I. INTRODUCTION
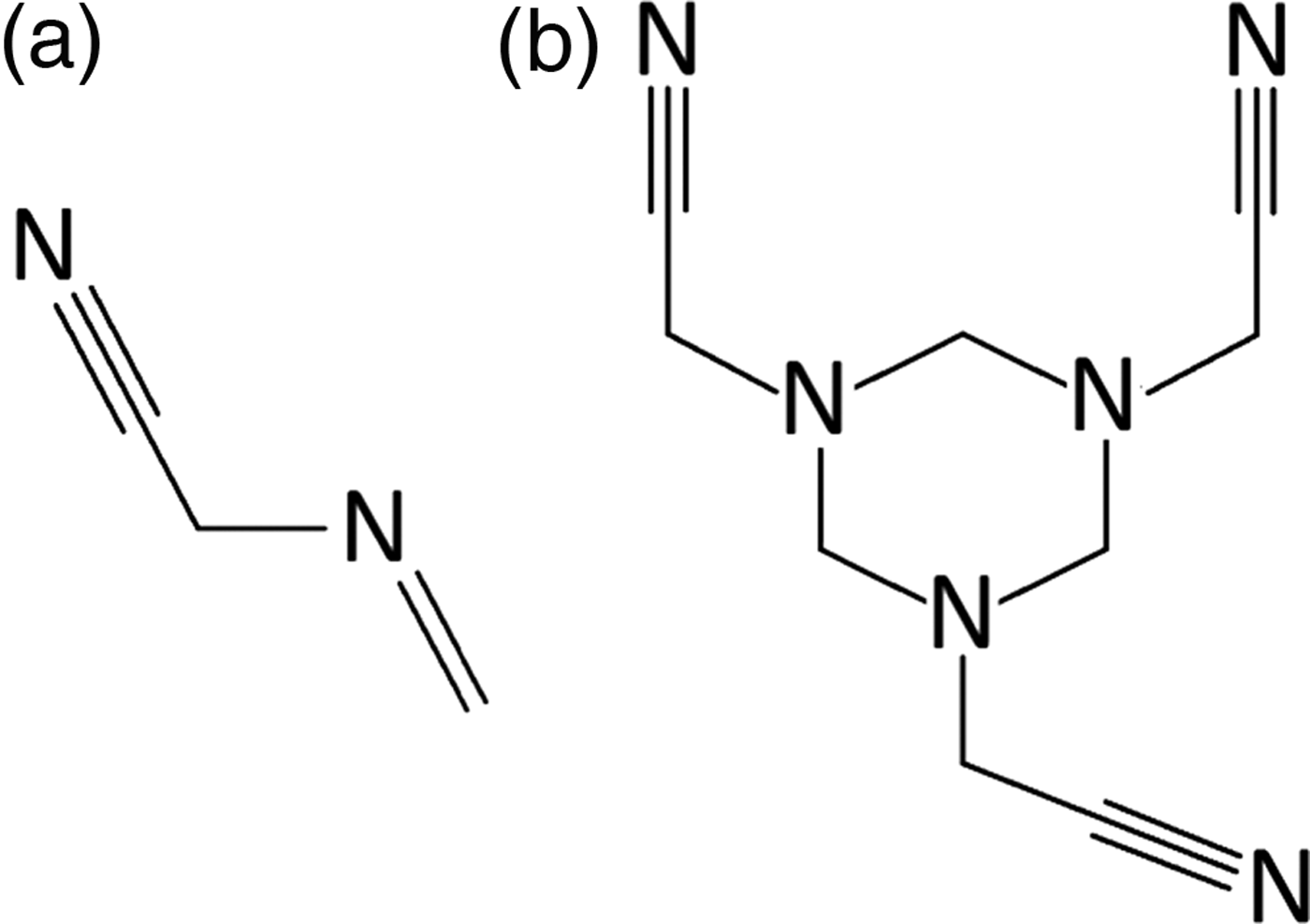
2-(methylideneamino)acetonitrile (MAAN) has raised some interest in the 1970s, owing to its reactivity with amino acids and the formation of peptides (Subbaraman et al., Reference Subbaraman, Kazi, Choughuley and Chadha1975; Kawashiro et al., Reference Kawashiro, Nishiguchi and Nara1989). Reports on its chemical properties included its ability to polymerize as a (cyclic) trimer (Figure 1). Several years later, Xiang et al. (Reference Xiang, Drenkard, Baumann, Hickey and Eschenmoser1994) had suggested a synthetic route in which the MAAN monomer is obtained by decomposition of the trimer, with a yield of 19%. Although both materials are reported to be crystalline, at the point of this study, no evidence for structural analysis of either compound was found. Material characterization, based on infrared spectroscopy (IR), nuclear magnetic resonance (NMR), or Mass Spectrometry, was performed by various workers and deposited in several databases. Although these results are interpreted as the monomer MAAN, some inconsistencies were noticed (Kalinovčić, Reference Kalinovčić2012). This suggested that reliable monomer–trimer disambiguation cannot rely solely on a single analytical technique. (i) In case of systems with similar functional groups (such as polymers) it may be challenging to discern one from the other, using IR data. (ii) Mass spectrometry in this case can be a useful technique, provided that a molecular ion can be obtained. However, neither of these techniques offers reliable identification of mixtures or exact molecular structure (conformation). (iii) NMR spectroscopy, on the other hand, is more straightforward, but may become quite complex, depending on the symmetry of the molecule, the method used (1H, 13C-NMR), and the purity of the sample. Moreover, the molecular conformation cannot be determined. In this study, based on a MAAN sample, the problem of polymer disambiguation is addressed by combining analytical techniques (IR, XRPD) with computational methods (DFT). In conclusion, some general guidelines for a full and reliable analysis of molecular crystals are presented.
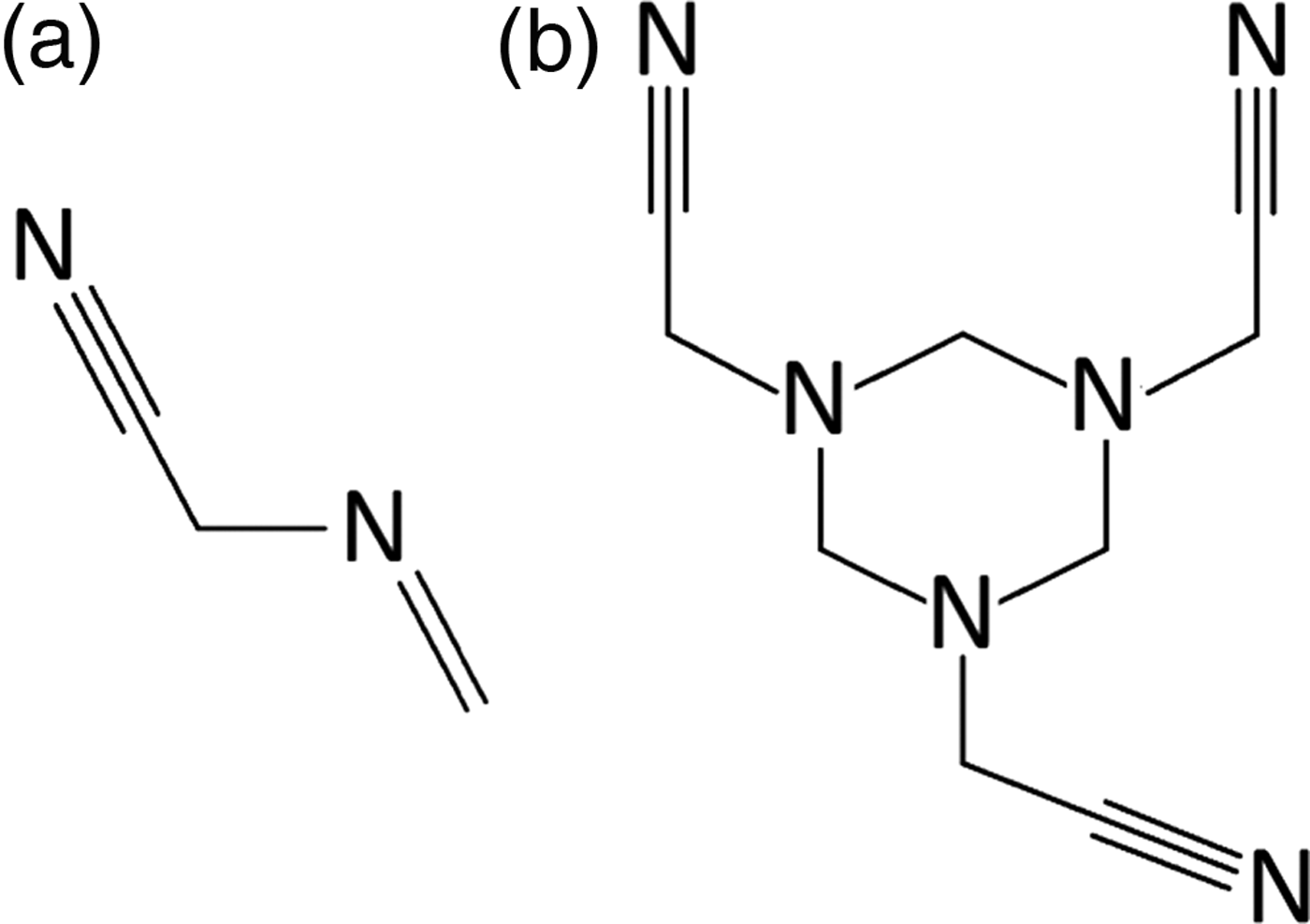
Figure 1. Two-dimensional molecular diagrams for (a) monomeric and (b) trimeric MAAN.
II. EXPERIMENTAL
A. Constructing an ideal molecular conformation
Given that very little was known about the molecular structures of both the monomer and trimer, the first part of the present study addresses possible conformations of these two species. For each molecule, a conformational search was carried out using a stochastic approach, implemented in the program qcc (Hrenar; Primožič et al., Reference Primožič, Hrenar, Baumann, Krišto, Križić and Tomić2014). Minima on the energetic landscape were assigned as the most probable conformations, and these geometries were then optimized using DFT calculations. All quantum-chemical calculations were performed using the Gaussian 09 program (Frisch et al., Reference Frisch, Trucks, Schlegel, Scuseria, Robb, Cheeseman, Scalmani, Barone, Mennucci, Petersson, Nakatsuji, Caricato, Li, Hratchian, Izmaylov, Bloino, Zheng, Sonnenberg, Hada, Ehara, Toyota, Fukuda, Hasegawa, Ishida, Nakajima, Honda, Kitao, Nakai, Vreven, Montgomery, Peralta, Ogliaro, Bearpark, Heyd, Brothers, Kudin, Staroverov, Keith, Kobayashi, Normand, Raghavachari, Rendell, Burant, Iyengar, Tomasi, Cossi, Rega, Millam, Klene, Knox, Cross, Bakken, Adamo, Jaramillo, Gomperts, Stratmann, Yazyev, Austin, Cammi, Pomelli, Ochterski, Martin, Morokuma, Zakrzewski, Voth, Salvador, Dannenberg, Dapprich, Daniels, Farkas, Foresman, Ortiz, Cioslowski and Fox2013).
1. Methodology
A conformational search for monomeric MAAN compounds was performed via a potential energy surface (PES) scan (0–360°), obtained by varying the torsional coordinate φ 1 using the automatic conformational generator implemented in program qcc. The step size was 10°, giving a total of 37 single-point calculations. These single-point calculations were carried out using DFT at the B3LYP/6-31G(d) level of the theory.
Data from the PES scan were arranged in a one-way array. A parallelized combinatorial optimization algorithm for the arbitrary number of ways (dimensions), implemented in program moonee (Hrenar, Reference Hrenar2014a, Reference Hrenar2014b), was used to determine local minima on the investigated PES. These minima were then re-optimized at the B3LYP/cc-pVTZ level of the theory. To ensure that the geometries obtained were indeed local minima, harmonic frequency calculations were performed. The standard Gibbs energies were calculated at T = 298.15 K and p = 101 325 Pa.
The conformational search for the trimeric form of MAAN was performed using a stochastic search, conducted within the multilevel scheme that uses random generation of structures. Each randomly generated structure was optimized at the molecular mechanics level, and clustering of conformers so obtained was performed. In the next stage, clustered geometries were optimized using a higher level of the theory (B3LYP/3-21G), followed by additional clustering. Local minima were re-optimized at the B3LYP/cc-pVTZ level, and harmonic frequency calculations were performed.
2. Results and discussion
For the monomer, the conformational space was found to comprise three conformers (Figures 2 and 3). Conformer 1 has C s symmetry, whereas conformers 2 and 3 (C 1 symmetry group) represent an enantiomeric pair. The relative Gibbs energy of conformers 2 and 3 is found to be 6.41 kJ mol−1 higher than for conformer 1. According to the Boltzmann distribution, the ratio of conformers at the room temperature is 93:7 in favor of conformer 1.

Figure 2. (Colour online) One-dimensional PES scan spanned by torsional coordinate φ 1 and calculated using the B3LYP/6-31G(d) method.

Figure 3. (Colour online) Monomeric MAAN: (a) minimal energy conformers with symmetry C s, and (b) enantiomeric pair of high-energy conformers with symmetry C 1.
For trimeric MAAN, out of 137 local minima found at the molecular mechanics level, DFT re-optimization and clustering revealed that the lowest energy conformer exhibits two neighboring cyanomethyl groups in equatorial positions, whereas the third one was in an axial position on the six-membered ring (Figure 4).

Figure 4. (Colour online) Lowest-energy conformer of trimeric MAAN (two orientations).
B. IR analysis of a commercial sample
A commercially available sample, purchased under the name MAAN, was analyzed. Its IR spectrum (4000–400 cm−1) was measured using the KBr pellet technique on a Bruker Equinox 55 spectrometer [Figure 5(a)]. Investigation of the spectra obtained revealed that the vibrational band at 1650 cm−1 is either not present or is very weak. This suggested that the sample does not contain a C = N group, that is, the molecule in question is not a MAAN monomer. In order to confirm this, DFT calculations for the lowest energy conformers of the monomer (93:7) and trimer were performed. Anharmonic wavenumbers were calculated using the second-order vibrational perturbational theory. Comparison of the calculated and measured spectra confirmed that the molecule in question is most likely a trimer [Figure 5(b)]. However, it was still not clear (i) if the sample is a mixture, and (ii) if the molecular structure of the trimer obtained from DFT calculations corresponds to the one in the crystal (solid-state sample).

Figure 5. (Colour online) IR spectra for trimeric MAAN (a) measured and (b) calculated.
C. XRPD analysis of a commercial sample
Question (i) about the purity of the commercial sample arose from its ability to polymerize (tendency of C = N bonds to react), and from the fact that one of the possible synthetic routes involves conversion of the trimer to a monomer. Question (ii) about the exact trimer configuration in the solid state originated from the fact that the ideal molecular conformation obtained by the DFT analysis was calculated for a single molecule, rather than a crystal. These two issues were addressed by XRPD analysis.
1. Data collection and structure determination
A ground sample was measured on a Stoe STADI MP diffractometer, in Stoe's application laboratory in Darmstadt (Table I). The XRPD pattern was indexed using the CMPR program package (Toby, Reference Toby2005), revealing a monoclinic cell. The systematic absences were indicative of P21/n symmetry. In order to avoid model bias, the powder charge flipping algorithm was used (Baerlocher et al., Reference Baerlocher, McCusker and Palatinus2007) for structure determination. As this attempt failed, a direct-space approach had to be used. Success for these methods relies on the accuracy of the starting model: this is particularly important for cyclic molecules, as major changes in ring conformation are usually not accepted in direct-space optimization (Černý and Favre-Nicolin, Reference Černý and Favre-Nicolin2004). Hence in this case, information obtained from IR/DFT data was crucial, as the stochastic search resulted in 137 possible conformers of trimeric MAAN. Therefore, the lowest energy conformer of the trimer was optimized using the simulated annealing algorithm with parallel tempering in program FOX (Favre-Nicolin and Černý, Reference Favre-Nicolin and Černý2002). Given that the molecular geometry was believed to be close to the correct one, direct-space optimization was carried out using soft geometrical restraints. Rietveld refinement of the resulting structure was performed by the XRS suite of programs (Baerlocher and Hepp, Reference Baerlocher and Hepp1982), using geometric restraints from the DFT model. In order to allow maximum flexibility of the molecular geometry, throughout the refinement procedure these geometric restraints were kept, but the weight factor was progressively decreased. Finally, positions of hydrogen atoms were calculated by the program Mercury (Macrae et al., Reference Macrae, Edgington, McCabe, Pidcock, Shields, Taylor, Towler and van de Streek2006). For this final model, the scale was adjusted, but the positions of hydrogen atoms were not refined. The details of the refinement are presented in Table II and Figure 6.

Figure 6. (Colour online) Last cycle of the Rietveld refinement: measured pattern (black), calculated (red), and difference curve (blue).
Table I. Details of the XRPD measurement of MAAN commercial sample.
Table II. Details of Rietveld refinement for the MAAN trimer crystal structure.

2. Results and discussion
The crystal structure is governed by the interactions between the hydrogens of the methylene groups and nitrogens in the cyano groups or in the ring. Such an arrangement of interactions results in tubular assemblies, where molecules are stacked on top of each other along the c-axis [Figure 7(a)].

Figure 7. (Colour online) (a) Crystal structure viewed along the c-axis (b) comparison of molecular structure (blue) to the molecular structure obtained from DFT calculations (red).
As expected, the absence of classical (strong) hydrogen bonds in the crystal resulted in minor geometrical distortions of the ideal molecular structure from that obtained from DFT calculation [Figure 6(b)].
After this work was presented at EPDIC, it became apparent that the structure of trimeric MAAN was already solved from single-crystal data. Results were published in Russian (Bushmarinov et al., Reference Bushmarinov, Golovanov and Lyssenko2013). Structure 2, reported in this paper, was compared to the structure obtained from XRPD data (Table III and Figure 8). In particular, the lengths of all the three axes obtained from XRPD data were found to be longer than the corresponding axes obtained from single-crystal data. This difference is most likely caused by different temperatures at which samples were measured: single-crystal data were collected at 100 K, and the XRPD measurement was carried out at room temperature. Moreover, the expansion caused by long irradiation (14 h in the laboratory diffractometer) cannot be excluded. Although XRPD patterns were calculated for both the structures, they could not be compared in a straightforward manner, because of the differences in peak positions. Instead, molecular structures obtained from the XRPD and single-crystal data were compared, and were found to be in good agreement (Figure 8).

Figure 8. Comparison of molecular structure (red) to the molecular structure obtained from single-crystal data (blue).
Table III. Comparison between the structures obtained from the XRPD and single-crystal data.

III. CONCLUSION
The study of MAAN addressed the following: (i) problems in the identification of monomer/trimer species in organic compounds (commercial sample) and (ii) the strategy for resolving this ambiguity. As the analysis of such samples using a single technique was found to be neither complete nor reliable, this work suggests an approach where two experimental analytical techniques (IR, XRPD) and DFT calculations are combined. Such a combination of methods and techniques resulted in a full characterization: purity of the sample, identification of the polymer in question, and its crystal and molecular structures.
ACKNOWLEDGEMENTS
We thank the author, Dr. Bushmarinov, for providing the English version of the paper, re-published after the Russian version, and for scientific discussions on molecular interactions. XRPD data used in this work were measured at STOE Application Laboratory in Darmstadt, Germany. The authors thank the entire STOE team, especially Dr. Thomas Hartmann for excellent technical support and scientific discussions.