I. INTRODUCTION
As a deoxycytidine analog that interferes with DNA synthesis, gemcitabine (Figure 1), systematic name 4-amino-1-[3,3-difluoro-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2-one, is currently one of the most widely used and efficacious anticancer drugs. (Mini et al., Reference Mini, Nobili, Caciagli, Landini and Mazzei2006). Gemcitabine combined with cytotoxins or molecularly targeted agents has been used in various carcinomas, including lung, pancreatic, bladder, and breast cancer (Plunkett et al., Reference Plunkett, Huang and Gandhi1995; Moysan et al., Reference Moysan, Bastiat and Benoit2013).
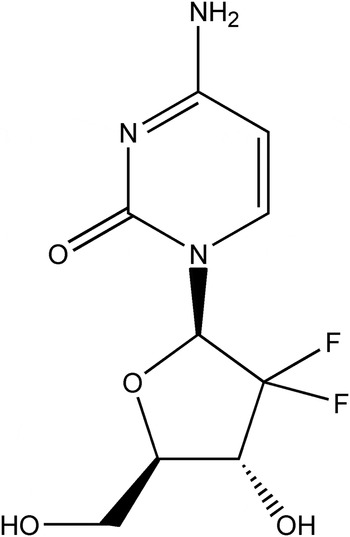
Figure 1. Structural formula of gemcitabine.
The crystal structure of gemcitabine has not been reported so far.
II. EXPERIMENTAL
A. Sample preparation
The title compound was purchased from J&K Chemical Co., Ltd., China. It was re-crystallized in methanol, dried, and ground into powder. The structure of gemcitabine was characterized by high-performance liquid chromatography (HPLC), ultraviolet (UV), and infrared (IR).
B. Diffraction data collection and reduction
The diffraction pattern for the title compound was collected at room temperature using an X'Pert PRO diffractometer (PANalytical Co., Ltd., Netherlands) with an X'celerator detector and CuKα 1 radiation (λ = 1.540 56 Å, generator setting: 40 kV and 40 mA). The diffraction data were collected over the angular range from 5° to 50° 2θ with a step size of 0.013 13° 2θ and a counting time of 30 ms step−1.
All the structure solution work was performed using the software package Material Studio 4.2 (Accelrys Co., Ltd. USA) and the powder diffraction pattern was pre-treated (Wu et al., Reference Wu, Li, Xu, Zhang, Wu and Li2014). Indexing was carried out using peak positions obtained from the powder diffraction profiles by the X-Cell method. Then the best indexing results with 1074 for the value of figure of merit were refined using Pawley refinement (Pan et al., Reference Pan, Guo, Duan, Cheng and Li2012). MC/SA search algorithm in the Powder Solve package (Engel et al., Reference Engel, Wilke, König, Harris and Leusen1999) was used to constantly adjust the conformation, position, and orientation of the trial model in a unit cell of gemcitabine. The result of Powder Solve (R wp = 6.59%) was refined by Rietveld refinement techniques based on the experimental X-ray powder diffraction pattern. In the Rietveld refinement (Young, Reference Young1993; Li et al., Reference Li, Wu, Pan, Cheng and Li2014), variables defining the structural model and the powder diffraction profiles were refined by the least-squares methods for obtaining an optimal fit between the experimental pattern and the calculated pattern. After the Rietveld refinement, the final R wp was 8.34%.
III. RESULTS
The experimental powder diffraction pattern is depicted in Figure 2. Indexing results confirmed that gemcitabine is orthorthombic with space group Pmna and unit-cell parameters after Pawley refinement: a = 17.641(8) Å, b = 6.985(1) Å, c = 18.653(2) Å, α = β = γ = 90°, unit-cell volume V = 2298.61 Å3, Z = 8. The values of 2θ obs, d obs, I obs, h, k, l, 2θ cal, d cal, and Δ2θ are listed in Table I.

Figure 2. X-ray powder diffraction pattern of the gemcitabine, using CuKα 1 radiation (λ = 1.540 56 Å).
Table I. Indexed X-ray powder diffraction data of gemcitabine, C9H11F2N3O4. Only the peaks with I rel of 1 or greater are reported [a = 17.641(8) Å, b = 6.985(1) Å, c = 18.653(2) Å, α = β = γ = 90°, unit-cell volume V = 2298.61 Å3, Z = 8, and space group Pmna]. All measured lines were indexed and are consistent with the Pmna space group. The d-values were calculated using CuK α 1 radiation (λ = 1.540 56 Å).

SUPPLEMENTARY MATERIALS AND METHODS
The supplementary material for this article can be found at http://www.journals.cambridge.org/PDJ






