Introduction
The Kohat Plateau (Fig. S1, available online) is part of the southern deformed fold and thrust belt (one of five tectono-stratigraphic landforms) in the Pakistani Himalaya (Shah, Reference Shah2003). Human impacts, including population migrations, urbanization and overexploitation of medicinal plants for commercial purposes, have led to severe fragmentation of populations of medicinal plants, particularly in the Kohat Basin of the plateau (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009). Recent fragmentation of Withania coagulans and Justicia adhatoda populations on the Kohat Plateau caused by human pressure has also occurred (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009, Reference Gilani, Fujii, Kikuchi, Shinwari and Watanabe2011).
The vegetation of the plateau ranges from dry-subtropical to dry-temperate vegetation, depending on the elevation. More than 100 medicinal plant species have been found in the Kohat District (Ilahi, Reference Ilahi2008). These include Rhazya stricta, which forms a community with W. coagulans on the plateau, and is also being exploited for medicinal purposes as well as to provide fuelwood, fodder and other commercial materials sold in local markets as herbal medicines. R. stricta is a perennial dwarf shrub that grows in dry subtropical regions. Its distribution is confined to South Asia and the Middle East, where it is sparsely distributed in its natural habitat (Bataher, Reference Bataher2004; Gilani et al., Reference Gilani, Kikuchi, Shinwari, Khattak and Watanabe2007; Gilani, Shinwari and Watanabe, Reference Gilani, Shinwari and Watanabe2007). As a result, it is a potentially endangered species.
Population fragmentation can lead to genetic divergence of populations that are reproductively isolated from each other as a result of limited gene flow among the populations (Frankham et al., Reference Frankham, Ballou and Briscoe2007). However, genetic divergence is not always easily visible (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009; Winkler et al., Reference Winkler, Koch and Heitz2011), particularly if it has begun recently. These fragmented populations will urgently require conservation measures. Before formulating a conservation strategy for in situ and ex situ conservations, it is important to understand the population structure of the species and information about its habitat requirements. In the present study, we evaluated the population structure of R. stricta populations from the Kohat Plateau, with the goal of identifying populations that should be prioritized for conservation and sustainable commercial utilization. Conservation and harvesting of medicinal plants are two opposite perspectives, but for the benefit of the local people, we feel that it is appropriate to integrate them. We hypothesized that populations of R. stricta may be threatened by habitat fragmentation rather than by genetic divergence.
In the present study, we used cytochrome P450-based analogue (PBA) markers and amplified fragment length polymorphism (AFLP) markers to assess the population structure. Our laboratory developed the PBA markers in previous research (Yamanaka et al., Reference Yamanaka, Suzuki, Tanaka, Takeda, Watanabe and Watanabe2003). PBA markers have shown high genetic diversity in intraspecific and interspecific comparisons (Yamanaka et al., Reference Yamanaka, Suzuki, Tanaka, Takeda, Watanabe and Watanabe2003; Wan et al., Reference Wan, Watanabe, Yi, Htai, Win, Yamanaka, Nakamura and Watanabe2005) and in comparisons among populations (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009).
Materials and methods
Plant collection
For our genetic diversity studies, we collected the youngest fully expanded leaves from 16 individuals per population from six randomly selected populations on the plateau (Table S1, available online). Three populations of R. stricta were collected from the Karak District (Wanki Siraj Khel, Takht Nasrati and Soordag) and three from the Kohat District (Kander, KDA and Sanda Fateh Khan). The leaves were collected separately in paper bags, which were then packed in a zipped plastic bag containing a silica gel desiccant.
Environmental factors
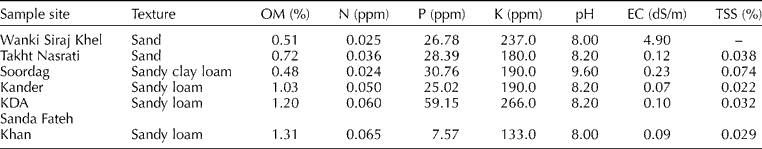
We collected three soil samples per population (at random locations but separated by at least 3 m) at a depth of c. 30 cm using a 5-cm-diamter cylindrical soil sampler from each of the six populations, and bulked the samples for analysis to provide a single composite value for each population. The samples were sent for analysis to the soil testing laboratories of the Agricultural Research Institute, Tarnab, Peshawar, Pakistan, or the Barani Area Research Station, Kohat, Pakistan. Table 1 summarizes the results of the soil analysis.
Table 1 Results of the soil analysis for the six sample sites
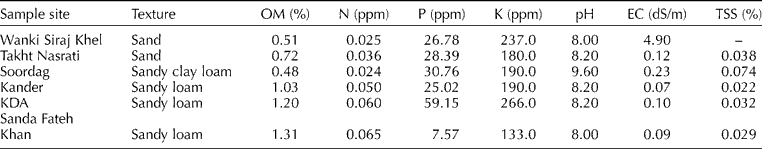
OM, organic matter; EC, electrical conductivity; TSS, total soluble salts.
Altitudes, latitudes and longitudes were determined using a GPSMAP 60CSx GPS receiver (Garmin, Olathe, KS, USA), with a two-dimensional accuracy of three satellites and a three-dimensional accuracy of four satellites. The coordinates of the populations were plotted using DIVA-GIS software, version 5.2.0.2 to plot (Hijmans et al., Reference Hijmans, Guarino, Cruz and Rojas2001). The minimum and maximum annual temperatures in the Kohat and Karak districts of the Kohat Plateau were 7 and 29°C, respectively, with a total annual precipitation of 546 mm and a mean annual relative humidity of 48% (Anonymous, 1999, 2000).
Genetic diversity
For DNA extraction, we used 96 individuals from the six populations, following the method of Doyle and Doyle (Reference Doyle and Doyle1990) with minor modifications (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009). We tested for the presence of 15 PBA markers (Table 2) following the protocol of Yamanaka et al. (Reference Yamanaka, Suzuki, Tanaka, Takeda, Watanabe and Watanabe2003). These markers consisted of three forward and five reverse primers.
Table 2 PBA and AFLP primer sets (F, forward; R, reverse) amplifying polymorphic bands from 96 individuals from the six populations of R. stricta

T m, melting temperature; PB, polymorphic bands; PPB, percentage of polymorphic bands.
a Yamanaka et al. (Reference Yamanaka, Suzuki, Tanaka, Takeda, Watanabe and Watanabe2003); E = EcoRI 5′-GACTGCGTACCAATTC-3′; M = MseI 5′-GATGAGTCCTGAGTAA-3′.
For AFLP analysis, we followed the protocol of Vos et al. (Reference Vos, Hogers, Bleeker, van de Lee, Hornes, Friters, Pot, Paleman, Kuiper and Zabeau1995), with modifications (Hirano et al., Reference Hirano, Kikuchi, Kawase and Watanabe2008, Reference Hirano, Jatoi, Kawase, Kikuchi and Watanabe2009). We added two internal controls for each primer set during the selective amplification stage, one at each end of a 96-well plate. We used different combinations of the EcoRI and MseI primer sets in a 15 μl reaction volume (Table 2). AFLP fragments were detected using a CEQ 8000 genetic analysis system (Beckman Coulter, Brea, CA, USA), following the manufacturer's protocol. Before scoring the peaks using GeneMarker software, version 1.80 (Softgenetics LLC, State College, PA, USA), we identified and manually checked peaks for their reproducibility and sharpness. The peaks were scored as present (1) or absent (0).
Statistical analysis
To calculate the observed number of alleles (n a), the effective number of alleles (n e), Nei's (Reference Nei1973) genetic diversity (H e), Shannon's information index (I) and the percentage of polymorphic bands (PPB), we used GenAlEx statistical software, version 6 (Peakall and Smouse, Reference Peakall and Smouse2006) based on the assumption that the populations were in Hardy–Weinberg equilibrium. To estimate the variations within and among the populations, we used analysis of molecular variance (AMOVA). We estimated the level of gene flow (N m) and the population structure (using the fixation index, F ST) following the methods of Lynch and Milligan (Reference Lynch and Milligan1994). We estimated the gene flow among the populations using the formula N m= ((1/F ST) − F ST)/4 (Frankham et al., Reference Frankham, Ballou and Briscoe2007). We used principal coordinate analysis and Mantel tests to evaluate the correlation between genetic and geographic distances among the populations. We constructed a dendrogram using the unweighted pair group method with arithmetic mean (UPGMA) cluster analysis (Lynch and Milligan, Reference Lynch and Milligan1994) using the algorithm provided by NTSYSpc software, version 2.1 (http://www.exetersoftware.com/cat/ntsyspc/ntsyspc.html) after transforming the data matrix of Nei's genetic distances into the NTSYS format.
For our correlation analysis, we converted the genetic distances and environmental factors into the matrix form. We used GenAlEx to analyse the data matrices by applying Mantel correlation tests (Peakall and Smouse, Reference Peakall and Smouse2006).
Bar plots were calculated using STRUCTURE. The optimal value of K was estimated by calculating the probability Pr(X/K), following the method of Pritchard et al. (Reference Pritchard, Stephens and Donnelly2000) and of Evanno et al. (Reference Evanno, Regnaut and Goudet2005).
Results
All the populations were geographically isolated by at least 20 km and had fewer than 50 individuals per population in the Karak District and fewer than 100 individuals in the Kohat District (Table S1, available online). The habitats ranged in altitude from 347 to 581 masl.
Environmental factors
Soil analysis at the sample sites showed variations in all of the measured parameters (Table 1). The soil texture ranged from sand to sandy clay loam in the Karak District, but was a sandy loam at all sites in the Kohat District (Table 1). The organic matter content of the soils was low at all sites, ranging from 0.48 to 1.31%, indicating the unfertile nature of the region's soils. The nitrogen (N) level was also very low, ranging from 0.024 to 0.065 ppm. However, the phosphorus (P) and potassium (K) contents were high at all sites. The P content ranged from 7.57 to 59.15 ppm, and the K content ranged from 133 to 266 ppm. pH was slightly alkaline in the Kohat District (ranging from 8 to 8.2), whereas that in the Karak District was more alkaline (ranging from 8.0 to 9.6). The electrical conductivity (EC) and total soluble salts (TSS) were also low, except for a high EC (4.9 dS/m) at Wanki Siraj Khel. We observed similar results in our previous research (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009).
Genetic diversity within and among the populations using PBA markers
Of the 15 PBA primer sets, 13 amplified PCR products. We identified a total of 169 loci in the six populations of R. stricta, of which 88.2% were polymorphic (Table 2). The CYP2C19F/CYP2B6R primer pair produced the highest PPB (100%), whereas the CYP2B6F/heme2B6 primer pair produced the lowest percentage (41.7%). The percentage for the remaining 11 primers ranged from 77.8 to 94.1%. Table 2 shows that the percentage was highest in the Kander population (75.7%) and lowest in the Takht Nasrati population (45.0%).
Table 3 summarizes the genetic variation data based on the PBA markers. Expected heterozygosity (H e) was highest in the Kander population (0.296) and lowest in the Takht Nasrati population (0.153). Shannon's information index followed the same pattern, with the lowest value in the Takht Nasrati population (0.223) and the highest value in the Kander population (0.423). The effective number of alleles was also lowest in the Takht Nasrati population (1.3) and highest in the Kander population (1.5). All these results showed that the Kander population possessed the highest genetic diversity in our sample, whereas the Takht Nasrati population had the lowest genetic diversity.
Table 3 Genetic variations within the populations of R. stricta using the PBA and AFLP markers

n= sample size; n a= observed number of alleles; n e= effective number of alleles; H e= Nei's (Reference Nei1973) genetic diversity; I= Shannon's information index; PPB = percentage of polymorphic bands.
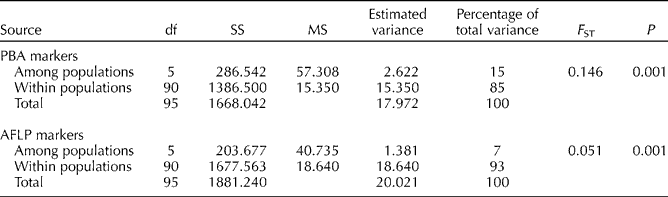
AMOVA provided the details of the population genetic structure of R. stricta (Table 4). There was higher genetic diversity within the populations (85%) than among the populations (15%).
Table 4 Summary of the AMOVA results for the within- and among-population genetic structures of R. stricta using the PBA and AFLP markers

SS, sum of squares; MS, mean square; F ST, fixation index.
Although the genetic diversity was lower among the populations, the genetic differentiation within the populations was also low, with an F ST value of only 0.146 (Table 4). When the level of gene flow was calculated using the formula N m= ((1/F ST)–1)/4, it was higher (1.46) than the value (0.22) for outcrossed animal-pollinated species (Hamrick and Godt, Reference Hamrick, Godt, Brown, Clegg and Kahler1989) and for outcrossed wind-pollinated species (0.10; Hamrick and Godt, Reference Hamrick, Godt, Brown, Clegg and Kahler1989).
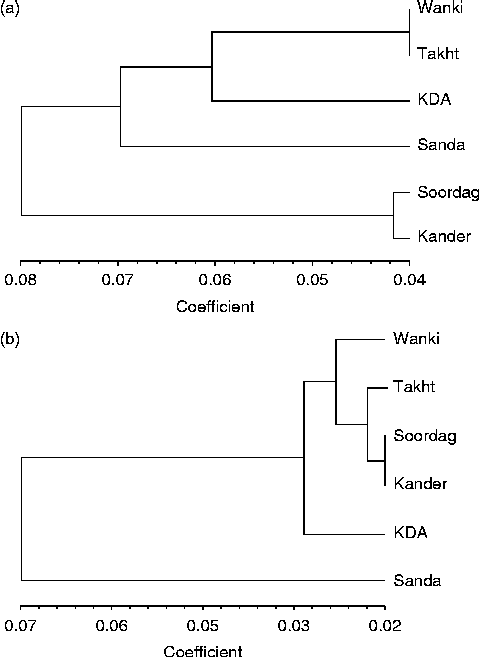
Cluster analysis of the six populations of R. stricta using the UPGMA method divided the populations into two major clusters: one with four populations and the other with the Soordag and Kander populations (Fig. 1), and this result was confirmed by the principal coordinate analysis (data not shown) and by bar plot analysis (Fig. 2). The first coordinate accounted for 31.9% of the variation and the second coordinate accounted for 23.2%. The bar plot analysis with an optimal value of K= 2 clusters showed an admixture of individuals in each population. However, lesser admixtures were observed in the KDA population, which remained distinct from the other populations. High admixture was found in the Soordag and Kander populations (Fig. 2).
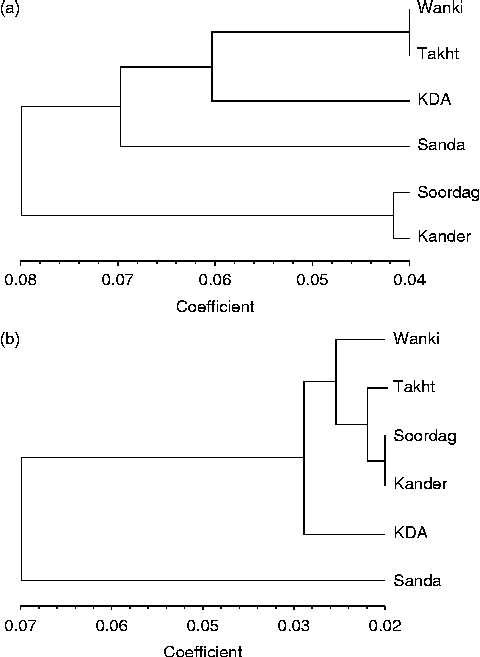
Fig. 1 UPGMA cluster analysis of the R. stricta populations based on genetic distances using (a) the PBA markers and (b) the AFLP data.

Fig. 2 Structure analysis of the six populations of R. stricta using (a) the PBA and (b) AFLP markers. The bar plot analysis was conducted with an optimal value of K= 2 clusters.
Genetic diversity within and among the populations using AFLP markers
All of the six AFLP primer pairs gave visible peaks and amplified a total of 173 loci (Table 2). The PPB was high in most cases (ranging from 95 to 100%), and moderately high for the E40/M60 pair (75%). The PPB was highest (80.9%) in the Sanda Fateh Khan population and lowest (60.1%) in the Takht Nasrati population (Table 2). Nei's expected heterozygosity (H e) was highest (0.250) in the Sanda Fateh Khan population and lowest (0.182) in the Wanki Siraj Khel population, where the PPB was 17.9% points lower than in the Sanda Fateh Khan population. Nei's expected heterozygosity occurred due to the sampling of the loci (VarL%) but not due to the sampling of individuals (VarI%) (data not shown).
AMOVA showed that the genetic diversity was high (93%) within the populations and low (7%) among the populations (Table 4). We confirmed this result using the Lynch and Milligan (Reference Lynch and Milligan1994) method, which showed high genetic diversity (0.265) within the population and low diversity (0.014) among the populations. Similar AMOVA results were observed in poplar populations (Smulders et al., Reference Smulders, Cottrell, Lefèvre, van der Schoot, Arens, Vosman, Tabbener, Grassi, Fossati, Castiglione, Krystufek, Fluch, Burg, Vornam, Pohl, Gebhardt, Alba, Agúndez, Maestro, Notivol, Volosyanchuk, Pospíšková, Bordács, Bovenschen, van Dam, Koelewijn, Halfmaerten, Ivens, van Slycken, Vanden Broeck, Storme and Boerjan2008), which had higher genetic diversity (72.9%) within the populations than among the populations (27.1%) based on the AFLP markers.
In addition to the lower diversity among the populations, the genetic differentiation was low, with a low F ST value (0.051). When the level of gene flow was calculated using the formula N m= ((1/F ST)–1)/4, it was 4.623, which is much higher than the value (0.22) for outcrossed animal-pollinated species (Hamrick and Godt, Reference Hamrick, Godt, Brown, Clegg and Kahler1989) and the value (0.10) for outcrossed wind-pollinated species (Hamrick and Godt, Reference Hamrick, Godt, Brown, Clegg and Kahler1989).
The cluster analysis of the six populations based on the AFLP markers using the UPGMA method divided the populations into two major clusters, one with five populations and the other with only the Sanda Fateh Khan population (Fig. 1). This result was confirmed by the principal coordinate analysis, in which the first coordinate accounted for 80.9% of the variation and the second coordinate accounted for 10.6% of the variation. The bar plots of all 96 individuals were drawn to estimate the admixture of the populations using an optimal value of K= 2 as the number of clusters (Fig. 2). These plots also confirmed the pattern with two clusters revealed by the cluster analysis (Fig. 1). In addition, none of the genetic or environmental factors was significantly correlated with each other based on the results of Mantel tests (Table S2, available online).
Discussion
Our genetic diversity analysis of R. stricta showed that there was little genetic differentiation among the populations, though with moderate polymorphism and heterozygosity (Tables 3 and 4). All of the populations thus seemed to be subunits of a single population (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009). Under natural conditions, cross-pollination occurs in R. stricta by both wind and insects such as honeybees and beetles (Gilani et al., Reference Gilani, Kikuchi, Shinwari, Khattak and Watanabe2007; Gilani, Shinwari and Watanabe, Reference Gilani, Shinwari and Watanabe2007). Modes of reproduction are not well studied in R. stricta (2n= 22); however, vegetative propagation by stem cuttings is possible while seed dispersal is through winds (Gilani et al., Reference Gilani, Kikuchi, Shinwari, Khattak and Watanabe2007; Gilani, Shinwari and Watanabe, Reference Gilani, Shinwari and Watanabe2007). It is therefore reasonable to hypothesize that the wind-pollinated seeds were dispersed throughout the study area, resulting in a similar population structure at all six sites, but the number of individuals per population was small ( < 100; Table S1, available online), and there should therefore have been higher genetic differentiation among the populations. Because R. stricta is a long-lived perennial species that predominantly outcrosses, it should have higher genetic diversity within populations, whereas short-lived perennials or self-pollinated species will have lower genetic diversity within their populations than among populations (Hamrick and Godt, Reference Hamrick and Godt1996; Nybom, Reference Nybom2004; Straub and Doyle, Reference Straub and Doyle2009). Thus, even with recent population fragmentation due to human pressure, the species maintained higher genetic diversity within populations than among populations. However, the signs of genetic drift may be visible in the future if conservation managers do not implement proper conservation measures to protect R. stricta.
The Kander population remained distinct from the others, with the highest genetic diversity in the PBA analysis and high diversity in the AFLP analysis, whereas the lowest diversity was observed in the Takht Nasrati population in the PBA analysis, with a low diversity in the AFLP analysis (Table 2). The Kander population is in the eastern part of the Kohat Plateau, where the human impact is minimal. In contrast, the Takht Nasrati population is in a settled area where human impacts are high; the plants were growing near a college and near the road used for transportation by the local villagers. Geographic isolation and decreased human impacts have protected the Kander population. However, the Takht Nasrati population, which has lower genetic diversity, is more vulnerable to human impacts, and particularly to increasing infrastructure construction. Therefore, the Takht Nasrati population should be given priority for conservation. The W. coagulans population in this area was also separated from other populations in a cluster analysis (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009). A model of isolation by distance revealed no correlation between genetic and geographic distances for W. coagulans. Therefore, our results confirm that populations may be more threatened by the consequences of rapid habitat loss due to fragmentation rather than by genetic consequences. We hypothesize that fragmentation of R. stricta populations has occurred primarily due to human impacts, including habitat destruction, rather than due to geographic isolation or barriers to pollination.
All of the six populations were smaller than 100 individuals, although the populations in the Karak District were fewer than 50 individuals (Table S1, available online). In theory, such population fragmentation combined with a smaller number of individuals should lead to higher differentiation among populations (Frankham et al., Reference Frankham, Ballou and Briscoe2007). This was not true in our study, in which genetic differentiation and the resulting gene flow among the populations were higher than those within the populations. This confirms our hypothesis that ecological consequences may cause rapid destruction of populations. It is possible that population decreases may have occurred relatively recently in the Kohat District.
Previous researchers have hypothesized that larger populations of R. stricta formed in the Kohat Plateau in the Eocene period (Meissner et al., Reference Meissner, Master, Rashid and Hussain1974; Shah, Reference Shah2003). These larger populations were subsequently reduced into large fragments that have recently been further reduced into the small populations currently observed on the Kohat Plateau. Most probably, the harsh and semi-arid climate and rapid urbanization of the plateau might explain this reduction in population sizes. The plant is used locally in Pakistan for medicinal purposes as a cooling agent, antidiabetic, blood purifier and cure for toothaches and other pains (Gilani et al., Reference Gilani, Kikuchi, Shinwari, Khattak and Watanabe2007; Gilani, Shinwari and Watanabe, Reference Gilani, Shinwari and Watanabe2007). The dried leaves are sold commercially in herbal markets in Pakistan (Gilani et al., Reference Gilani, Kikuchi, Shinwari, Khattak and Watanabe2007; Gilani, Shinwari and Watanabe, Reference Gilani, Shinwari and Watanabe2007). Thus, unsustainable harvesting of the leaves may also explain the population decline.
Local population fragmentation often results from past disturbances (Silverton and Charlesworth, Reference Silverton and Charlesworth2001), and one of the main reasons for rapid fragmentation is habitat destruction (Channel and Lomolino, Reference Channel and Lomolino2000). Although we observed a high rate of gene flow among the populations in our study, all six populations had fewer than 100 individuals. In our previous research (Gilani et al., Reference Gilani, Kikuchi and Watanabe2009), W. coagulans from the same region have shown a similar pattern. The two studies have therefore suggested that populations of these two species, and possibly other species that live in similar habitats, are at risk of local extinction if their populations continue to decrease. Urbanization will play a key role in determining the fate of the populations of R. stricta. The average polymorphism and heterozygosity in R. stricta populations detected by PBA and AFLP can be considered moderate. Therefore, it is suggested that conservation intervention for the habitat and species types is required.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S147926211300052X
Acknowledgements
This research was supported in part by Grant-in-Aid # 25257416 from Japan Society for the Promotion of Science. This work was conducted at the University of Tsukuba, Tsukuba, Japan as a part of the doctoral degree research of the principal author. The principal author was financially supported by a Japanese government MEXT scholarship during his whole stay in Japan. All the travel and field expenses were sponsored by Kohat University of Science and Technology (KUST) under the joint collaborative project titled ‘Establishment of Linkages with University of Tsukuba (Japan) for Joint Research and Education Program’, between KUST and University of Tsukuba, which was funded by Higher Education Commission Pakistan.