Introduction
Melon (Cucumis melo L.) is an economically important horticultural crop of the Cucurbitaceae family. It is diploid (n = 12) with genome size of 450 Mb (Arumuganathan and Earle, Reference Arumuganathan and Earle1991) and with wide diversity in fruit characteristics, plant type, floral structure and sex expression (Robinson and Decker-Walters, Reference Robinson and Decker-Walters1997).
Fossilized melon seeds from excavations near the ancient Greek colony Chersonese, located at the outskirts of the Sevastopol, Crimean peninsula of Ukraine (founded approximately 2500 years ago on the shore of the Black Sea), were dated back to the 2nd century BC (Pangalo, Reference Pangalo1958). Melons were known to be cultivated in the territory of Ukraine and Russia for about 15 centuries, and the genealogy records of these melon types have been traced to eastern and central Asian lineages (Pyzhenkov and Malinina, Reference Pyzhenkov and Malinina1994). Environmental differences across a wide spread agroecological zone diverged this group from the other melon morphotypes grown in western and southern Europe (Pyzhenkov and Malinina, Reference Pyzhenkov and Malinina1994; Urina et al., Reference Urina, Pivovarov and Balashova1998). Melon production is predominantly spread throughout the large river basins: Don, Volga, and Dnepr and the coast of Azov and Black seas. The diverse melon germplasm that exists in Ukraine and the neighbouring countries Russia, Uzbekistan, Tajikistan and Kazakhstan evolved as one of the secondary centres of melon diversity (Fig. 1; Pangalo, Reference Pangalo1958; Robinson and Decker-Walters, Reference Robinson and Decker-Walters1997). Currently, this particular market group of melons occupies about 50,000 ha throughout eastern Europe.

Fig. 1 Distribution of secondary centre of melon diversity across the agroecological zones of Ukraine and neighbouring countries (a colour version of this figure can be found at journals.cambridge.org/pgr).
A number of studies have used molecular markers to examine genetic diversity of melons from the USA, western Europe, Africa, India and Japan (Silberstein et al., Reference Silberstein, Kovalski, Ruguo, Anagnostou, Jahn and Perl-Treves1999; Stepansky et al., Reference Stepansky, Kovalski and Perl-Treves1999; Oliver et al., Reference Oliver, Garcia-Mas, Morales, Dolcet-Sanjuan, Vicente, Gómez, Leeuwen, Monfort, Puigdomenech, Arús, Katzir and Paris2000; Mliki et al., Reference Mliki, Staub, Zhangyong and Ghorbel2001; Decker-Walters et al., Reference Decker-Walters, Chung, Staub, Quemada and López- Sesé2002; López-Sesé et al., Reference López-Sesé, Staub, Katzir and Gómez-Guillamón2002, Reference López-Sesé, Staub and Gómez-Guillamón2003; Monforte et al., Reference Monforte, Garcia-Mas and Arús2003; Ritschel et al., Reference Ritschel, De Lima Lins, Tristan, Cortopassi Buso, Buso and Ferrira2004; Staub et al., Reference Staub, Danin-Poleg, Fazio, Horejsi, Reis and Katzir2000, Reference Staub, López-Sesé and Fanourakis2004; Nakata et al., Reference Nakata, Staub, López-Sesé and Katzir2005; Szabó et al., Reference Szabó, Gyulai, Humphreys, Horváth, Bittsánszky, Lágler and Heszky2005; Yashiro et al., Reference Yashiro, Iwata, Akashi, Tomita, Kuzuya, Tsumura and Kato2005; Dhillon et al., Reference Dhillon, Ranjana, Singh, Eduardo, Monforte, Pitrat, Dhillon and Singh2007; Sensoy et al., Reference Sensoy, Büyükalaca and Abak2007; Tanaka et al., Reference Tanaka, Nishitani, Akashi, Sakata, Nishida, Yoshino and Kato2007). On the other hand, there is little information about melon germplasm from east Europe, particularly from Ukraine.
The eastern European melon varieties were classified under the convar Europeus, which is also known as adana (Pangalo, Reference Pangalo1958; Pyzhenkov and Malinina, Reference Pyzhenkov and Malinina1994). The convar Europeus, is further divided into three different morphotypes: var. europeus – Skorospelka (early melon); var. aestivalis – Letniai (summer melon); and var. hiemalis – Zimniai (winter melon). Our objective of this study was to estimate genetic diversity in the Ukrainian melon collection using the polymorphisms generated by the amplified fragment length polymorphisms (AFLP) and simple sequence repeats (SSRs) and to further determine whether this molecular diversity corroborates the existing classification.
Materials and methods
The source of seed includes various accessions collected from the melon breeding program at Dnepropetrovsk State Agrarian University (DSAU) and from the Institute of Vegetable and Melon Crops at Ukrainian Academy of Agrarian Science. The list of pertinent accessions is presented in Table 1.
Table 1 List of the Ukrainian melon collections

a According to Pyzhenkov and Malinina (Reference Pyzhenkov and Malinina1994).
DNA was extracted from frozen leaf tissues using the method described in the DNeasy plant mini kit (Qiagen, Hilden, Germany). AFLP analysis was carried out using the protocols and kits developed by LI-COR Biosciences, Lincoln, NE, USA (www.licor.com). The EcoRI and MseI enzyme-digested products were ligated to respective restriction-site-specific adapters and diluted 10-fold. Diluted adapter ligated templates were preamplified using adapter-specific primers with overhangs of A and C for EcoRI and MseI, respectively. Preamplified products were further diluted 20-fold and subjected to selective amplification using IR-700 or IR-800 labelled EcoRI-AXX primers and unlabelled MseI-CXX primers using standard touchdown polymerase chain reaction (PCR) conditions (Vos et al., Reference Vos, Hogers, Bleeker, Reijans, Van de Lee, Hornes, Fritjers, Pot, Peleman, Kuiper and Zabeau1995). Amplified products were denatured and resolved on a LICOR-4500 genotyper. New SSRs were developed using the enrichment procedure developed by Connell et al. (Reference Connell, Pammi, Iqbal, Huizinga and Reddy1998). Genomic DNA was digested with a set of restriction enzymes. Purified digested DNA was ligated to AP-11 and AP-12 adapters and hybridized with biotin labelled oligos, which contain the repeat motifs. DNA fragments that are hybridized with the repeat oligos were separated using streptavidin beads. These repeat-motif-enriched fragments were separated from the beads in an alkaline buffer for purification using a QIAGEN PCR purification kit. These enriched fragments were cloned (TOPO cloning kit; Invitrogen, Carlsbad, CA, USA) and 96 randomly picked clones were sequenced. The sequences with repeat motifs were identified and used for designing SSR primer pairs. PCR conditions for SSRs were used as per Reddy et al. (Reference Reddy, Pepper, Abdurakhmonov, Saha, Jenkins, Brooks and BoelkYEl-Zik2001) and gel electrophoresis was carried out using SFR high-resolution agarose (www.amresco-inc.com).
Minor AFLP polymorphisms that were not uniformly amplified, such as being faint or not distinct in some genotypes, were eliminated from the analysis. Similarly, the stutter and background bands were not considered, while scoring SSR markers. The presence or absence of each fragment was scored as a binary unit character (1 = present and 0 = absent). Genetic similarities based on Jaccard's coefficients (Jaccard, Reference Jaccard1908) were calculated using the SIMQUAL program of the Numerical Taxonomy Multivariate Analysis System (NTSYS-pc) Version 2.0 software package (Rohlf, Reference Rohlf1998). The resulting genetic similarity indices were used to generate a tree using the neighbour joining method (Saitou and Nei, Reference Saitou and Nei1987). The robustness of the clustering was verified by bootstrapping (Felsenstein, Reference Felsenstein1985) using PAUP*4.0. Principal component analysis (PCA) based on the genetic similarity matrices were performed using DCENTER and EIGEN algorithms of the NTSYS-pc software package.
Results
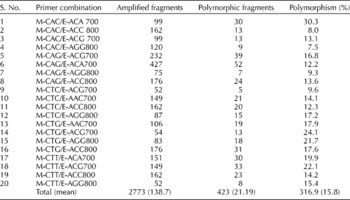
Twenty AFLP primer combinations collectively amplified 2773 bands, out of which 423 bands were polymorphic among the Ukrainian melon collections. The polymorphism level of AFLPs among the collections in the current study was 15.8%. On average, 138 bands were amplified per primer combination. The range of polymorphisms for various primer combinations was from 7.5 to 30.3%. Polymorphic bands ranged from 5 to 52 for various primer combinations. Information pertaining to primer-pairwise amplification pattern and the respective number of polymorphic bands is presented in Table 2.
Table 2 Total number of amplified and polymorphic fragments generated using 20 AFLP primer combinations
We made an SSR-enriched library and sequenced 96 clones and have obtained 40 SSR containing sequences. When we amplified, 12 of these were found to be polymorphic among the Ukrainian collections. Twelve polymorphic SSRs amplified a total of 42 alleles (Table 3). A diversity analysis using these 42 alleles produced a dendogram that did not resolve varietal relationships (tree not shown). Therefore, these 42 alleles were added to 423 AFLPs and a combined diversity analysis was carried out (Fig. 2).
Table 3 Details of microsatellites generated and used to amplify melon collections in the current study


Fig. 2 Phenogram obtained with the joint analysis of amplified fragment length polymorphisms and simple sequence repeats. The numbers adjacent to some nodes indicate bootstrap confidence values (1000 bootstrap replicates).
Genetic diversity among the Ukrainian cultivars ranged from 0 to 38%. Similarity indices within the groups of europeus, aestivalis and hiemalis were 88, 74 and 85%, respectively. The phylogenetic tree contained three distinct clusters (Fig. 2). We estimated the bootstrap values (BV), and the clusters that were supported with 50 or above BV were considered as well supported. The first two clusters were on the top of the phenogram contained the aestivalis and europeus groups, respectively, and these clusters were supported by BV of 74 and 65, respectively, indicate a robust clustering pattern. A basal mixed group mainly consisted of the hiemalis along with the other exotic collections and was not supported by BV. The aestivalis cluster further resolved into six sub-clusters. Some of these sub-clusters were highly supported with BVs ranging from 51 to 98. A sub-cluster containing of Desertnaia 5, Lipneva, Samarskaia, Zlata Kubanka 93 and Pepsha were aestivalis types with furrowed fruit types. This was supported with the highest BV of 98. Two other sub-clusters were supported with BVs of 81 and 60 and contained aestivalis types with netted fruits. The fourth sub-cluster of aestivalis with the varieties of half-sibs Lada and Tavrichanka was supported with the BV of 52. A fifth sub-cluster of aestivalis varieties were L22/1–25, Musa, G-14 and Bronzovka, which have high sucrose content. The sixth sub-cluster of aestivalis types L20/1 and Bereginia was rooted from the bottom.
The second major clade containing europeus types resolved into sub-clusters based mainly on their maturity or fruit type. For example, the heirlooms KRL, Krinichanka and Rannia 133 were in one cluster and had similar fruit types, which were oval shape, white flesh colour, yellow exocarp and netted exocarp surface. Second sub-cluster had many early maturing landraces of Titovka along with Ingulka, a core collection of the early type from the melon breeding station Khersone, Ukraine.
Third major clade of hiemalis along with some exotic collections resolved into two sub-clusters of several diverse melon types (Blue sweet from Taiwan, Kynpou from Japan and Koy Bash from Uzbekistan) and hiemalis (Dianna, Promitei, Dneprianka 163). Morphological marker accessions for Bush type KZhT and virescent marker ZhZl were also clustered with the mixed group of melons. The collection Ineia, which is a typical aestivalis group, clustered with the mixed group of melons as an exception from the other aestivalis collections. The collections N38 and Gruntovaia gribovskaia are mixed-melon types, which appear to have undergone extensive introgression with the exotic melon types.
To corroborate the results of diversity analysis, we also carried out a PCA using the first three eigen vectors that cumulatively absorbed 66.04% of the variation (vector I = 32.15, vector II = 21.63 and vector III = 12.39). The extent of the cumulative variation absorbed by the first three vectors indicates robustness of the analysis. A three-dimensional graph of PCA was made using these three vectors (Fig. 3). Interestingly, this analysis also resolved three groups with the boundaries defining the classical morphotypes with few exceptions and more or less in agreement with the neighbor joining (NJ) analysis. A group of aestivalis types were clustered with an exception of Rannia and KRL, which are of typical europeus types. Another exception being two of the aestivalis types Kubanka 93 and Zlata, which were clustered with the mixed-melon group at the bottom of the PCA graph. All the europeus types were grouped together on top of the PCA graph. A cluster of mixed melons were at the bottom of the groups as topologies resolved in the NJ tree.

Fig. 3 Principal component analysis depicting relationships of the Ukrainian melon collections.
Discussion
Our study identified a robust set of AFLP polymorphisms that defined morphotype boundaries within the Ukrainian collections and was in agreement with previous studies. Therefore, the AFLPs are very informative in melon genome analysis. In melons, Garcia-Mas et al. (Reference Garcia-Mas, Oliver, Gómez-Paniagua and De Vicente2000), after using random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP) and AFLPs, indicated that the AFLP markers are highly polymorphic and more informative than the other marker systems. Périn et al. (Reference Périn, Hagen, De Conto, Katzir, Danin-Poleg, Portnoy, Baudracco-Arnas, Chadoeuf, Dogimont and Pitrat2002) used 346 AFLPs to generate a genetic map using melon RIL populations, indicating that the AFLPs are definitely useful for understanding cultivar relationships and mapping endeavours in melons.
The SSRs reported in the present study should prove to be very useful for the Ukrainian melon breeding program. They are simple to use in laboratories, which are equipped with horizontal gel electrophoresis but where no high-throughput facilities available. Several melon geneticists have used SSRs for understanding genetic diversity, species relationships, synteny with the other cucurbit species, quantitative trait loci (QTL) identification and genetic mapping (Staub et al., Reference Staub, Danin-Poleg, Fazio, Horejsi, Reis and Katzir2000; Danin-Poleg et al., Reference Danin-Poleg, Reis, Tzuri and Katzir2001; Decker-Walters et al., Reference Decker-Walters, Chung, Staub, Quemada and López- Sesé2002; López-Sesé et al., Reference López-Sesé, Staub, Katzir and Gómez-Guillamón2002; Monforte et al., Reference Monforte, Garcia-Mas and Arús2003; Garcia-Mas et al., Reference Garcia-Mas, Monforte and Arús2004; Ritschel et al., Reference Ritschel, De Lima Lins, Tristan, Cortopassi Buso, Buso and Ferrira2004; Gonzalo et al., Reference Gonzalo, Oliver, Garcia-Mas, Monfort, Dolcet-Sanjuan, Katzir, Arús and Monforte2005). Three-hundred and twenty-four SSRs are currently available from various studies for public use to date. It is important to develop more SSR markers as these are technically facile and highly informative markers for various genetic studies. The small number of clones (n = 96) sequenced in this study yielded a high percentage of SSRs (n = 40), suggesting that significantly more SSRs might be obtained by sequencing a larger portion of the library.
In the current study, the phenogram resolved aestivalis and europeus groups with the strong support of significant BV. All the collections in the aestivalis group (cluster I) had smooth fruit surfaces except for five varieties (Desetnaia 5, Dachnitza, Lipneva, Pepsha and Samarskaia) which had fruits with a furrowed surface. The europeus group (cluster II) had accessions with predominantly early types. The fruit colour, while still immature varied from the green to dark green for the collections in cluster II. Further, when these fruits attained maturity, the colour changed to yellowish brown in all the accessions. The europeus varieties Ananas, Titovka original, Diana, Samarskaia, Gruntovaia gribovskaia, Koy Bash and Titovka Zaporojie had netted fruit surfaces with white coloured flesh. Ukrainian consumers prefer melons with white coloured flesh (Tomason, Reference Tomason2002). The polymorphisms generated in the current study, which are specific to the grouping of fruit types and days to maturity would be very useful to pursue further genetic studies and marker-assisted selections.
The third major clade of hiemalis, along with some exotic collections resolved into two sub-clusters of several diverse melon types (Blue sweet from Taiwan, Kynpou from Japan and Koy Bash from Uzbaekistan) and hiemalis (Dianna, Promitei and Dneprianka 163). The morphotype hiemalis has non-climacteric fruits and hence long shelf life. Another distinguishing feature among these groups is the differences in the sex expression. The sex expression in europeus and hiemalis groups is andromonoecious or monoecious, whereas in aestivalis, a majority collections are andromonoecious except in two collections. The two collections – L20/1 and L22/1–25 that are exceptions in sex expression from the rest of the aestivalis group are interestingly gynomonoecious (Tomason, Reference Tomason2002).
Our study provides useful information pertaining to morphological and classical morphotype characterizations of melons specific to Ukrainian collections. However, the current analysis would not shed any light on the cultivar relationships in the broader perspective of collections from other regions of the world. It would be very interesting to extend this study along with the existing international reference collections to draw relationships with the other melon morphotypes such as agrestis, flexuosus, conomon, cantalupensis, inodorus, chito, dudaim and momordica (Munger and Robinson, Reference Munger and Robinson1991; Robinson and Decker-Walters, Reference Robinson and Decker-Walters1997, Greuter et al., Reference Greuter, Mcneil, Barrie, Burdet, Demoulin, Filgueiras, Nicolson, Silva, Skog, Trehane, Turland and Hawsworth2000, Staub et al., Reference Staub, Danin-Poleg, Fazio, Horejsi, Reis and Katzir2000, Reference Staub, López-Sesé and Fanourakis2004). The relationships of various Ukrainian collections and knowledge about the extent of genetic diversity among them would aid planning the melon improvement programs in the Ukraine.
Acknowledgements
Authors would like to profusely thank NATO Science Foundation (Expert Visit: Ref. 982309) for funding the visit of Dr Yan Tomason and USDA-CSREES Research Grant (2007-03 466) Agreement Number (2007-38 814-18 472) for the funding support. We acknowledge Drs Mark Chatfield, Robert Harris and Gerald Hankins for their critical comments.








