I. INTRODUCTION
Minocycline hydrochloride dihydrate (brand names Minocin® oral suspension and Dynacin® anhydrous tablets) is a semisynthetic tetracycline that functions primarily as a bacteriostatic antibiotic (Bstatic), which is a biochemical agent that exerts its antimicrobial effects through the inhibition of protein synthesis, DNA replication, and/or other phases of bacterial cellular metabolism; thus, inhibiting the bacterial growth and reproductive cycle, without killing the bacteria. Minocin is commonly used in the treatment of bacterial infections (Gram-positive, Gram-negative, and other bacterial microorganisms), rheumatoid arthritis, skin or soft tissue infection, trachoma (eye infection), and several other bacterial-causing diseases. Minocin often is considered when penicillin is contraindicated and is utilized as an alternative option only to treat or prevent infections caused by susceptible strains of microorganisms. The IUPAC name (CAS Registry number 128420-71-3) is (4S,4aS,5aR,12aS)-4,7-bis(dimethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide hydrochloride dihydrate. A two-dimensional molecular diagram is shown in Figure 1.
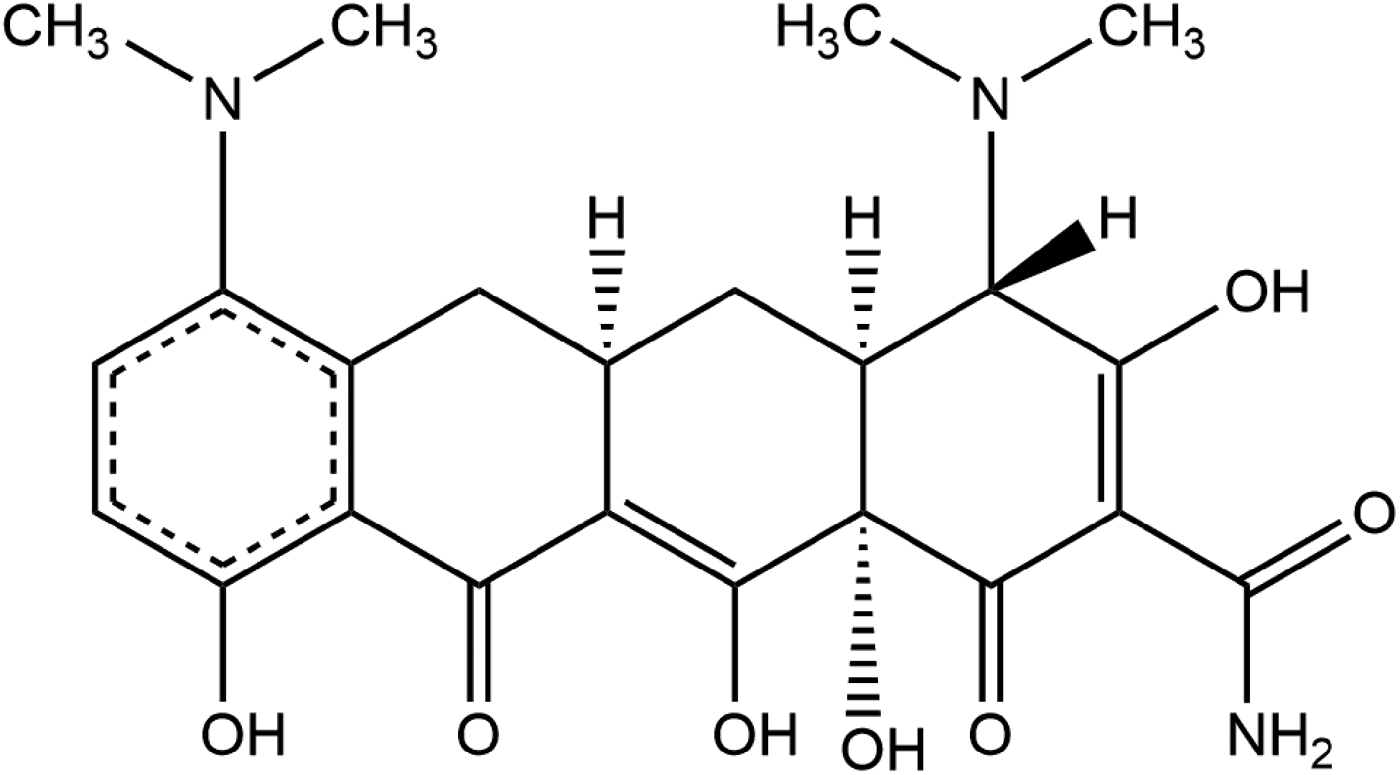
Figure 1. The molecular structure of minocycline (PDB ligand MIY).
Three crystalline polymorphs of minocycline base, as well as a process for producing amorphous minocycline, are claimed in US patent application 2010/0286417 (Mendes et al., Reference Mendes, Antunes, Marto and Heggie2010). Chinese patent CN101693669B (Xiurong et al., Reference Xiurong, Jianming and Linschen2012) claims Form A of minocycline hydrochloride hydrate, and discloses Form B. Form A crystallizes in P212121, with a = 7.405(3), b = 14.452(5), c = 22.315(8) Å, V = 2388.28 Å3, and Z = 4. Form B crystallizes in P3121 with a = 13.143, c = 27.417(7) Å, V = 4101.85 Å3, and Z = 6. A powder pattern for “α-minocycline hydrochloride” (which corresponds to Form A of the Chinese patent) is reported in Rodrigues et al. (Reference Rodrigues, Tiago, Padrela, Matos, Nunes, Pinheiro, Almeida and Gomes de Azavedo2014), as well as a new β polymorph.
This work was carried out as part of a project (Kaduk et al., Reference Kaduk, Crowder, Zhong, Fawcett and Suchomel2014) to determine the crystal structures of large-volume commercial pharmaceuticals, and include high-quality powder diffraction data for these pharmaceuticals in the Powder Diffraction File (Fawcett et al., Reference Fawcett, Kabekkodu, Blanton and Blanton2017).
II. EXPERIMENTAL
Minocycline hydrochloride dihydrate was a commercial reagent, purchased from USP (Lot #R01680), and was used as-received. The yellow powder was packed into a 1.5 mm diameter Kapton capillary, and rotated during the measurement at ~50 Hz. The powder pattern was measured at 295 K at beam line 11-BM (Lee et al., Reference Lee, Shu, Ramanathan, Preissner, Wang, Beno, Von Dreele, Ribaud, Kurtz, Antao, Jiao and Toby2008; Wang et al., Reference Wang, Toby, Lee, Ribaud, Antao, Kurtz, Ramanathan, Von Dreele and Beno2008) of the Advanced Photon Source at Argonne National Laboratory using a wavelength of 0.414163 Å from 0.5 to 50° 2θ with a step size of 0.001° and a counting time of 0.1 s step−1.
The pattern was indexed on a primitive orthorhombic unit cell with a = 7.405, b = 14.456, c = 22.340, V = 2391.4 Å3, and Z = 4 using DICVOL as incorporated into FOX (Favre-Nicolin and Černý, Reference Favre-Nicolin and Černý2002). A reduced cell search in the Cambridge Structural Database (Groom et al., Reference Groom, Bruno, Lightfoot and Ward2016) combined with the chemistry C, H, N, O, and Cl only yielded one hit, but no crystal structure for a minocycline derivative. Analysis of the systematic absences suggested the space group P212121 (#19), which was confirmed by successful solution and refinement of the structure. The structure was solved by direct methods using EXPO2014 (Altomare et al., Reference Altomare, Cuocci, Giacovazzo, Moliterni, Rizzi, Corriero and Falcicchio2013). Some atom types were reassigned manually based on the known connectivity of the molecule. The structure solution yielded the opposite stereoisomer to the known structure. The molecule was inverted using EXPGUI (Toby, Reference Toby2001).
Rietveld refinement was carried out using GSAS (Toby, Reference Toby2001; Larson and Von Dreele, Reference Larson and Von Dreele2004). Only the 1.8–25.0° portion of the pattern was included in the refinement (d min = 0.957 Å). All non-H bond distances and angles were subjected to restraints, based on a Mercury/Mogul Geometry Check (Bruno et al., Reference Bruno, Cole, Kessler, Luo, Motherwell, Purkis, Smith, Taylor, Cooper, Harris and Orpen2004; Sykes et al., Reference Sykes, McCabe, Allen, Battle, Bruno and Wood2011) of the molecule. The Mogul average and standard deviation for each quantity were used as the restraint parameters. The restraints contributed 0.75% to the final χ 2. The hydrogen atoms were included in calculated positions, which were recalculated during the refinement using Materials Studio (Dassault, 2016). Positions of the active hydrogens were derived by analysis of potential hydrogen bonding patterns. A common U iso was refined for the non-H atoms of the fused-ring system, another U iso for the ring-substituent oxygen atoms, another for the nitrogen and carbons for the other substituents, and a common U iso for the water molecules. The U iso for O7 and N12 of the amide group were refined independently. The Uiso for each hydrogen atom was constrained to be 1.3× that of the heavy atom to which it is attached. The peak profiles were described using profile function #4 (Thompson et al., Reference Thompson, Cox and Hastings1987; Finger et al., Reference Finger, Cox and Jephcoat1994), which includes the Stephens (Reference Stephens1999) anisotropic strain broadening model. The background was modeled using a two-term shifted Chebyshev polynomial, with a seven-term diffuse scattering function to model the Kapton capillary and any amorphous component.
The structure was also solved with FOX, using the MIY ligand from the Protein Data Bank (Berman et al., Reference Berman, Westbrook, Feng, Gilliland, Bhat, Weissig, Shindyalov and Bourne2000) and a Cl atom as fragments. This structure (experimental from complexes with proteins) was used because of the variability in the minocycline structures among the various online sources. The oxygen atoms of the two water molecules were located in a difference Fourier map. The conformation of the amide group was opposite to that from the direct methods solution. Refinements were carried out on both models. The simulated annealing model yielded poorer residuals (reduced χ 2 = 4.943 vs. 3.820 for the direct methods model) and an energy 16.0 kcal mol−1 higher, so this model was discarded.
The final refinement of 141 variables using 23293 observations (23200 data points and 93 restraints) yielded the residuals R wp = 0.0869, R p = 0.0708, and χ 2 = 3.820. The largest peak (1.55 Å from H46) and hole (1.63 Å from C23) in the difference Fourier map were 0.58 and −0.58 eÅ−3, respectively. The Rietveld plot is included as Figure 2. The largest errors in the fit are in the shapes of some of the strong, low angle peaks.

Figure 2. (Color online) The Rietveld plot for the refinement of minocycline hydrochloride dihydrate. The red crosses represent the observed data points, and the green line is the calculated pattern. The blue curve is the difference pattern, plotted at the same vertical scale as the other patterns. The vertical scale has been multiplied by a factor of 5 for 2θ > 7.5°, and by a factor of 20 for 2θ > 15.7°.
A density functional geometry optimization (fixed experimental unit cell) was carried out using VASP (Kresse and Furthmüller, Reference Kresse and Furthmüller1996). The calculation used the GGA-PBE functional, a plane wave cut-off energy of 400.0 eV, and a k-point spacing of 0.5 Å−1 leading to a 2 × 1 × 1 mesh. A density functional geometry optimization was also carried out using CRYSTAL14 (Dovesi et al., Reference Dovesi, Orlando, Erba, Zicovich-Wilson, Civalleri, Casassa, Maschio, Ferrabone, De La Pierre, D-Arco, Noël, Causà and Kirtman2014). The basis sets for the H, C, N, and O atoms were those of Gatti et al. (Reference Gatti, Saunders and Roetti1994), and the basis set for chlorine was that of Peintinger et al. (Reference Peintinger, Vilela Oliveira and Bredow2013). The calculation was run on eight 2.1 Ghz Xeon cores (each with 6 GB RAM) of a 304-core Dell Linux cluster at Illinois Institute of Technology, using 8 k-points and the B3LYP functional, and took ~82 h.
III. RESULTS AND DISCUSSION
The experimental powder pattern of minocycline hydrochloride dihydrate matches those of Form A from Xiurong et al. (Reference Xiurong, Jianming and Linschen2012) and that of “α-minocycline hydrochloride” from Rodrigues et al. (Reference Rodrigues, Tiago, Padrela, Matos, Nunes, Pinheiro, Almeida and Gomes de Azavedo2014) well enough to conclude that the materials are the same (Figure 3). The material studied by Rodrigues et al. (Reference Rodrigues, Tiago, Padrela, Matos, Nunes, Pinheiro, Almeida and Gomes de Azavedo2014) was also a commercial sample, so we can conclude that the material studied here is representative of that actually used in the market.

Figure 3. (Color online) Comparison of the synchrotron pattern to that of minocycline hydrochloride dihydrate from Figure 1 of Chinese Patent Application101693669B (reported as hydrate) and Figure 4 from Rodrigues et al. (Reference Rodrigues, Tiago, Padrela, Matos, Nunes, Pinheiro, Almeida and Gomes de Azavedo2014). The Literature patterns (measured using CuK α radiation) were digitized using UN-SCAN-IT, and re-scaled to the synchrotron wavelength of 0.414163 Å using Jade 9.8.
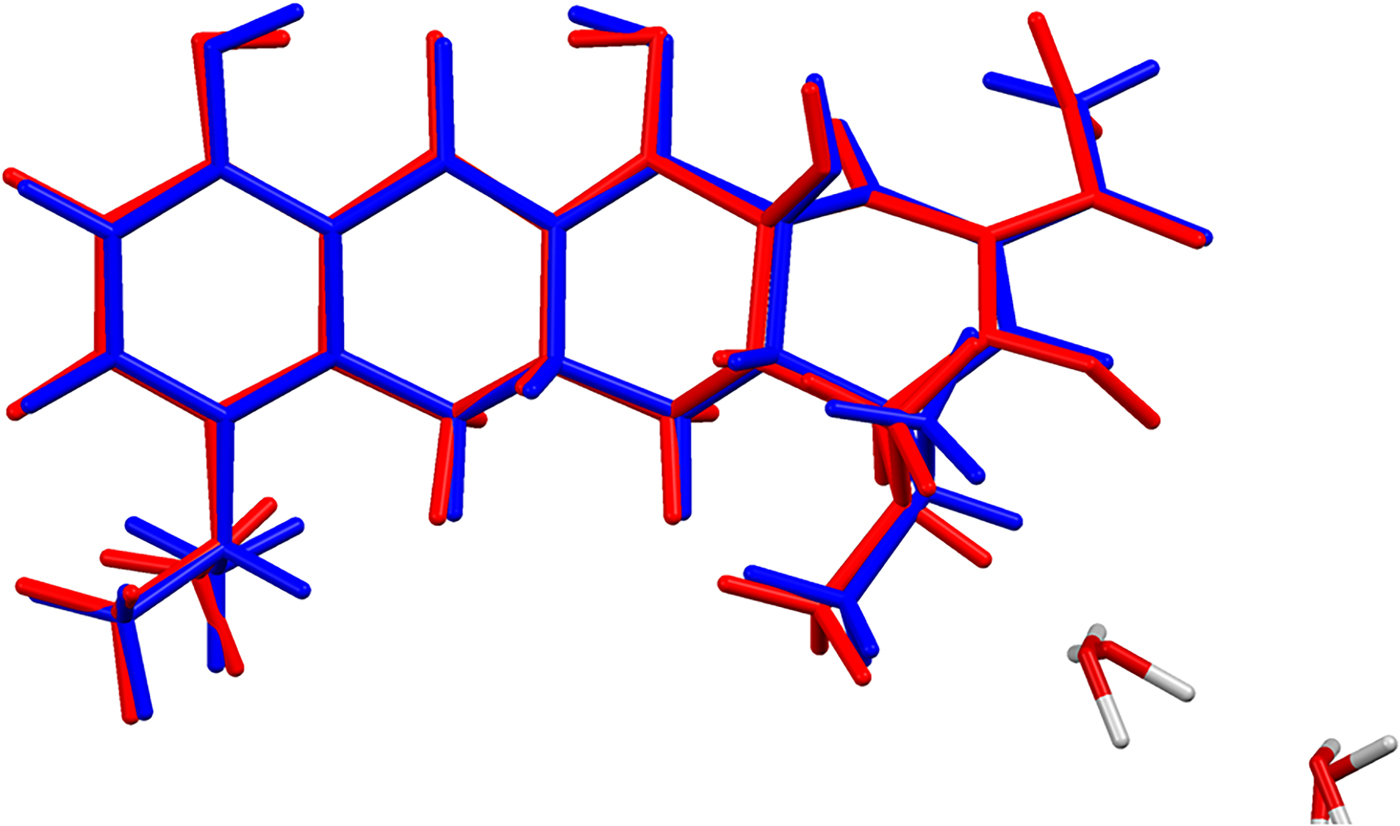
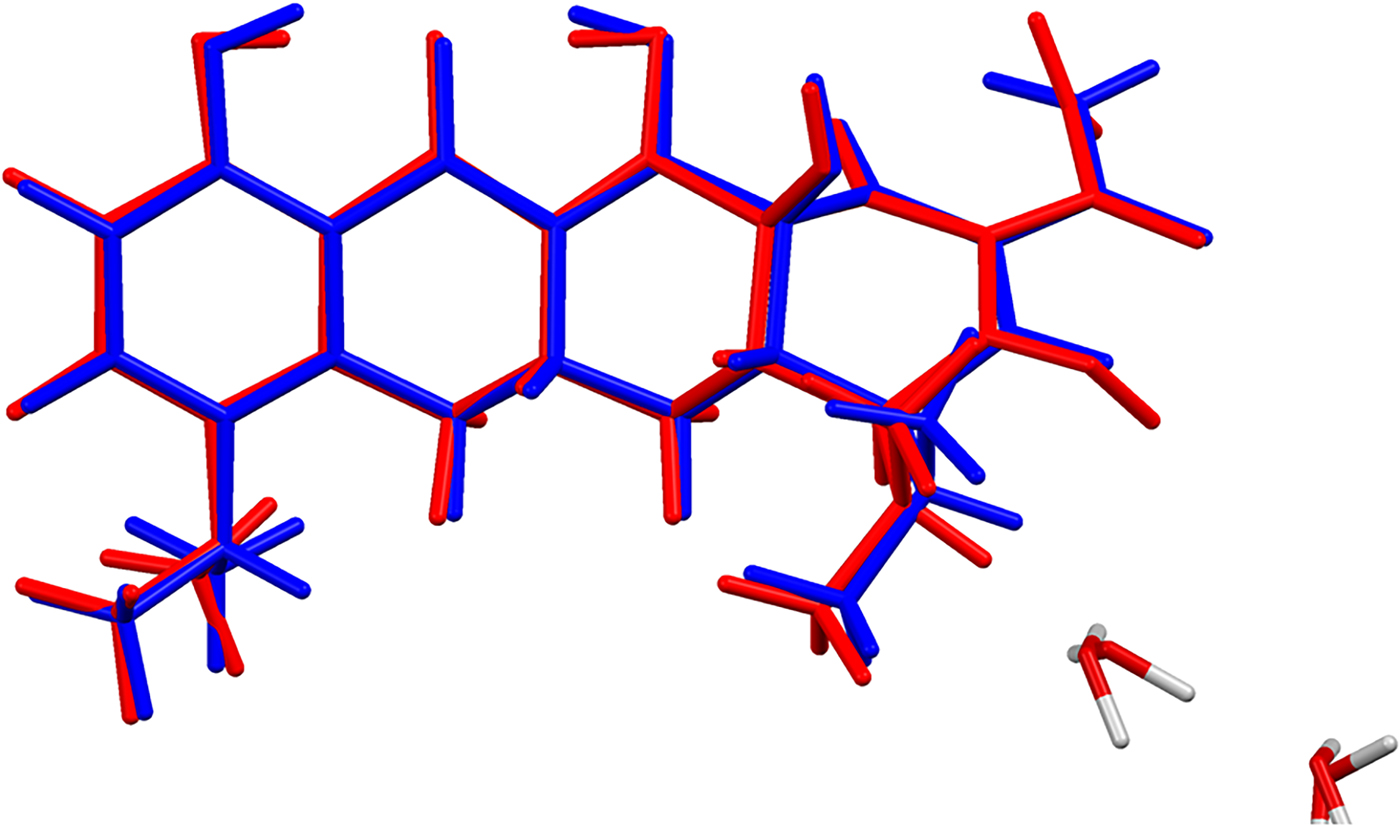
Figure 4. (Color online) Comparison of the refined and optimized (CRYSTAL14) structures of the cation in minocycline hydrochloride dihydrate. The Rietveld refined structure is in red, and the DFT-optimized structure is in blue.
The refined atom coordinates of minocycline hydrochloride dihydrate and the coordinates from the DFT optimizations are reported in the CIFs attached as Supplementary Material. The root-mean-square Cartesian displacement of the non-hydrogen atoms in the minocycline cations is 0.094 Å (Figure 4). The largest deviation is 0.210 Å at C30. The excellent agreement between the refined and optimized structures is evidence that the experimental structure is correct (van de Streek and Neumann, Reference van de Streek and Neumann2014). The optimized structures from VASP and CRYSTAL14 are compared in Figure 5. The structures are almost identical. The RMSD is only 0.0456 Å and the maximum displacement is 0.1339 Å. The major difference was in the orientation of the methyl group C34. This discussion concentrates on the CRYSTAL14-optimized structure. The asymmetric unit (with atom numbering) is illustrated in Figure 6, and the crystal structure is presented in Figure 7.
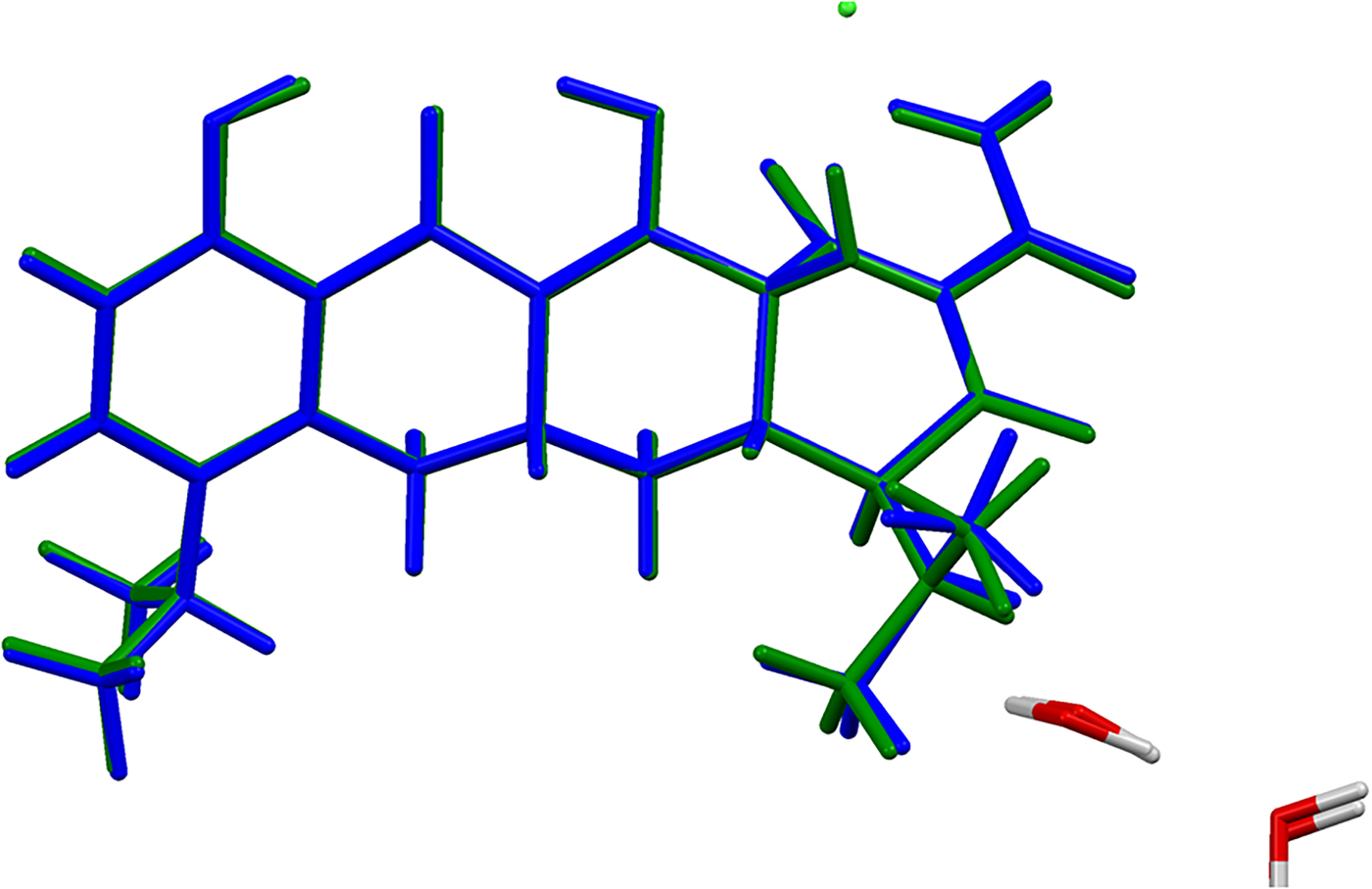
Figure 5. (Color online) Comparison of the VASP (green) and CRYSTAL14 (blue) optimized structures of the minocycline cation.

Figure 6. (Color online) The asymmetric unit of minocycline hydrochloride dihydrate, with the atom numbering. The atoms are represented by 50% probability spheroids.
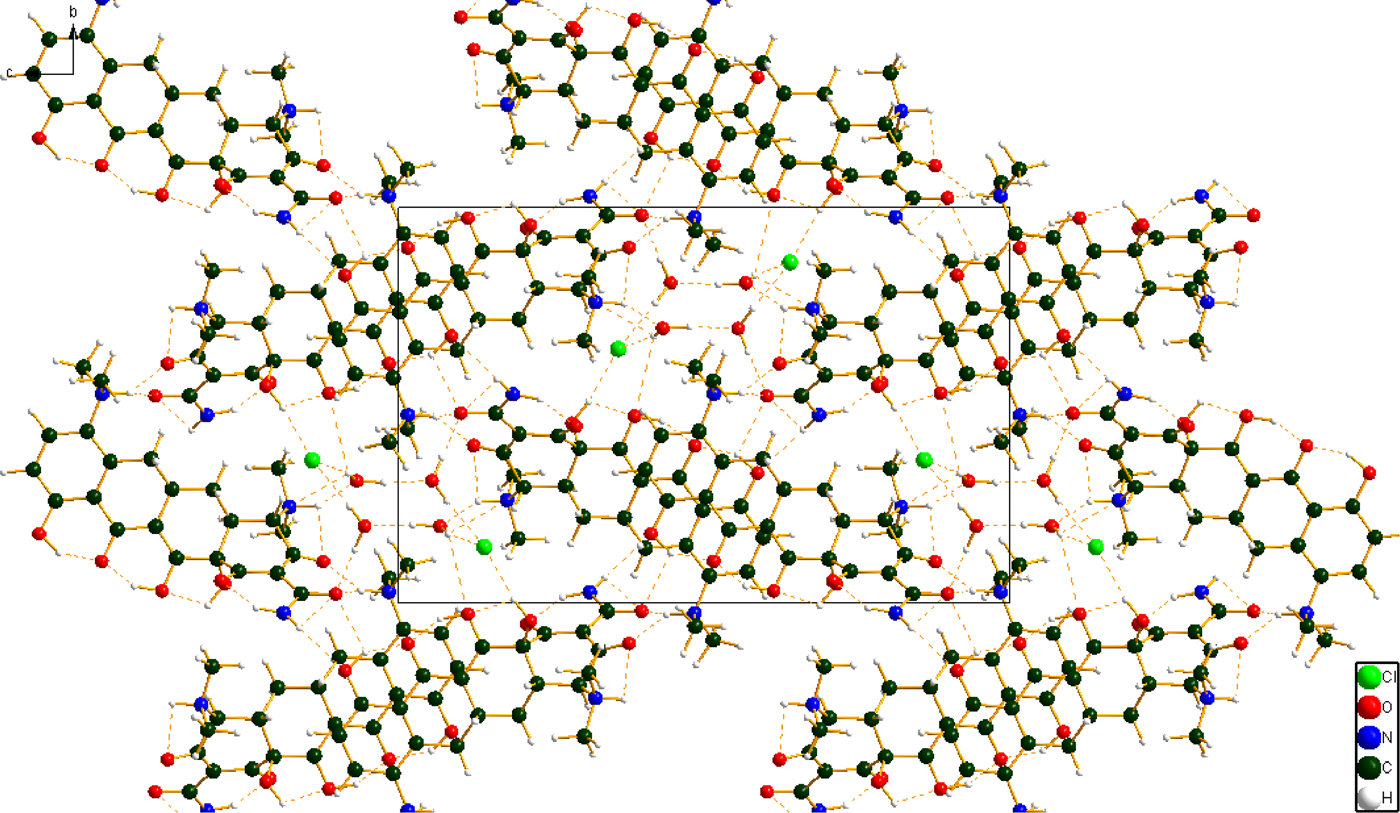
Figure 7. (Color online) The crystal structure of minocycline hydrochloride dihydrate, viewed down the a-axis.
In MIY, O6 is identified as a hydroxyl group, and it was included in the initial Rietveld refinement as O6–H67. The O6⋯N9 intermolecular distance was 2.608 Å, ideally oriented to form a hydrogen bond. In both DFT calculations, H67 moved to protonate N9. Rodrigues et al. (Reference Rodrigues, Tiago, Padrela, Matos, Nunes, Pinheiro, Almeida and Gomes de Azavedo2014) indicate that both dimethylamine groups are protonated in α-minocycline hydrochloride dihydrate, consistent with this observation. In the solid state, the minocycline cation occurs in a different configuration than is usually pictured: the cation is a zwitterion. The C33–O6 distance (1.254 Å from VASP and 1.247 Å from CRYSTAL14) is smaller than the average phenyl–O− distance of 1.304(44) Å from 4519 hits in the CSD (Groom et al., Reference Groom, Bruno, Lightfoot and Ward2016) (Figure 8). The difference is only −1.2 esd, so the distance is reasonable.

Figure 8. (Color online) Histogram of the phenyl–O− distances in the Cambridge Structural Database. The C33–O6 distance (1.254 Å from VASP and 1.247 Å from CRYSTAL14) is smaller than the average phenyl–O− distance of 1.304(44) Å from 4519 hits in the CSD (Groom et al., Reference Groom, Bruno, Lightfoot and Ward2016). The difference is only −1.2 esd, so the distance is reasonable.
There is a potential ambiguity in the orientation of the amide group O7/N12. Direct methods suggested the orientation presented here, while simulated annealing yielded the opposite orientation. The current orientation yielded lower refinement residuals and a lower DFT energy. The displacement coefficients of O7 and N12 were refined independently to values of 0.0708(12) and 0.0662(27), respectively. The difference is only 1.6 esd helping to confirm the atom identifications.
Almost all of the bond distances, bond angles, and torsion angles fall within the normal ranges indicated by a Mercury Mogul Geometry Check (Macrae et al., Reference Macrae, Bruno, Chisholm, Edington, McCabe, Pidcock, Rodriguez-Monge, Taylor, van de Streek and Wood2008). The O3–C17 distance of 1.296 Å [average = 1.228(21), Z-score = 3.2] is flagged as unusual. This carbonyl group acts as an acceptor in two intramolecular O–H···O hydrogen bonds, and is longer than normal, but is not an unreasonable distance. The O5–C27–C22 angle of 106.3° [average = 110.1(12), Z-score = 3.1] is flagged as unusual. The Z-score is the result of the small ESD on the average.
Quantum chemical geometry optimizations (DFT/6-31G*/water) using Spartan ‘16 (Wavefunction, 2017) indicated that the observed conformation of minocycline in minocycline hydrochloride dihydrate is 8.1 kcal mole−1 higher in energy than the local minimum energy conformation. The differences are in the orientations of the protonated dimethylamine groups and the amide suggesting that intermolecular interactions contribute to the observed conformation of the molecule. Attempts to perform molecular mechanics conformational analysis yielded “folded” conformations with the tetracycline bent around the C19–C21 axis.
Analysis of the contributions to the total crystal energy using the Forcite module of Materials Studio (Dassault, 2016) suggests that angle, bond, and torsion distortion terms are significant in the intramolecular deformation energy, as might be expected from a fused ring system. The intermolecular energy contains significant contributions from van der Waals repulsions and electrostatic attractions, which in this force-field-based analysis include hydrogen bonds. The hydrogen bonds are better analyzed using the results of the DFT calculation.
Both water molecules act as donors in O–H···Cl hydrogen bonds to the chloride anion (Table I). The energies of these hydrogen bonds were calculated using the correlation from Kaduk (Reference Kaduk2002). The water molecule O35 acts as a donor to water molecule O36. The energy of this and other O–H···O hydrogen bonds were calculated using the correlation from Rammohan and Kaduk (Reference Rammohan and Kaduk2018). The water molecule O36 acts as a donor to O7, the carbonyl oxygen of the amide group. Both protonated dimethyl amine groups N10 and N9 act as donors in N–H···O hydrogen bonds to the ionized oxygen O6. The energies of these N–H···O hydrogen bonds were calculated using the correlation from Wheatley and Kaduk (Reference Wheatley and Kaduk2018). The bond involving N10–H68 is bifurcated both to O6 and the water molecule O35. Both hydroxyl groups O2 and O4 act as donors in intramolecular hydrogen bonds to the carbonyl oxygen atom O3. The hydroxyl group O5–H60 forms bifurcated hydrogen bonds to the chloride anion and (intramolecular) to the hydroxyl oxygen O2. The hydroxyl group O4–H59 also acts as a donor in an intermolecular hydrogen bond to the O7, the carbonyl oxygen atom of the amide group.
Table I. Hydrogen bonds (CRYSTAL14) in minocycline hydrochloride dihydrate.

*Intramolecular.
The volume enclosed by the Hirshfeld surface (Figure 9; Hirshfeld, Reference Hirshfeld1977; Turner, et al., Reference Turner, McKinnon, Wolff, Grimwood, Spackman, Jayatilaka and Spackman2017) is 587.82 Å3, 98.36% of 1/4 the unit cell volume. The molecules are thus not tightly packed. All of the significant close contacts (red in Figure 9) involve the hydrogen bonds.
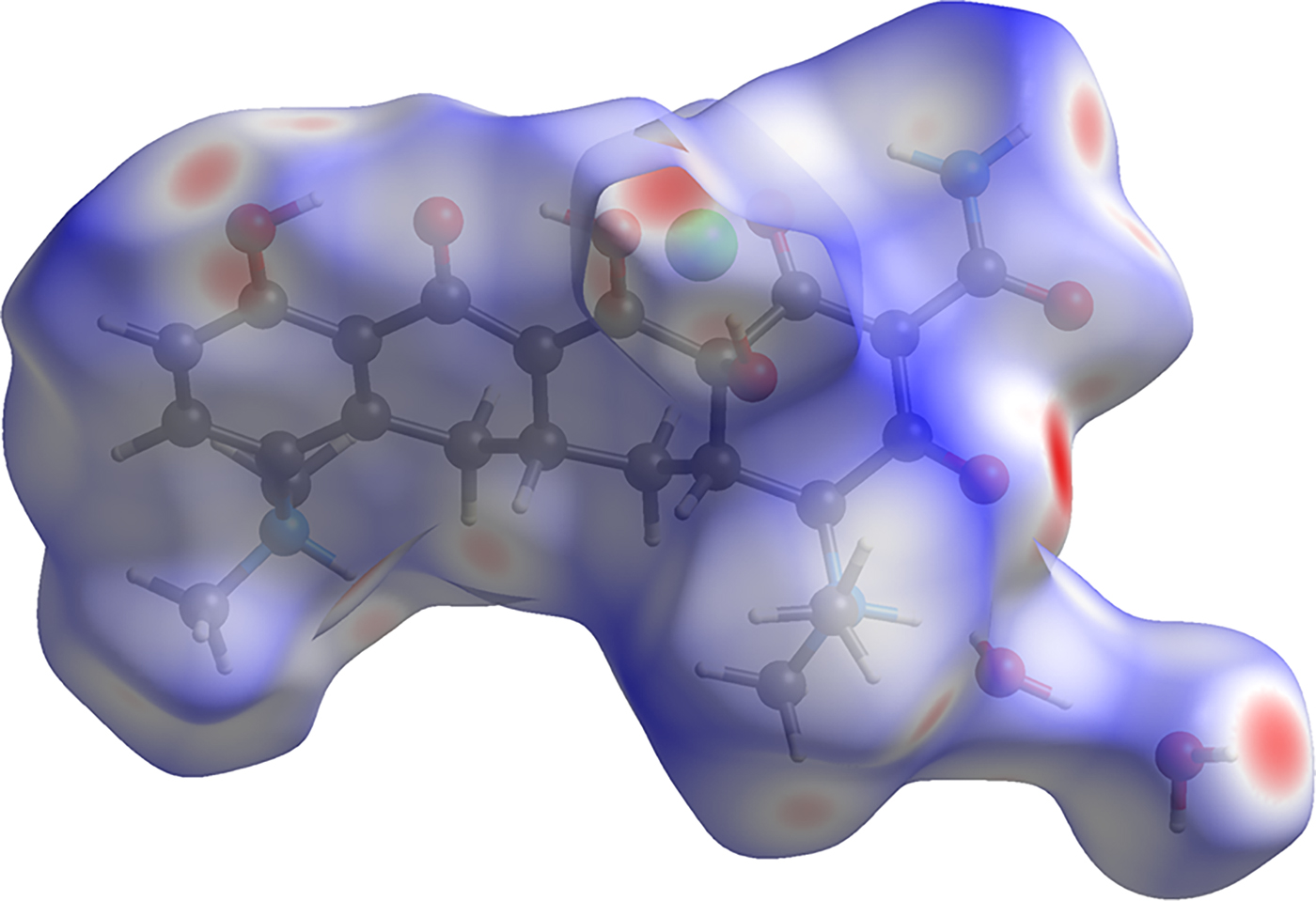
Figure 9. (Color online) The Hirshfeld surface of minocycline hydrochloride dihydrate. Intermolecular contacts longer than the sums of the van der Waals radii are colored blue, and contacts shorter than the sums of the radii are colored red. Contacts equal to the sums of radii are white.
The Bravais–Friedel–Donnay–Harker (Bravais, Reference Bravais1866; Friedel, Reference Friedel1907; Donnay and Harker, Reference Donnay and Harker1937) morphology suggests that we might expect elongated morphology for minocycline hydrochloride dihydrate, with {100} as the long axis. A fourth-order spherical harmonic preferred orientation model was included in the refinement; the texture index was 1.015, indicating that preferred orientation was not significant in this rotated capillary specimen. The powder pattern of minocycline hydrochloride dihydrate from this synchrotron data set is included in the Powder Diffraction File™ as entry 00-066-1606.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S0885715618000787.
ACKNOWLEDGEMENTS
Use of the Advanced Photon Source at Argonne National Laboratory was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was partially supported by the International Centre for Diffraction Data. The authors thank Lynn Ribaud and Saul Lapidus for their assistance in the data collection, and Andrey Rogachev for the use of computing resources at IIT.