystallized in methanol, this dehydration occurs at 105.50 °C as indicated in the DTA by an endotherm. A second endotherm indicates that the melting of the dehydrated material takes place at 141.83 °C, in agreement with the value reported in the Merck Index (O'Neil et al., Reference O'Neil, Heckelman, Merck Koch and Roman2006). The decomposition process begins at around 200 °C.
The powder diffraction patterns recorded for the samples recrystallized in acetone, methanol, and methanol/H2O were very similar among them and similar to the pattern recorded for raw clotrimazole. The pattern of raw fluconazole contained the diffraction maxima of the patterns recorded for the recrystallized samples and four additional maxima, which coincide with some of the most intense peaks of the unindexed experimental pattern reported in entry PDF 00-059-1308. The X-ray powder patterns obtained for clotrimazole and fluconazole, recrystallized in methanol, are shown in Figures 2 and 3, respectively. In both patterns the square root of the intensity is plotted against 2θ in order to better show the weak diffraction maxima.

Figure 2. Powder diffraction pattern of clotrimazole recrystallized in methanol.

Figure 3. Powder diffraction pattern of monohydrated fluconazole recrystallized in methanol.
The diffraction patterns of recrystallized clotrimazole matched a pattern reported in the PDF-4/Organics (entry: PDF 02-071-0743) which corresponds to a pattern calculated using the crystal structure data reported in entry with refcode PUVRIH of the CSD for anhydrous clotrimazole (Figure 4). This is in accordance with the TGA/DTA results. The indexing of the best pattern recorded, from the sample recrystallized in methanol, carried out with the computer program DICVOL06 (Boultif and Louër, Reference Boultif and Loüer2004), using the first 20 peaks, produced a triclinic unit cell. The analysis of all the 71 diffraction maxima registered, performed with NBS*AIDS83 (Mighell et al., Reference Mighell, Hubbard and Stalick1981), using the unit cell obtained by DICVOL06 led to the following unit cell parameters: a = 8.776(1) Å, b = 10.571(2) Å, c = 10.622(3) Å, α = 114.08(2)°, β = 96.87(2)°, γ = 97.61(2)°, V = 875.2(2) Å3. The de Wolff (de Wolff, Reference de Wolff1968) and Smith–Snyder (Smith and Snyder, Reference Smith and Snyder1979) figures of merit obtained were M 20 = 22.2 and F 30 = 40.5(0.0098, 76), respectively. The results are presented in Table I. The diffraction pattern recorded for the sample recrystallized in glacial acetic acid was notably different from the other patterns registered. Single-crystal data are being collected in order to determine the structure of this potentially new phase.

Figure 4. Comparison of the powder diffraction pattern of clotrimazole recrystallized in methanol (blue) with the pattern calculated in PDF 02-071-0743 (red).
Table I. X-ray powder diffraction data of clotrimazole recrystallized in methanol.

In the case of fluconazole, the patterns of the recrystallized samples are very similar to the pattern reported in entry PDF 02-085-0757, calculated from entry IVUQIZ of the CSD, which corresponds to a monohydrated fluconazole. As it was the case for clotrimazole, for fluconazole, the sample obtained after recrystallization in methanol produced the best diffraction pattern. The comparison of this pattern with the calculated pattern of PDF 02-085-0757 is shown in Figure 5. The best result obtained in the indexing of the first 20 peaks of this pattern, carried out with DICVOL06 (Boultif and Louër, Reference Boultif and Loüer2004), was a triclinic unit cell. The analysis, using this triclinic unit cell, carried out with NBS*AIDS83 (Mighell et al., Reference Mighell, Hubbard and Stalick1981) for the 80 diffraction maxima recorded, produced the following unit cell parameters:
a
= 5.6353(4) Å,
b
= 11.753(1) Å,
c
= 12.326(1) Å, α = 71.220(8)°, β = 79.896(9)°, γ = 84.35(1)°, and V = 760.13(9) Å3. The de Wolff (de Wolff, Reference de Wolff1968) and Smith–Snyder (Smith and Snyder, Reference Smith and Snyder1979) figures of merit obtained were M
20 = 48.2 and F
30 = 117.8(0.0065, 39), respectively. The results of this analysis are presented in Table II. Both compounds crystallize in the triclinic system, with space group P
![]() ${\bar 1}$
, and Z = 2, as indicated in the Cambridge Structural Database.
${\bar 1}$
, and Z = 2, as indicated in the Cambridge Structural Database.

Figure 5. Comparison of the powder diffraction pattern of monohydrated fluconazole recrystallized in methanol (blue) with the pattern calculated in PDF 02-085-0757 (red).
Table II. X-ray powder diffraction data of fluconazole recrystallized in methanol.

ACKNOWLEDGEMENTS
This work was made possible thanks to grant LAB-97000821 from FONACIT-Venezuela for Laboratorio de Cristalografía-LNDRX and by a grant for Laboratorio de Difracción de Rayos-X, PTG, Universidad Industrial de Santander, Bucaramanga, Colombia. The authors thank Marlin Villarroel for technical assistance with the TGA/DSC analysis.