Introduction
Systematic classification of Musa began in 1778 when Linnaeus established the Musa genus. In 1887, Sagot (Reference Sagot1887) suggested that this genus is classified into different sections, namely giant bananas, fleshy and edible bananas, and ornamental bananas. Baker adopted Sagot's suggestion and formally divided Musa into three subgenera: Physocaulis, Eumusa and Rhodochlamys (Baker, Reference Baker1893). Cheesman further divided Musa into four sections based on morphology and number of chromosomes: Eumusa (2n= 22), Rhodochlamys (2n= 22), Callimusa (2n= 20) and Australimusa (2n= 20) (Cheesman, Reference Cheesman1947). A new section, Incertae sedis, was added by Simmonds (Reference Simmonds1960), and was proposed to include two species: Musa ingens Simmonds (2n= 14) and Musa beccarii Simmonds (2n= 18). Simmonds also established the morphological taxonomy of wild Musa germplasm (Simmonds, Reference Simmonds1960). Argent (Reference Argent1976) placed M. ingens into the new section Ingentimusa. Häkkinen classified M. beccarii into section Callimusa (Häkkinen et al., Reference Häkkinen, Suleiman and Gisil2005). With the support of several molecular analyses, Häkkinen (Reference Häkkinen2013) restructured Musa species into two sections: Musa and Callimusa. To date, approximately 70 Musa species have been reported (Häkkinen, Reference Häkkinen2013).
Classification of bananas by morphology is limited, and molecular markers are increasingly used in classifying banana germplasm. Previous reports have used molecular markers such as restriction length fragment polymorphisms (RFLP) (Gawel et al., Reference Gawel, Jarret and Whittemore1992), random amplified polymorphic DNA (Howell et al., Reference Howell, Newbury, Swennen, Withers and Ford-Lloyd1997), amplified fragment length polymorphisms (AFLP) (Ude et al., Reference Ude, Pillay, Nwakanma and Tenkouano2002; Wong et al., Reference Wong, Argent, Kiew, Set and Gan2002) and PCR-RFLP (Nwakanma et al., Reference Nwakanma, Pillay, Okoli and Tenkouano2003) to evaluate the genetic diversity of Musa. DNA sequencing techniques have also been widely used in phylogenetic studies of this genus. These techniques include nuclear ribosomal internal transcribed spacers and chloroplast DNA fragments such as trnL-F (Li et al., Reference Li, Häkkinen, Yuan, Hao and Ge2010; Liu et al., Reference Liu, Kress and Li2010), atpB-rbcL, rps16 (Li et al., Reference Li, Häkkinen, Yuan, Hao and Ge2010) and trnT-trnF (Bekele and Shigeta, Reference Bekele and Shigeta2011).
Simple sequence repeat (SSR) markers pinpoint polymorphisms in numbers of repeats for stretches of consecutively repeated small units (one to six nucleotides) (Condit and Hubbell, Reference Condit and Hubbell1991). The mutation rate (approximately 10− 3) is higher than that for point mutations (approximately 10− 9). Consequently, SSRs can generate many more polymorphic markers compared with other methods and can detect recent polymorphisms between closely related accessions (Perrier et al., Reference Perrier, Bakry, Carreel, Jenny, Horry, Lebot and Hippolyte2009). SSRs are spread across the genome and are easy to generate; because of their high levels of polymorphism, co-dominance, efficiency and cost-effectiveness, they are widely used in genetic studies. SSR length polymorphism analysis can detect high levels of polymorphism between individuals of Musa breeding populations (Crouch et al., Reference Crouch, Crouch, Jarret, Cregan and Ortiz1998, Reference Crouch, Ortiz and Crouch2000).
Here, SSRs were used to analyse the phylogeny of wild Musa species. A total of 53 wild Musa species, including 25 species and 28 Musa acuminata subsp. were analysed. This intensive sampling and analysis can improve the understanding of the phylogeny of the M. acuminata complex and the genus. Our specific objectives were to (1) provide new molecular evidence for the classification of wild Musa germplasm resources and (2) to provide valuable information for germplasm collections for cultivated bananas.
Materials and methods
Plant materials and genomic DNA extraction
We analysed accessions of 53 wild species consisting of 25 Musa species and 28 M. acuminata subsp. Of these accessions, 40 were obtained from Bioversity International's ITC (Supplementary Table S1, available online) and the others were collected from China (Table 1). Identification of the wild Musa specimens was performed according to the descriptions of morphological characters provided by Bioversity International (INIBAP/CIRAD, 1996). Genomic DNA was extracted from young leaves using the cetyltrimethylammonium bromide protocol (Paterson et al., Reference Paterson, Brubaker and Wendel1993). Total genomic DNA samples were diluted to 20 ng/μl with sterile H2O.
Table 1 Plant materials of the genus Musa collected from China

SSR analysis
In total, 12 pairs of SSR primers (Supplementary Table S2, available online), synthesized by Invitrogen, Shanghai, were used to analyse the genetic diversity of the Musa samples. SSR assays were carried out in a 20 μl reaction mixture containing 0.125 mM of each deoxy-ribonucleoside triphosphate (dNTP), 10 mM Tris–HCl (pH 8.8), 2 mM MgCl2, 50 mM KCl, 0.08% NP-40, 0.5 μM of each primer, 40 ng genomic DNA and 0.5 U Taq DNA polymerase (Shanghai Sangon, China). Amplification was performed using a Biometra T1 thermocycler (Whatman Biometra, Göttingen, Germany). The PCR reactions were performed as follows: initial denaturation at 94°C for 2 min followed by 30 cycles of denaturation at 94°C for 30 s; annealing at 53–62°C for 45 s and extension at 72°C for 1 min, with a final extension at 72°C for 7 min. The amplified products were separated on 8% polyacrylamide gels in Tris-base/boric acid/Ethylene Diamine Tetraacetie Acid (EDTA) buffer. The gels were stained with silver nitrate as described by Zhang et al. (Reference Zhang, Wu, Guo and Zhang2000). PCR analyses were repeated at least twice to ensure repeatability.
Data analysis
The SSR gel images were analysed with BandScan Software version 5.0 (Glyko Inc., Novato, CA, USA; http://www.glyko.com) and confirmed manually. SSR bands were sized and binary coded with 1 or 0 for their presence or absence in each locus/allele, excluding smeared or weak bands. Polymorphism information content (PIC) was calculated for each primer as follows:
where p is the relative frequency of the jth pattern of SSR marker i (Botstein et al., Reference Botstein, White, Skolnick and Davis1980). Genetic similarity coefficients were determined using the index presented in Nei and Li (Reference Nei and Li1979), and the unweighted pair-group method using arithmetic mean (UPGMA) cluster analysis was performed using the Numerical Taxonomy System version 2.1 (NTSYS-pc) program (Applied Biostatistics Inc., New York, NY, USA; Rohlf, Reference Rohlf2000). The clustering was also tested by bootstrap analysis using the WinBoot program (Yap and Nelson, Reference Yap and Nelson1996, International Rice Research Institute, Manila, Philippines) with 1000 iterations.
Results
Polymorphism of SSR primers
The 12 primer pairs generated polymorphic bands with good repeatability; 91 polymorphic bands were detected. The number of bands generated from each primer ranged from 4 to 15 (average = 7.5). The EST-SSR34 and AGMI67/68 primers generated the largest numbers of bands (11), whereas MA19 produced the fewest bands (4). The PIC was highest in primer EST-SSR34 and lowest in MA19 (average = 0.7226). All primer pairs detected high levels of polymorphism among the samples (Supplementary Table S2, available online).
Phylogenetic analysis of Musa
UPGMA clustering assigned the 53 accessions to two significantly different clusters based on a similarity coefficient of 0.63 (Fig. 1). The first cluster included the accessions of sect. Musa and Musa paracoccinea, which belong to sect. Callimusa with basic chromosome number X= 10. The first cluster splits into four branches near a genetic similarity of 0.78. Branch I comprised 18 subspecies of M. acuminata. Branch II included eight M. acuminata subsp. and four species of sect. Musa that have erect inflorescences and are distributed in China. Branch III consisted of 13 species of sect. Musa (Musa itinerans, Musa balbisiana, Musa nagensium, Musa basjoo, Musa schizocarpa subsp. 502, Musa schizocarpa subsp. 507, Musa balbisiana× Musa textilis, Musa yunnanensis, Musa ornata, Musa acuminata var. chinensis, Musa laterita, Musa velutina and Musa tongbiguanensis) and one species of sect. Callimusa (M. paracoccinea). Musa boman (X= 9) was separated as a single cluster in branch IV.
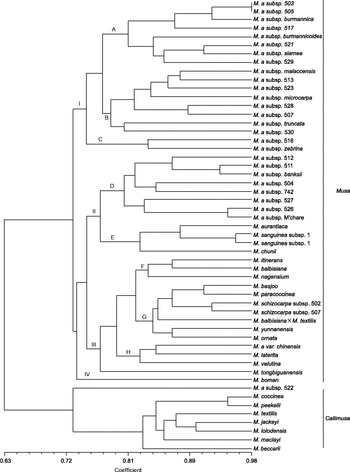
Fig. 1 Dendrogram of genus Musa generated with 12 SSR markers using the unweighted pair-group method with arithmetic mean. M. a, M. acuminata.
The second cluster included seven species of sect. Callimusa, and ‘Marges Elargies’ (M. acuminata subsp. 522) formed a single branch on the dendrogram with a similarity of 0.74.
Phylogenetic analysis of Musa acuminata complex
The UPGMA cluster analysis of 28 accessions of M. acuminata complex constructed with SSR markers separated the accessions into six clusters based on a similarity coefficient of 0.77 (Fig. 2): (1) ‘Tavoy’ (subsp. burmannica), ‘Calcutta 4’ (subsp. burmannicoides), ‘Khae (Phrae)’ (subsp. siamea), ‘Type 3 × ,’ ‘Type 2 × ,’ ‘Pahang IRFA,’ ‘Pa (Musore) no. 3,’ ‘Pisang Cici Alas’; (2) ‘Malaccensis’ (subsp. malaccensis), ‘Borneo’ (subsp. microcarpa), ‘Truncata’ (subsp. truncata), ‘Hybrid 513,’ ‘Selangor 2,’ ‘Higa,’ ‘Hybrid 507,’ ‘Pa (Songkhla)’; (3) ‘Zebrina’ (subsp. zebrina), ‘Pisang Cici’; (4) ‘Banksii’ (subsp. banksii), ‘A3617/9,’ ‘Hybrid 511,’ ‘Agutay,’ ‘Rung Hoa Xoan,’ ‘THA018,’ ‘Vietnam no. 5,’ ‘Makyughu II’; (5) ‘Xiao Guo Ye Jiao’ (var. chinensis); and (6) ‘Marges Elargies.’

Fig. 2 Dendrogram of Musa acuminata (M. a) complex generated with 12 SSR markers using the unweighted pair-group method with arithmetic mean.
Discussion
Infrageneric phylogeny and classification of Musa
The genus Musa was previously separated into five sections (Eumusa, Rhodochlamys, Callimusa, Australimusa and Ingentimusa) based on basic chromosome numbers and morphological characters (Cheesman, Reference Cheesman1947; Simmonds, Reference Simmonds1960, Reference Simmonds1962; Argent, Reference Argent1976; Simmonds and Weatherup, Reference Simmonds and Weatherup1990). With support from several molecular analyses, Häkkinen (Reference Häkkinen2013) restructured Musa species into two sections, Musa and Callimusa. Our results are largely congruent with those of previous molecular studies (Gawel et al., Reference Gawel, Jarret and Whittemore1992; Wong et al., Reference Wong, Argent, Kiew, Set and Gan2002; Nwakanma et al., Reference Nwakanma, Pillay, Okoli and Tenkouano2003; Liu et al., Reference Liu, Kress and Li2010; Li et al., Reference Li, Häkkinen, Yuan, Hao and Ge2010; Bekele and Shigeta, Reference Bekele and Shigeta2011; Christelová et al., Reference Christelová, Valárik, Hřibová, de Langhe and Doležel2011). By SSR-marker UPGMA cluster analysis, 53 accessions were grouped into two significantly different clusters. One cluster comprised species of sect. Musa with basic chromosome number of X= 11; most species of sect. Callimusa (X= 10/9) formed the other cluster. These results provide new molecular evidence for sectional relationships in the genus Musa.
Relationships within sect. Musa
Although the basic chromosome number of both Musa and Rhodochlamys was X= 11; these sections were separated by differences in morphological characteristics and edibility of fruit. The inflorescences of Musa are pendent or semi-pendent; the bracts are commonly green, brown, or dull violet; and the fruit are usually edible. In contrast, inflorescences of Rhodochlamys are erect, the bracts are always bright in colour (often red), and the fruit are usually inedible (Cheesman, Reference Cheesman1947). Despite these differences, however, there is no significant evidence to support separate classification. SSR analysis revealed cross-clustering of Musa and Rhodochlamys species. This result was consistent with that of Ude et al. (Reference Ude, Pillay, Nwakanma and Tenkouano2002); however, Ude proposed that Rhodochlamys be considered a separate category. In contrast, we agree with the suggestions of Wong et al. (Reference Wong, Argent, Kiew, Set and Gan2002) and Christelová et al. (Reference Christelová, Valárik, Hřibová, de Langhe and Doležel2011) that sections Musa and Rhodochlamys be considered as one category.
Musa chunii, a recently described species (Häkkinen and Hong, Reference Häkkinen and Hong2007), has a close relationship with Musa aurantiaca and Musa sanguinea (subclade E); each of these species has erect inflorescences and brightly coloured bracts and is distributed in Yunnan, China. M. laterita and M. velutina, both from sect. Musa and having erect inflorescences and brightly coloured bracts, cluster with M. acuminata var. chinensis (subclade H). Ude et al. (Reference Ude, Pillay, Nwakanma and Tenkouano2002) maintained that M. laterita was closely related to M. acuminata and had the closest relationship with M. acuminata subsp. burmannica. In this study, we examined numerous M. acuminata subspecies, including the new variety of M. acuminata (M. acuminata var. chinensis) discovered by Häkkinen and Hong (Reference Häkkinen and Hong2007) in China. We observed that M. laterita had the closest genetic relationship to M. acuminata var. chinensis (similarity coefficient = 0.84); the similarity coefficient for M. laterita and M. acuminata subsp. burmannica was only 0.73. M. ornata, another species previously of sect. Rhodochlamys, has a close relationship with M. schizocarpa (subclade G), which is considered to have the S genome for cultivated bananas (Ude et al., Reference Ude, Pillay, Nwakanma and Tenkouano2002). This affinity among M. laterita, M. velutina, M. ornata, M. acuminata and M. schizocarpa suggests that they may constitute a secondary gene pool for the improvement of cultivated bananas.
M. balbisiana, M. itinerans and M. nagensium, which form subclade F, are closely related. M. nagensium is unique in seed shape and inflorescence structure and has limited distribution in southern Yunnan and northern Myanmar (Liu et al., Reference Liu, Li and Li2002). M. itinerans is unique for having long rhizomes and commonly grows in southern China. Although M. yunnanensis and M. itinerans have similar-shaped seeds and partly overlap in their distribution ranges (Häkkinen et al., Reference Häkkinen, Hong and Ge2008), they clustered in different subclades. M. yunnanensis clustered with M. ornata, M. basjoo, M. paracoccinea, and M. schizocarpa in subclade G. This is largely in agreement with Li et al. (Reference Li, Häkkinen, Yuan, Hao and Ge2010).
M. tongbiguanensis is a new species of Musa that we observed and described in Yunnan, China (Chen et al., Reference Chen, Feng and Wu2008). The vernacular name of M. tongbiguanensis is ‘lubajiao’ (green banana); its main characteristics are large fruit and seeds (seeds are approximately two-fold larger than those of M. acuminata var. chinensis), and it is only distributed in the Tongbiguan Nature Reserve of Yunnan. Our molecular data indicate that M. tongbiguanensis is a distinct, new species. M. boman formed a single branch; it was placed in sect. Australimusa by Argent (Reference Argent1976) and was confirmed by Gawel et al. (Reference Gawel, Jarret and Whittemore1992) based on RFLP analysis. However, our results showed that M. boman was closer to sect. Musa than to sect. Callimusa.
Furthermore, M. acuminata, M. balbisiana and M. schizocarpa, which have the A, B and S genome for cultivated bananas, respectively, were placed into different subclades. The M. acuminata complex clustered into subclades A, B, C, D and H; M. balbisiana clustered into subclade F; and M. schizocarpa clustered into subclade G. Li et al. (Reference Li, Häkkinen, Yuan, Hao and Ge2010) also observed wide hybridization and suggested that all wild species of this clade should be used as genetic resources for banana breeding. Our results are largely in agreement with that suggestion.
Relationships within sect. Callimusa
Cheesman (Reference Cheesman1947) suggested that Australimusa and Callimusa were distinct sections because of significant differences in their seeds. However, we found that these two groups were genetically indistinguishable. The basal position of M. beccarii could indicate that it is an independent branch because of unique chromosome numbers (X= 9). RFLP analysis of chloroplast DNA showed that M. beccarii was more similar to the M. acuminata complex (Gawel and Jarret, Reference Gawel and Jarret1991). Based on AFLP data, Wong et al. (Reference Wong, Argent, Kiew, Set and Gan2002) found that M. beccarii has a close relationship with Australimusa species. Our results showed that although M. beccarii formed an independent branch, it is closely related to Callimusa species.
The relatively close genetic relationship between M. textilis and M. balbisiana is supported by previous analysis of Chloroplast Deoxyribonucleic Acid (cpDNA) (Gawel and Jarret, Reference Gawel and Jarret1991). Our result showed that the natural hybrid of M. balbisiana× M. textilis clustered with M. schizocarpa; however, M. textilis is closely related to other Callimusa species, Musa jackeyi and Musa lolodensis.
A close relationship was also observed between Musa coccinea and Musa peekelii subsp. angustigemma (similarity coefficient = 0.94).
The grouping of M. paracoccinea with sect. Musa was unexpected and contrary to previous morphological data (Liu et al., Reference Liu, Li and Li2002) and molecular data (Liu et al., Reference Liu, Kress and Li2010), which showed that M. paracoccinea was closely related to M. coccinea. However, we found the closest relationship between M. paracoccinea and M. basjoo (similarity coefficient = 0.87), whereas the similarity between M. paracoccinea and M. coccinea was only 0.74.
With a few exceptions, our results were in general agreement with previously published data. Some species (e.g. M. paracoccinea) were classified differently than expected, suggesting the need for further investigation. The numbers of Callimusa species were also limited in our study, and some recently described species were absent from this analysis. Thus, further study of additional species and morphological characters will be undertaken.
Relationships among Musa acuminata accessions
The most prominent species of Musa are M. acuminata and M. balbisiana, which are the wild progenitors donating the A and B genomes, respectively, to banana cultivars (Perrier et al., Reference Perrier, Langhe, Donohue, Lentfer, Vrydaghs, Bakry, Carreel, Hippolyte, Horry, Jenny, Lebot, Risterucci, Tomekpe, Doutrelepont, Ball, Manwaring, Maret and Denham2011). M. acuminata includes abundant genetic diversity. Totally, ten subspecies and varieties have been reported based on morphological characters: M. acuminata subsp. banksii, errans, malaccensis, zebrina, truncata, halabanensis, siamea, microcarpa, burmannica, burmannicoides and var. chinensis (Feng et al., Reference Feng, Chen and Deng2009). Our results grouped M. acuminata subsp. into the following six clusters: (1) burmannica–burmannicoides–siamea; (2) malaccensis–microcarpa–truncata; (3) zebrina; (4) banksii; (5) var. chinensis and (6) ‘Marges Elargies.’ Our SSR results were close to those of Perrier et al. (Reference Perrier, Langhe, Donohue, Lentfer, Vrydaghs, Bakry, Carreel, Hippolyte, Horry, Jenny, Lebot, Risterucci, Tomekpe, Doutrelepont, Ball, Manwaring, Maret and Denham2011) who reported the geographical distribution of subspecies of M. acuminata: (1) banksii: New Guinea; (2) malaccensis: Malayan Peninsula; (3) burmannica–burmannicoides–siamea complex: South China, Thailand, Myanmar, Bangladesh and northeastern India with sporadic populations southward to Sri Lanka (complex is genetically closer to malaccensis); (4) zebrina/zebrina–microcarpa complex: Java/Sabah; (5) truncata: Malaysian Peninsula; (6) errans: Philippines.
Our SSR results showed that M. acuminata subsp. malaccensis, subsp. microcarpa and subsp. truncata clustered together but were well separated from subsp. truncata. This is largely in agreement with Sequence Related Amplified Polymorphism (SRAP) and AFLP analysis by Muhammad et al. (Reference Muhammad, Andrew, Renata, Rodomiro and Rosa2011). By the geographical distribution, subsp. malaccensis and subsp. truncata overlap and are close to subsp. microcarpa (Perrier et al., Reference Perrier, Langhe, Donohue, Lentfer, Vrydaghs, Bakry, Carreel, Hippolyte, Horry, Jenny, Lebot, Risterucci, Tomekpe, Doutrelepont, Ball, Manwaring, Maret and Denham2011).
M. acuminata var. chinensis, a new variety of M. acuminata, was identified by Häkkinen and is distributed only within China (Häkkinen and Hong, Reference Häkkinen and Hong2007). The vernacular name of M. acuminata var. chinensis is ‘Xiao Guo Ye Jiao’ (small-fruited wild banana); it showed a distant genetic relationship to other reported M. acuminata subsp. complex.
Our SSR data also showed that M. acuminata subsp. 522 was genetically distant from other M. acuminata subsp. The local name of M. acuminata subsp. 522 is ‘Marges Elargies’; this subspecies originated from India and was donated to the Bioversity International ITC in 1988 by France. There was no further information about this subspecies in the Musa Germplasm Information System database. In future, we will resample M. acuminata subsp. 522 from Bioversity International ITC and will grow it out to identify its morphological characteristics and determine its classification.
These findings suggest that it is important to collect and protect M. acuminata var. chinensis and M. acuminata subsp. 522.
Because of the maternal inheritance of the chloroplast genome and the paternal inheritance of the mitochondrial genome in bananas (Lebot, Reference Lebot1999), further in-depth research on the chloroplast and mitochondrial DNA of Musa species is necessary to provide useful information for improving cultivated bananas.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S1479262115000222
Acknowledgements
This work was financially supported by the Protection Project of Tropical Crop Germplasm Resources of China (14RZZY-46), the National Natural Science Foundation of China (grant no. 31440075) and the National Natural Science Foundation of Hainan (grant no. 314083).
The authors declare no conflicts of interest.





