Introduction
Castor (Ricinus communis L.) is a non-edible and industrially important oilseed crop. It is the only species (monotypic) classified under genus Ricinus belonging to Euphorbiaceae family. It is well adapted to arid and semi-arid regions. East Africa is considered the probable origin of castor based on the prevalence of diversity (Vavilov, Reference Vavilov, Chester and Waltham1951); however, it is widespread in several countries. India, China, Mozambique and Brazil are the major castor growing countries. India leads the castor production with approximately 1.7 million tonnes (FAOSTAT, 2015) and meets more than 80% of the global demand of castor oil. The castor oil and its derivatives are used in manufacturing of various industrial products including paints, lubricants, cosmetics, nylon, pharmaceuticals, plastics and textiles (Ogunniyi, Reference Ogunniyi2006; Mutlu and Meier, Reference Mutlu and Meier2010).
Wilt disease caused by Fusarium oxysporum f. sp. ricini (Nanda and Prasad, Reference Nanda and Prasad1974) is the most important disease of castor in India (Desai et al., Reference Desai, Dange, Patel and Patel2003). The pathogen is predominantly a soil borne fungus; however, seed borne nature has also been reported (Naik, Reference Naik1994). The disease occurs across seasons and causes yield losses up to 77% depending on the stage at which the plants wilt (Pushpawathi et al., Reference Pushpawathi, Sarwar, Raoof and Babu1998). Cultural and chemical control of wilt disease in castor has been ineffective due to vascular spread of the disease and soil borne nature of the pathogen (Dange et al., Reference Dange, Desai and Patel2006).
Use of resistant cultivars is the most effective and simplest way for the management of wilt disease problem in castor production. Excellent sources of resistance to wilt have been reported in castor germplasm (Raoof and Rao, Reference Raoof and Rao1996; Anjani et al., Reference Anjani, Raoof and Desai2014). Knowledge on the inheritance pattern of the wilt resistance in these germplasm sources would help to identify diverse genes and to pyramid them in the background of improved parental lines, which would eventually lead to development of cultivars with durable resistance to wilt in castor.
Classical genetic studies hitherto conducted on wilt resistance in castor have indicated the involvement of recessive genes (Lavanya et al., Reference Lavanya, Raoof and Prasad2011), dominant genes (Singh et al., Reference Singh, Chaudhuri, Mandal and Chaudhuri2011), duplicate genes (Anjani and Raoof, Reference Anjani and Raoof2014), complimentary genes (Gourishankar et al., Reference Gourishankar, Rao and Reddy2010) and polygenes (Patel and Pathak, Reference Patel and Pathak2011). These observations suggest the possibility of Mendelian and/or quantitative genetic basis of wilt resistance in castor. Till date, none of those genes has been defined at genetic or molecular level. Nevertheless, breeding for wilt resistance has been successful and a number of cultivars have been released for cultivation. But those cultivars lack information on genes they carry. Breakdown of wilt resistance in hybrid GCH4 and variety DCS9 has been observed (Anjani et al., Reference Anjani, Raoof, Reddy and Rao2004; Lavanya et al., Reference Lavanya, Raoof and Prasad2011), which underscores the need for a systematic study on genetics of resistance to wilt in castor in order to identify diverse genes, establish allelic relationships and deploy them carefully. In view of this, the present study was carried out to study the inheritance of wilt resistance in a set of resistant castor inbred lines, which have the potential to contribute to breeding programmes.
Materials and methods
Plant material
Choice of parents for crossing work
A set of 12 castor inbred lines were selected for the study, which included four resistant lines namely 48–1, CI-1, AP42 and AP48 and eight susceptible lines namely JI35, JC12, AP3, AP52, AP72, AP130, AP134 and AP306. The pedigree and origin of these lines are given in Supplementary Table S1. The selected lines showed consistent reaction to three isolates of Fusarium oxysporum f. sp. ricini in field and glass house-based screenings (Shaw et al., Reference Shaw, Shaik, Mir, Prasad, Prasad and Senthilvel2016).
Development of segregating populations
A total of nine crosses were made using four resistant and eight susceptible lines as parents. The details of the crosses are as follows: JI35 × 48–1, 48–1 × JI35, JC12 × 48–1, AP72 × CI-1, AP3 × CI-1, AP130 × CI-1, AP306 × CI-1, AP52 × AP48 and AP134 × AP42. Further, two backcross populations (BC1) of the cross JI35 × 48–1 were derived by crossing the F 1 plant with both the susceptible (JI35) and resistant (48–1) parents, which were designated as [JI35 × (JI35 × 48–1)] and [48–1 × (JI35 × 48–1)].
Crosses were made in field during winter season of 2014–2015 at IIOR. While crossing, enough precaution was taken to prevent contamination by natural outcrossing. The inflorescence on the selected female parents were emasculated before anthesis and covered with butter paper bag. Pollination was carried out by hand-transferring the pollen collected from the covered inflorescence of selected male parent to the stigmatic surface of the female parent. The F 1 plants were raised in the field and the hybridity was confirmed by observing the morphological traits. The F 1s of all the crosses except 48–1 × JI35 were selfed by covering the inflorescence before opening of flowers. The F 2 seeds were harvested from the selected single F 1 plant. Overall, eight F 2 populations and two BC1F1 populations were available for wilt screening purpose.
Screening of segregating populations for resistance to wilt
Fungal isolation and inoculum preparation
The initial inoculum of Fusarium oxysporum f. sp. ricini was prepared by isolating the pathogen from the infected root of susceptible castor genotype, JI35 (grown at research farm of IIOR, Hyderabad) and by culturing it on potato dextrose agar (PDA) medium. The fungal culture was purified by single spore isolation technique and maintained in paraffin oil at −20°C as described by Nakasone et al. (Reference Nakasone, Peterson, Foster, Mueller, Bills and Foster2004). Mass multiplication of the pathogen was carried out on sorghum (Sorghum bicolor) grains as substrate. Semi-cooked sorghum grains (100 g in 250 ml of conical flask) were sterilized at 15 psi for 20 min at 121°C. The flasks were inoculated by actively growing 7-d old fungal culture (grown on PDA) and incubated at 25 ± 2°C in an incubator for 15 d. The flasks were hand shaken daily to ensure complete fungal colonization of the sorghum grains. The 15 d old fungal culture on sorghum substrate was used to inoculate the pots.
Sick pot screening design
A high throughput sick pot screening method (Shaw et al., Reference Shaw, Shaik, Mir, Prasad, Prasad and Senthilvel2016) was used for screening the parents, F 1s, F 2 and BC1 F 1 plants of the crosses for reaction to Fusarium oxysporum f. sp. ricini. The potting mixture was prepared by mixing red soil, black soil and farm yard manure in the proportion of 5:3:1 and autoclaved. Approximately 4 kg of potting mixture was filled in plastic pots (30 × 15 × 13 cm3). Then, 12 g of inoculum (mass multiplied on sorghum) was thoroughly mixed with the sterile soil at the rate of 3 g of inoculum per kg of sterile soil under aseptic conditions. Each inoculated pot was watered and kept for incubation for 24 h before sowing.
For the screening of F 1s, six seeds each of resistant parent, susceptible parent and the F 1 of the corresponding crosses were sown in separate rows in a single pot. Three seeds of JI35 (susceptible check) were sown in each pot on both the sides of the test rows. The set up was repeated three times. For the screening of F 2 populations, 15 seeds were sown in each pot in three rows (five seeds/row). Three seeds of respective parents of the cross were sown on both sides of the F 2 rows. Number of seeds per F 2 population ranged from 97 to 146. Similar sowing arrangement was used for BC1 F 1 seeds as well. The sick pots were kept in rain-out shelter at ambient temperature (28 ± 2°C) and watered regularly.
Disease scoring
The seeds germinated after about 12 d and the seedlings were observed regularly for wilt symptoms. Scoring of plant's reaction to wilt was done based on ‘days to wilt’ criterion as proposed by Shaw et al. (Reference Shaw, Shaik, Mir, Prasad, Prasad and Senthilvel2016). As per this scoring system, the level of resistance of test plants was categorized into 1–4 scale based on days to wilting of plant from the date of sowing. The scale 1 (susceptible) was assigned to plants that wilted within 30 d after sowing, 2 (moderate) for plants that wilted between 31 and 50 d after sowing, 3 (resistant) for plants that wilted between 51 and 65 d after sowing and 4 (highly resistant) for plants that survived beyond 65 d after sowing without any disease symptoms.
Data analysis
Chi-square test was used to check goodness of fit of the wilt resistance scores of F 2 and BC1 F 1 populations to various classical Mendelian ratios. The Chi-square value was calculated as per the standard formula χ2 = ∑ (O–E)2/E, where O = observed frequency of resistant/susceptible plants and E = expected frequency of resistant/susceptible plants. The deviation between observed and expected ratios was considered non-significant, if the calculated χ2 value was lesser than the tabular value at P = 0.05 for (n−1) degrees of freedom, where ‘n’ is the total number of phenotypic categories/classes.
Results
Reaction of parents and F1s to Fusarium infection
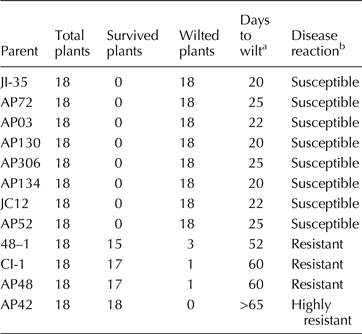
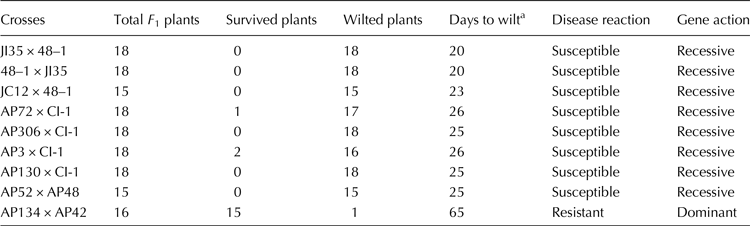
The reaction of parents and F 1s to Fusarium infection is presented in Tables 1 and 2, respectively. The parents JI35, JC12, AP3, AP52, AP72 API30, API34 and AP306 were susceptible (wilted between 20 and 25 d); 48–1, CI-1 and AP48 were resistant (wilted between 52 and 65 d) and AP42 was highly resistant (did not wilt beyond 65 d) as per the scoring system of Shaw et al. (Reference Shaw, Shaik, Mir, Prasad, Prasad and Senthilvel2016). The F 1s of the crosses involving 48–1, CI-1 and AP48 showed susceptibility (wilted between 20 and 26 d) indicating that the nature of resistance in these sources could be recessive. The reaction of resistant parent, 48–1, susceptible parent, JI35 and their F 1 (susceptible) is shown in Fig. 1. The F 1 of AP134 × AP42 cross was resistant (Fig. 2) indicating the resistance to wilt in AP42 could be dominant.

Fig. 1. Susceptible reaction of F 1s of the cross: JI35 × 48–1 indicating recessive nature of resistance to wilt in 48–1.

Fig. 2. Resistant reaction of F 1s of the cross: AP134 × AP42 indicating dominant nature of resistance to wilt in AP42.
Table 1. Reaction of parental lines to Fusarium infection in sick pot screening
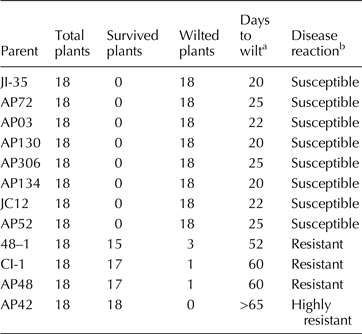
a Days after sowing in which 80% of plants wilted.
b Scoring system: susceptible (<30 d to wilt), resistant (51–65 d to wilt) and highly resistant (did not wilt beyond 65 d) as per Shaw et al. (Reference Shaw, Shaik, Mir, Prasad, Prasad and Senthilvel2016).
Table 2. Reaction of F 1s to Fusarium infection in sick pot screening
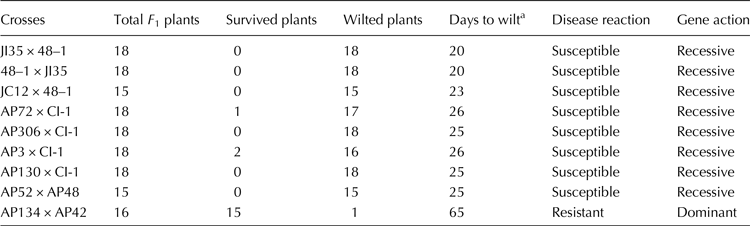
a Days after sowing in which 80% of plants wilted.
Reaction of F2 and BC1F1 populations to Fusarium infection
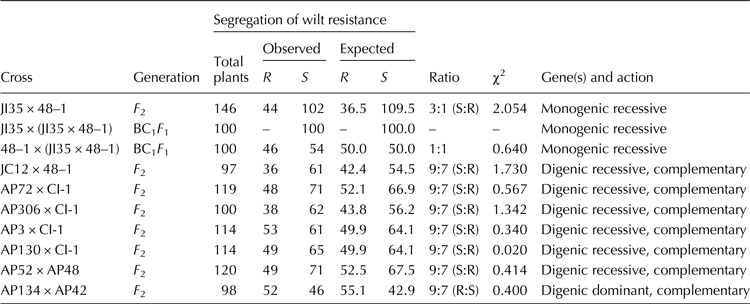
The segregation pattern of F 2 population for wilt resistance in all the eight crosses is given in Table 3. The F 2 plants of the crosses namely JI35 × 48–1 segregated in the ratio of 3 (Susceptible):1 (Resistant) showing the role of single recessive gene. As expected, all plants of BC1 F 1 population: JI35 × (JI35 × 48–1) showed susceptibility and the BC1 F 1 plants of the cross: 48–1 × (JI35 × 48–1) segregated in 1:1 ratio. The F 2 plants of the crosses namely JC12 × 48–1, AP52 × AP48, AP72 × CI-1, AP3 × CI-1, AP130 × CI-1, AP306 × CI-1, segregated in the ratio of 9 (Susceptible):7 (Resistant) and in the cross AP134 × AP42, the F 2 plants segregated in the ratio of 9 (Resistant):7 (Susceptible) showing complementary gene interaction. Overall, the inheritance results showed that resistance to wilt is conferred by a single locus (only in case of JI35 × 48–1 cross) or mostly two loci with complementary interaction. The calculated χ2 values for all the crosses were lesser than the table value (3.84, P = 0.05 at 1 df) indicating that the deviation between the observed ratios and the expected Mendelian ratios was not significant.
Table 3. Chi-square test for Mendelian segregation of wilt resistance in segregating populations
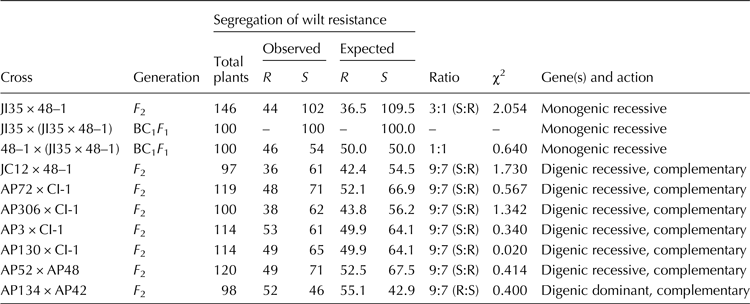
R, resistant; S, susceptible.
χ2 (table) for 1 df = 3.841 at P = 0.05.
Prediction of probable allelic combinations of parental genotypes for wilt resistance
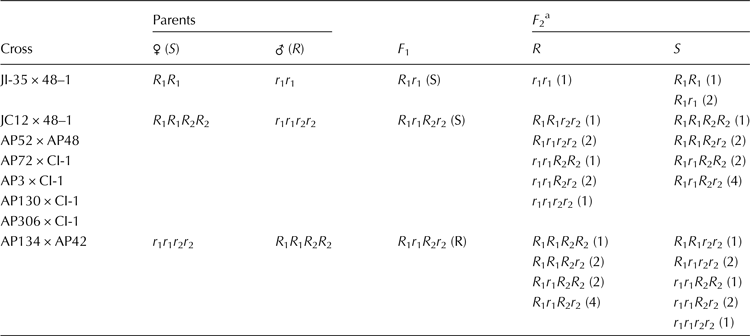
Based on the inheritance of resistance in F 2 progenies, probable genotypes of parents as well as the F 2 progenies at wilt resistance loci were predicted and presented in Table 4. The susceptible and resistant parents of the crosses, JI35 × 48–1, which showed monogenic inheritance with the ratio of 3 (Susceptible):1 (Resistant) were assigned with allelic combination of R 1 R 1 (JI35) and r 1 r 1 (48–1). The susceptible and resistant parents of crosses, JC12 × 48–1, AP52 × AP48, AP3 × CI-1, AP72 × CI-1, AP130 × CI-1 and AP306 × CI-1, which showed recessive and complementary gene interaction with the ratio of 9 (Susceptible):7 (Resistant) were assigned with allelic combination of R 1 R 1 R 2 R 2 (JC12, AP52, AP3, AP72, AP130 and AP306) and r 1 r 1 r 2 r 2 (48–1, AP48 and CI-1), respectively. The susceptible and resistant parents of the cross AP134 × AP42, which showed dominant and complementary interaction with the ratio of 9 (Resistant):7 (Susceptible) were assigned with allelic combinations of r 1 r 1 r 2 r 2 (AP134) and R 1 R 1 R 2 R 2 (AP42), respectively.
Table 4. Probable allelic combinations of parents, F 1 and F 2 genotypes of castor crosses studied for resistance to wilt
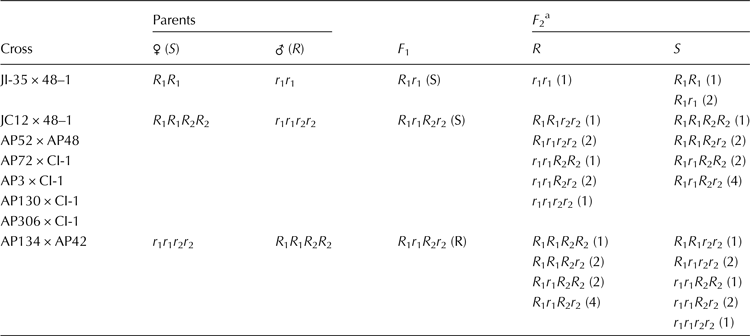
S, susceptible; R, resistant.
a Values in parenthesis indicate frequency of genotypes having the particular allelic combination.
Discussion
Genetic resources with resistance to wilt disease are critical in castor breeding for development of improved cultivars with higher productivity. In this study, a set of four castor inbred lines (48–1, CI-1, AP42 and AP48) has been characterized for Mendelian inheritance of the resistance to wilt through studying F 2 and backcross populations. Among the inbred lines, AP42 showed better resistance than 48–1, CI-1 and AP48 based on ‘days to wilt’ criterion. All the plants of 48–1, CI-1 and AP48 survived more than 50 d after inoculation. They started wilting from 52nd day onwards and completely wilted within 60 d. Only highly resistant genotype AP42 survived beyond 65 d after inoculation. It is presumed that the resistant parents (48–1, AP42 and AP48) are genetically diverse except CI-1, which has been derived from the cross involving 48–1 as parent; therefore, they would perhaps represent diversity of resistance to wilt available in castor germplasm. Genetic characterization studies on castor germplasm accessions with resistance to wilt are limited. Lavanya et al. (Reference Lavanya, Raoof and Prasad2011) studied wilt resistance in 48–1 and RG297. Anjani and Raoof (Reference Anjani and Raoof2014) studied wilt resistance in RG2758, RG2822, RG109 and RG2529. The genotypes, CI-1, AP42 and AP48 are the newly found sources of resistance to wilt in castor, which could be valuable in castor breeding.
Susceptibility of the F 1s of the crosses namely JI35 ×48–1, JC12 × 48–1, AP72 × CI-1, AP3 × CI-1, AP130 × CI-1, AP306 × CI-1 and AP52 × AP48 suggested that resistance to wilt in 48–1, CI-1 and AP48 are recessive in nature. The recessive nature of wilt resistance in castor has been reported by previous researchers as well. Lavanya et al. (Reference Lavanya, Raoof and Prasad2011) reported that resistance to wilt in 48–1 was a recessive trait. The current study further supported the results of Lavanya et al. (Reference Lavanya, Raoof and Prasad2011) on 48–1. Also, susceptibility of the F 1 of the reciprocal cross 48–1 × JI35 suggested that resistance to wilt in 48–1 is governed by nuclear genes and there was no maternal influence. On the other hand, the F 1 of the cross AP134 × AP42 was resistant indicating that the resistance was a dominant trait in AP42. The dominant nature of resistance to wilt in castor has also been reported by several authors (Rao et al., Reference Rao, Raoof and Lavanya2005; Gourishankar et al., Reference Gourishankar, Rao and Reddy2010; Reddy et al., Reference Reddy, Janila, Rao, Ahammed, Reddy, Shankar and Singh2010, Reference Reddy, Sujatha, Reddy and Reddy2011; Singh et al., Reference Singh, Chaudhuri, Mandal and Chaudhuri2011).
The segregation pattern of F 2 populations showed that either one or mostly two loci are involved for resistance to wilt in four resistant genotypes examined in this study. Inheritance of resistance to wilt in the cross, JI35 × 48–1 was found to be monogenic with the ratio of 3 (Susceptible):1 (Resistant), which is an indicative of single recessive gene action. In this case, it is hypothesized that wilt resistance is governed by one locus (designated as R 1). The presence of dominant allele of ‘R 1’ would result in susceptible phenotype. Only those plants that carry recessive allele in homozygous condition (r 1 r 1) would show resistant phenotype and the heterozygous combination (R 1 r 1) would show susceptible phenotype (Table 4). Inheritance results of the backcross populations of the cross JI35 × 48–1 also supported the F 2 results. All the plants of the backcross involving the susceptible F 1 plant and the susceptible parent JI35 (F 1 × JI35) were susceptible, as expected because both the F 1 and JI35 carried the dominant allele of gene ‘R 1’ resulting in all susceptible genotypes. In the other backcross, where the susceptible F 1 plant was crossed with the resistant parent 48–1, half of the population showed resistance reaction and the other half was susceptible. The susceptible F 1 plant carried the dominant allele of the gene ‘R 1’, which is responsible for susceptibility and the resistant parent carried the recessive allele of the ‘r 1’. So out of 100 BC1 F 1 plants screened, 54 plants carrying the dominant allele of gene (R 1) showed susceptible reaction and 46 plants carrying recessive allele of the gene (r 1) showed resistant reaction. Similar pattern of single recessive gene inheritance for Fusarium wilt resistance with 3(Susceptible):1(Resistant) has been reported in pigeon pea (Odeny et al., Reference Odeny, Githiri and Kimani2009; Patil et al., Reference Patil, Singh, Dhar, Chaudhar, Datta, Chaturvedi and Nadarajan2013), banana (Ssali et al., Reference Ssali, Kiggundu, Lorenzen, Karamura, Tushemereirwe and Viljoen2013) and chickpea (Kumar and Haware, Reference Kumar and Haware1982; Sindhu et al., Reference Sindhu, Singh and Slinkard1983; Tullu et al., Reference Tullu, Muehlbauer, Simon, Mayer, Kumar, Kaiser and Kratt1998; Sharma et al., Reference Sharma, Chen and Muehlbauer2005).
The crosses JC12 × 48–1, AP52 × AP48, AP72 × CI-1, AP3 × CI-1, AP130 × CI-1, AP306 × APCI-1 and AP134 ×AP42 revealed digenic inheritance with complementary gene interaction. The CI-1 showed same inheritance pattern [9 (Susceptible):7 (Resistant)] in four crosses involving different susceptible backgrounds suggesting the reliability of results. As the F 1s of the crosses involving 48–1 (JC12 × 48–1), AP48 (AP52 × AP48), CI-1 (AP72 × CI-1, AP3 × CI-1, AP130 × CI-1, AP306 × CI-1) were susceptible, it is presumed that resistance in 48–1, AP48 and CI-1 is governed by two recessive genes. Similar report of wilt resistance governed by two recessive genes involving complementary epistasis has been reported in castor (Sridhar, Reference Sridhar2007) and pigeon pea (Odeny et al., Reference Odeny, Githiri and Kimani2009; Ajay et al., Reference Ajay, Prasad, Gowda, Ganapathy, Gnanesh, Fiyaz, Veerakumari, Babu, Venkatesha and Ramya2013). The complementary gene action happens due to non-allelic gene interaction of two loci (designated as R 1 and R 2). As the F 1 of these crosses were susceptible, the presence of dominant alleles at both the loci in homozygous or heterozygous condition (R 1 R 1 R 2 R 2/R1R1R 2 r 2/R 1 r 1 R 2 R 2/R 1 r 1 R 2 r 2) would lead to susceptible reaction. It is hypothesized that resistance to wilt is expressed when one of the genes or both the genes are in homozygous recessive condition (R 1 R 1 r 2 r 2/r 1 r 1 R 2 R 2/r 1 r 1 r 2 r 2) (Table 4).
In case of the cross AP134 × AP42, segregation pattern with digenic ratio of 9 (Resistant):7 (Susceptible) was observed and the F 1 of this cross was resistant suggesting that two dominant genes (designated as R 1 and R 2) in homozygous or heterozygous condition could be responsible for resistance in AP42. It is hypothesized that resistance to wilt disease is expressed when both the dominant genes are in homozygous (R 1 R 1 R 2 R 2) or heterozygous (R 1 r 1 R 2 r 2, R 1 R 1 R 2 r 2, R 1 r 1 R 2 R 2) conditions (Table 4). Similar reports on the complementary gene action by two dominant genes for wilt resistance have been reported in castor (Rao et al., Reference Rao, Raoof and Lavanya2005; Sridhar, Reference Sridhar2007; Gourishankar et al., Reference Gourishankar, Rao and Reddy2010) and in other crops namely pigeon pea (Kumar et al., Reference Kumar, Varma, Suresh and Sreelakshmi2009; Changaya et al., Reference Changaya, Melis, Derera, Laing and Saka2012; Latha et al., Reference Latha, Kumar and Reddy2012; Singh et al., Reference Singh, Sinha, Rai, Singh, Singh, Kumar and Singh2016) and safflower (Shivani and Varaprasad, Reference Shivani and Varaprasad2016).
In this study, it was interesting to note different modes of inheritance when different susceptible parents were crossed with the same resistant source. Two susceptible parents namely JI35 and JC12 were crossed with the common resistant parent (48–1) in two crosses namely JI35 × 48–1 and JC12 × 48–1. In both the crosses, the F 1s showed susceptible reaction but the F 2 population showed different modes of inheritance namely monogenic [3 (Susceptible):1 (Resistant)] in the cross JI35 × 48–1 and digenic with complementary gene interaction [9 (Susceptible):7 (Resistant)] in the cross JC12 × 48–1. In both the crosses (JI35 × 48–1 and JC12 × 48–1), ‘r 1’ could be the common locus responsible for resistance in 48–1. In case of JC12 × 48–1 cross, it is likely that another locus ‘r 2 ’ (from 48–1) interacts with ‘r 1 ’ in complementary way in the background of susceptible parent, JC12. So, this observation signals the possible role of susceptible parent in determining the mode of inheritance for resistance in the progenies. Similar observation was made in pigeon pea for wilt resistance when the different susceptible parents were crossed with the same resistance source. Changaya et al. (Reference Changaya, Melis, Derera, Laing and Saka2012) crossed two susceptible parents AP 2 and AP 9 with the common resistant parent ICPL 87051 and found F 2 segregation ratio of 3 (Resistant):1 (Susceptible) in the cross AP 2 × ICPL 87051 and 9 (Resistant):7 (Susceptible) in the cross AP 9 × ICPL 87051. Sreelakshmi et al. (Reference Sreelakshmi, Shivani and Kumar2011) crossed the common resistant parent ICPL 87119 with five different susceptible backgrounds (LRG 30, MRG 66, ICPL 85063, LRG 41 and Nallakandi) in pigeon pea and found segregation ratio of 13 (Resistant):3 (Susceptible) in three crosses and 9 (Resistant):7 (Susceptible) in other two crosses. Similar results were reported by Kumar et al. (Reference Kumar, Varma, Suresh and Sreelakshmi2009) and Ajay et al. (Reference Ajay, Prasad, Gowda, Ganapathy, Gnanesh, Fiyaz, Veerakumari, Babu, Venkatesha and Ramya2013).
From the literature, it is noted that inheritance results from several studies on resistance to wilt in castor are inconsistent, which is a concern. For instance, we observed that either a single gene or two genes with complementary mode of inheritance are responsible for resistance to wilt in 48–1. In previous studies, resistance to wilt in 48–1 has been reported to be governed by single dominant gene (Reddy et al., Reference Reddy, Janila, Rao, Ahammed, Reddy, Shankar and Singh2010, Reference Reddy, Sujatha, Reddy and Reddy2011; Singh et al., Reference Singh, Chaudhuri, Mandal and Chaudhuri2011), two dominant complementary genes (Rao et al., Reference Rao, Raoof and Lavanya2005), two recessive genes with complementary epistasis (Sridhar, Reference Sridhar2007) and polygenic (Lavanya et al., Reference Lavanya, Raoof and Prasad2011; Patel and Pathak, Reference Patel and Pathak2011). The inconsistency in the inheritance results across studies could be attributed to some of the factors including heterogeneity of the genetic material, different susceptible genetic backgrounds and differences in the screening method. Castor being an outcrossing species and wind pollinated, it is really a challenge to maintain genetic purity of genotypes. It is also not uncommon that inheritance of a single resistant genotype varies in accordance with susceptible parent, which is used in the crosses (Ajay et al., Reference Ajay, Prasad, Gowda, Ganapathy, Gnanesh, Fiyaz, Veerakumari, Babu, Venkatesha and Ramya2013). The method of screening for resistance to wilt could be the most important factor that could have caused the inconsistency. Previous studies on inheritance of resistance to wilt in castor were based on percentage of wilt incidence in sick plot under field condition (Rao et al., Reference Rao, Raoof and Lavanya2005; Gourishankar et al., Reference Gourishankar, Rao and Reddy2010; Reddy et al., Reference Reddy, Janila, Rao, Ahammed, Reddy, Shankar and Singh2010; Lavanya et al., Reference Lavanya, Raoof and Prasad2011; Anjani and Raoof, Reference Anjani and Raoof2014), which has inherent drawbacks such as escapes from the pathogen infection. Shaw et al. (Reference Shaw, Shaik, Mir, Prasad, Prasad and Senthilvel2016) suggested that the ‘days to wilt in sick pot under artificial condition’ would be more effective for inheritance studies. This method of screening is high throughput, reproducible, causes no injury to the tissues and would eliminate the chances of escapes; hence, the discrepancy in disease scoring due to environmental variability can be minimized to a larger extent. This new scoring system also provides opportunity to record days to death of seedlings progressively; therefore, quantitative levels of resistance across genotypes could easily be resolved. For example, resistance level of CI-1 (60 d to wilt) was substantially higher than its parents 48–1 (52 d to wilt) suggesting the possible role of modifier gene for resistance to wilt in CI-1 in addition to two recessive genes. In this study, it was ensured that parental genotypes were genetically pure to the extent possible through maintenance by selfing. Furthermore, the improved method was used for screening the parents and progenies. Therefore, we believe that the inheritance results obtained in this study would be more dependable.
There is a concern that resistance to wilt found under artificial screening may not be reproducible in the field conditions due to involvement of plant, fungi and nematode complex. It is presumed that resistance to wilt in castor genotypes occurs possibly in two ways: by innate ability or by resistance to nematodes (injury caused by the nematodes present in the soil predisposes castor plants for wilt infection). Study on morphology or structure of the roots would possibly explain if resistance to nematode could have resulted in wilt resistance in the genotypes. But, the role of nematodes in influencing wilt reaction in castor is not yet well established. To date, the information available in the reports is not confirmatory, in general. For instance, it was found that nematode did not affect wilt reaction in castor genotypes 48–1 and VP-1 under artificial inoculation of both nematode and fungi (DOR, 2011). Furthermore, variation in the pathogen isolates could affect the results of field screening. Therefore, more intensive studies on interaction among plant, fungi and nematode are needed for understanding wilt resistance in castor.
Out of eight crosses tested in the present study, resistance to Fusarium wilt was dominant in only one F 1 (AP134 × AP42 cross). To confirm this dominance nature of resistance, AP42 was crossed with two other susceptible background namely PMC13 and AP39 and the F 1s were found to be resistant in sick pot as well as in the field screenings. The dominance nature of resistance in AP42 could be of greater interest to breeding for resistant cultivars. Dominant genes would be more desirable than recessive ones because transfer of dominant genes by backcrossing would be simpler and resistant hybrids can be quickly developed by incorporating them into any one of the parental lines. In contrast, transfer of recessive genes by backcrossing is time consuming as it would require selfing at each generation to find out homozygous resistant progenies. Furthermore, recessive genes are needed to be incorporated in both the parental lines, which is not advantageous in terms of cost and time.
Historically, qualitative and/or quantitative nature of resistance to diseases has been recognized in crop species. The qualitative resistance is governed by one or two genes that follow Mendelian inheritance pattern while the quantitative resistance is governed by polygenes. To date, the literature in castor suggests the predominance of Mendelian resistance against wilt though polygenic control cannot be ruled out (Lavanya et al., Reference Lavanya, Raoof and Prasad2011). Breeding for durable resistance to wilt in castor would require knowledge on pathogen diversity, race specificity and gene diversity in germplasm. Even though variability in Fusarium oxysporum f. sp. ricini isolates has been reported (Prasad et al., Reference Prasad, Sujatha and Raoof2008), race specificity of resistance to wilt is still unknown in castor. Therefore, diversity of resistance genes can only be established by means of either conventional allelism test or by trait mapping using molecular markers. Nevertheless, the sources of resistance to wilt reported in this study are new, which can be exploited in castor breeding. The resistance can be transferred into the backgrounds of parental lines through backcrossing. Though the plant breeders would prefer the dominant genes considering the simplicity of their use, achieving durable resistance would warrant deployment of both dominant and recessive genes judiciously. In future, identification of molecular markers linked to wilt resistance in the genotypes reported in this study would be helpful for pyramiding of genes through marker-assisted backcrossing.
Supplementary Material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262117000120
Acknowledgements
The financial support from Indian Council of Agricultural Research (ICAR), New Delhi is greatly acknowledged. The authors thank Mr Ilesh for his technical support for conducting the glasshouse experiment.