Published online by Cambridge University Press: 29 May 2003
There are strong biological, evolutionary and immunological arguments for predicting extensive polymorphism among helminth parasites, but relatively little data and few instances from which the selective forces acting on parasite diversity can be discerned. The paucity of information on intraspecific variation stands in contrast to the fine detail with which helminth species have been delineated by morphological techniques, accentuating a trend towards considering laboratory strains as representative of a relatively invariant organism. However, in the fast-moving evolutionary race between host and parasite one would predict a monomorphic species would be driven to extinction. We review the arena of intraspecific variation for the major helminth parasites, ranging from biological properties such as host or vector preference, to biochemical and immunological characteristics, as well as molecular markers such as DNA sequence variants. These data are summarized, before focusing in more detail on polymorphisms within protein-coding genes of potential relevance to the host-parasite relationship, such as vaccine candidates. In particular, we discuss the available data on a number of major antigens from the filarial nematode Brugia malayi. Information is currently too sparse to answer the question of whether there is antigenic variation in filariasis, but the indications are that proteins from the blood-borne microfilarial stage show significant intraspecific variability. Future work will define whether polymorphisms in these antigens may be driven by exposure to the host immune response or reflect some other facet of parasite biology.
As we proceed in this exciting era of genome sequencing, unwrapping details of thousands of new genes from pathogens large and small, it is becoming increasingly obvious that information from a single ‘type’ individual will not suffice. The complete genome sequence of any organism is indeed the gateway to the functional biology of novel genes, identification of new drug targets and vaccine antigens, and in establishing the fundamental physiology of highly evolved parasites with specialized lifestyles (Blaxter et al. 1999; Johnston et al. 1999). Here however, we wish to review where we stand on the question of polymorphism of helminth parasites, with the perspective that a genome sequence represents but one reference point against which many variant sequence types may be found within the same species. Within this context, our focus will be on protein-coding sequences, genes for functional proteins in which allelic variation may act to modify biological activity, or provide diversity in the face of specific host immune responses. Such analyses are conspicuously rare for any helminth parasite (Read & Viney, 1996).
The study of helminth protein polymorphisms has been hindered by three issues. First, there is the biological problem that helminth parasites are, in general, sexually reproducing animals which do not themselves multiply in their host. Thus clonal populations do not exist (except in a few circumscribed instances), requiring that the analysis of coding alleles be performed on individual worms. Second, in many species the quantities of protein (or even DNA) that can be recovered from one individual precludes many analyses. Thirdly, where field isolates require propagation through laboratory animals, it is likely that this very propagation reduces the variability in the parasite by selecting subpopulations best adapted to the unnatural host (LoVerde et al. 1985; Curtis & Minchella, 2000).
Why then should we study ‘wild’ helminths? There are both practical and theoretical imperatives to do so. The development of vaccine candidate antigens cannot proceed far without knowledge of the distribution and conservation of antigen genes among parasite populations worldwide. Moreover, the critical question of whether helminth parasites display any form of antigenic variation has not been systematically addressed despite the fundamental implications this would have on the whole of parasite immunology.
Equally important is the threat posed by drug-resistance alleles to the success of basic anthelmintic therapy (Sangster, 1996; Prichard, 2001). Conceptually, we need to understand the population genetic structure of helminth parasites to predict how such resistance alleles may spread and reach fixation in natural populations. This requires more data on allele frequency, interbreeding and molecular variation among all important helminth species (Nadler, 1995; Anderson, Blouin & Beech, 1998). In addition, there is the argument that natural variation in genes provides a spotlight on proteins with interesting functions in the host-parasite relationship. Furthermore, comparisons between isogenic organisms differing at only one locus can provide a far more conclusive link between gene and function than any number of analyses of newly-identified sequences in a single isolate.
In this review, we discuss the currently known polymorphisms in helminths generally, and then proceed to consider the filarial nematodes in particular. We illustrate this with examples from the literature and from our own recent work in Indonesia.
There are plenty of documented polymorphisms for helminth parasites, whether judged by biological characteristics (e.g. morphology, infectivity), biochemical (e.g. enzymatic and susceptibility to anthelmintic drugs), immunological (e.g. antibody reactivity) or molecular biological (e.g. DNA sequence) (McManus & Bowles, 1996; Read & Viney, 1996; Wakelin & Goyal, 1996). We discuss each of these issues briefly below.
Most contemporary molecular information involves non-coding sequences such as microsatellite dinucleotide repeats and spacing regions flanking mitochondrial genes. This field has been recently reviewed by Anderson et al. (1998) and so will not be discussed in detail here. A good example of the potential of these marker sequences is in Anderson's own work describing the separate population structures of human and swine Ascaris parasites in the Americas (Anderson, Romero-Abal & Jaenike, 1993; 1995). In a different manner, a particularly interesting sequence variation has emerged from Haemonchus contortus in which the random interpolation of a short transposable sequence has been noted (Hoekstra et al. 2000). These variants appear to have little functional consequence, but have proven invaluable markers to identify ‘strains’ and geographical isolates of the parasite.
Infectivity is perhaps the characteristic of most intrinsic importance to parasites, and has been carefully studied in most major species. Not only is there a great practical importance to knowing which parasite types may be most infective to target species, but variation in infectivity patterns can give insights into how the infection process itself is controlled. Several examples can be drawn from nematode parasites of humans and animals. Different isolates of the gastrointestinal nematode Trichuris muris vary in their ability to resist immune-mediated expulsion by mice (Bellaby et al. 1995). Parasite survival appears to be associated with the type of immune response stimulated in the host, as the most persistent isolate induces a stronger ‘Th1’ bias than the others (Bellaby, Robinson & Wakelin, 1996). The Th1 response is pro-inflammatory, typically through macrophage activation, and while well-adapted to eliminating intracellular micro-organisms is known to be ineffective against nematode parasites in the gut. Significantly, sharp differences in infectivity can be observed in sibling lines of T. muris derived from a single isolate, indicating that the parasite population in the wild is polymorphic in this respect (as described elsewhere in this supplement by D. Wakelin et al.). Similarly, Trichinella parasites from various geographical locales show markedly different infectivities to laboratory mice, as well as differences in the extent to which they elicit a protective ‘Th2’ immune response (Bolas-Fernandez & Wakelin, 1990; Wakelin & Goyal, 1996).
Many helminth parasites can be ‘adapted’ to laboratory animal species which are not their natural hosts. For example, Necator americanus, a human hookworm, can be ‘adapted’ to hamsters (Sen, 1972) and laboratory rat strains of Nippostrongylus brasiliensis to optimal infection of mice (Solomon & Haley, 1966; Wescott & Todd, 1966). A fascinating set of experiments were conducted by Dobson and co-workers, in which lines of Heligmosomoides polygyrus were propagated by serial passage through naive or immune mice, and the latter set of parasites shown to be less immunogenic (Su & Dobson, 1997). Similar selection experiments in other nematode species have had variable results (reviewed by Read & Viney, 1996), with adaptation evident after a single passage, or failing to be evident at all. A missing factor from these experiments is any genetic marker in order to establish that adaptation represents selection from a polymorphic ancestral population, and to exclude any heritable epigenetic mechanism.
A clearer indication that different host species select distinct subpopulations of parasites comes from studies on Schistosoma mansoni, (LoVerde et al. 1985). A single African isolate has been propagated separately for many years in baboons and mice. The baboon parasites are polymorphic at several loci, while single alleles have become fixed in the murine population. When baboon-derived parasites were ‘adapted’ by serial passage in mice, the resultant parasites were found to have fixed the same alleles as in the original mouse strain, indicating selective pressure against certain alleles is exerted in the mouse environment.
Biochemical polymorphisms are evident from almost all isoenzyme (or allozyme) analyses (Anderson et al. 1998), but their use in determining an average level of parasite genetic variability has been interpreted in different ways. The mean heterozygosity in any one individual Ascaris was found to be only 2 of 38 loci (6.6%), leading to the suggestion that gastrointestinal parasites were relatively monomorphic (Leslie et al. 1982). However, in other closely related ascarid worms such as Toxocara, Nadler (1986) found a much higher level (8.5–13.7%) mean heterozygosity, and reported that 22–39% of all loci tested showed some degree of allelic polymorphism across the population as a whole. These data led Nadler (1990) to reach a contrary conclusion, that ascarids do display considerable genetic diversity. Another proposal was that nematodes with direct life cycles were less polymorphic than those with indirect life histories, but again this association was not supported by an analysis of data from some 35 nematode species (Anderson et al. 1998). With expressed sequence tag (EST) data from many of these species now identifying the enzymes in question at the sequence level, it would be both possible and timely to quantify the heterozygosity and degree of sequence variation in these same enzymes to resolve these questions.
An example of the more precise information now available in the biochemical arena is the identification of alleles responsible for drug resistance in nematodes. Benzimidazole resistance in gastrointestinal nematodes of pastoral animals, a phenomenon of considerable concern (Sangster, 1996), can be attributed to 2 specific amino acid substitutions in the β-tubulin protein, which itself is encoded at two separate loci. The predominant susceptible allele encodes Tyr-200, and the resistant allele Phe-200, in both H. contortus (Kwa, Veenstra & Roos, 1994) and Teladorsagia circumcincta (Elard, Comes & Humbert, 1996). Moreover, resistance is enhanced if Phe-167 changes to Tyr or His (Prichard, 2001), and increasing degrees of resistance occur when these substitutions coincide, when they are present at both loci, and when either or both loci become homozygous for a resistance allele. The rapidity with which drug selection appears indicates that the resistant alleles are pre-existing in the wild populations.
An even greater degree of polymorphism is observed in the P-glycoprotein gene of H. contortus, thought to be important in multi-drug resistance (Blackhall et al. 1998). No fewer than 7 alleles, defined by restriction fragment length polymorphism, were found in just 30 individual worms from a single strain. A larger comparison of 5 different strains (3 of which had been selected through 17 generations of ivermectin- and moxidectin-treated sheep infection) identified some 31 RFLP alleles in a total of 180 individuals. Although individual alleles were not sequenced, this system perhaps represents the most polymorphic yet described for any helminth gene.
Immunological variation within individual helminth species has been the most elusive to identify at the genetic level. One may expect that the ceaseless ‘arms race’ between host and parasite has generated a wide variety of antigenic variants, but no conclusive evidence has yet been provided. One direct approach to this question is to screen parasite populations with specific antibodies. Ascaris larvae do vary in surface binding to human antibody, but this may be phenotypic as different individual worms could express quantitatively different levels of surface antigens, or may be out of phase in development (Fraser & Kennedy, 1991). Gilleard described a monoclonal antibody, generated to Dictyocaulus viviparus, which binds only to a subpopulation of Necator americanus larvae (Gilleard, Duncan & Tait, 1995). As with Ascaris, this enticing observation has yet to be linked to an inherited factor within the parasite population. If substantiated, such instances would have direct relevance to immune system recognition, immunity and parasite immune evasion mechanisms.
Some studies have addressed the critical question of whether vaccines and vaccine antigens from different isolates are equally efficacious. Irradiated cercariae of S. mansoni induce partial protection against subsequent challenge with undamaged parasites. When mice were vaccinated with irradiated S. mansoni from Egypt or Puerto Rico, the protection against the homologous isolate was broadly similar, but cross-protection was not reciprocal (Hackett et al. 1987). In S. japonicum, a Chinese isolate propagated in the laboratory since 1937 immunized effectively against challenge with 3 wild isolates taken 50 years later (Moloney, Hinchcliffe & Webbe, 1989), but failed to protect against a Phillipine strain (Moloney, Garcia & Webbe, 1985). In the case of Trichinella spiralis, isolates from Spain, Poland and London showed variable levels of protective immunization when tested against homologous challenge, but markedly poorer protection against heterologous infection in some combinations (Goyal & Wakelin, 1993). At the very least, antigens involved in protective immunity do not appear to be fully conserved within species. Studies such as these place greater urgency on defining the precise structural polymorphisms at the amino acid sequence level. As argued elsewhere, if vaccine antigen targets are in fact highly variable, this brings into question the utility of those antigens which may be so effective in the laboratory context.
A number of recent studies have explicitly sought intraspecific comparisons between potential alleles of coding genes (Table 1). One investigation focused on a candidate vaccine antigen from hookworm, termed Ancylostoma secreted protein (ASP) (Hawdon et al. 1996). ASP belongs to a large gene family represented in all nematodes so far studied, and with more distant relatives in all other taxa from plants to mammals. When Ancylostoma caninum samples derived from dogs in China and the USA were compared (Qiang et al. 2000), 10 amino acid substitutions were found in the 421-amino acid protein (2 of the changes being within the short N-terminal signal sequence); this is a relatively low level of diversity, and the applicability of ASP-1 for vaccination is promising as one domain of the protein showed only one amino acid difference. At the nucleotide level, there were a total of 30 nucleotide changes in ∼1300 bp, the ratio of coding to non-coding changes implying that all may be neutral.

It surely is essential that similar studies are undertaken on vaccine candidate antigens for other major helminth parasites. The paucity of species-wide data on sequence diversity or conservation of the antigens selected for development as schistosomiasis vaccine components has been commented upon elsewhere (Curtis & Minchella, 2000). One wonders whether an approach which identifies the most variable antigens across the species as a whole should not be a mandatory part of the vaccine development process?
While studies such as these deal with protein coding sequence allelisms, it is important to remember that many functional polymorphisms are coded in regulatory elements within non-coding DNA (e.g. promoter and enhancer sequences), in RNA elements, and within introns of protein-coding genes. Analyses of these regions is likely to receive more attention once individual proteins of known importance are analysed in sufficient detail.
Lymphatic filariasis is primarily caused by two closely related nematodes, Brugia malayi and Wuchereria bancrofti. The microfilarial (Mf) stages of B. malayi and W. bancrofti can be distinguished by the cell nuclei at the caudal tip visible in the former species, and adults differ in details of external morphology, such as the numbers of papillae. The principal distinction is size, the adults of W. bancrofti reaching a maximum about 50% larger than B. malayi. Although these may appear slender grounds for separation of B. malayi into a distinct genus (Buckley, 1960), it is generally felt that any further change in nomenclature would not serve the field well.
In addition to these two major species, there are additional members of each genus. The human parasite B. timori has a restricted distribution but causes disease very similar to B. malayi. The animal filaria B. pahangi is considered non-infective to humans, but is known to form fertile hybrids with B. malayi. These three Brugia species show extensive antigenic similarities (Maizels et al. 1983). There are at least 6 additional species of Brugia (Sasa, 1976), and one other known species of Wuchereria (W. kalimantani), found in the silvered leaf monkey Presbytis cristatus (Palmieri et al. 1980).
Within individual species, it is known that the adult male worms of B. malayi show significant morphological variation with respect to their posterior cuticular ornamentation (required for successful mating), and these features distinguish isolates from China, India, Indonesia/Malaysia and Korea (Bain et al. 1989). However, the most striking variation within the species of both B. malayi and W. bancrofti are those observed with respect to periodicity. This remarkable feature describes the circadian rhythm with which microfilarial numbers rise and fall in the peripheral blood, in apparent synchrony with the biting behaviour of the local mosquito species (Hawking, 1975). Most W. bancrofti infections are nocturnally periodic, with few Mf present in the peripheral blood during the daytime hours. Exceptionally, the Pacific variant is diurnally periodic in accordance with the daytime biting habits of the Aedes mosquitoes.
B. malayi shows a wider variety of periodicities, but most parasites are either nocturnally periodic or sub-periodic, a dichotomy within the species which was established by Wilson et al. in 1959. These authors reported that periodic B. malayi was typically Anopheles-transmitted, rarely found in animals, and had slightly longer Mf which typically cast their sheath when stained with Giemsa on a thin blood film. In contrast, the subperiodic parasites were generally Mansonia-transmitted, could be found naturally in, and readily transmitted to, cats, and had Mf which retained their sheath on Giemsa staining. This subdivision into periodic and sub-periodic is often still followed in the filariasis literature.
The question of periodicity and subdivision of B. malayi into biological types was thoroughly re-examined by Partono & Purnomo (1987). They identified several problems. First, periodicity indices (e.g. the ratio of night-time peak to the daytime nadir) drew an arbitrary borderline between the ‘periodic’ and ‘subperiodic’ types. Second, some isolates (e.g. East Kalimantan) were in fact aperiodic, although in all other respects such as infectivity to non-human hosts were like the subperiodic. Third, and most importantly, isolates such as Pekan Baru showed nocturnal periodicity but were otherwise ‘subperiodic’ with features such as Mansonia transmission and infectivity to laboratory animals. Reviewing the biological properties of some 10 isolates from diverse localities in Indonesia, Partono and Purnomo concluded that the key differentiating factor was infectivity to animals, thus reclassifying the ‘periodic’ type as anthropophilic and the ‘subperiodic’ as zoophilic. This revised division is one we endorse and, presenting it in Table 2, we suggest that it becomes the standard nomenclature.

Most B. malayi across South and East Asia is the periodic or anthropophilic type, while the zoophilic type is restricted to Indonesia, Malaysia and the Phillipines (Sasa, 1976). A variant of the anthrophilic type, termed the rocky beach strain, was described as being Aedes-transmitted in Korea and Japan, but has not been further characterized (Sasa, 1976). The strain of B. malayi used in most laboratories is a zoophilic type, originating in Pahang in West Malaysia (Edeson & Wharton, 1958). This strain is now propagated by TRS Labs Inc. of Georgia, USA, after which the strain is named. There is no explicit indication of any differential pathology between the different strains of B. malayi, in the manner distinguishing Bancroftian filariasis from the Brugian form of the disease (Partono, 1987).
Molecular polymorphisms among the B. malayi strains have been demonstrated by Underwood et al. (2000) using two microsatellite markers. BMsat1 shows that several zoophilic isolates from Indonesia have a microsatellite allele which is 2 bp longer than the allele from the one anthropophilic parasite tested, and 2 bp shorter than the allele present in the TRS strain. One Indonesian isolate also showed 6 bp longer form of the microsatellite BMsat2. Interestingly, this microsatellite is suggested to reside within a novel protein coding sequence, which is maintained in-frame by the 6 bp insertion.
Antigenic variation or diversity is an evolutionary strategy for pathogens to escape the effects of a specific immune response mediated by, for example, antibodies. Although the role of antibodies in human immunity is not established, there is an inverse relationship between detectable antibody binding to the surface of Mf, and the presence of Mf themselves (McGreevy et al. 1980) suggesting that antibodies mediate clearance of Mf. If so, then the antigens on the external sheath of Mf may be expected to vary within one filarial species.
Whether Mf surface antigens do indeed vary has been directly addressed in only one study (Ravindran, Satapathy & Sahoo, 1994). Stimulated by the observation that around 20% of Mf+ individuals do have antibodies that bind to the surface of Mf, and that in all published assays the Mf used for testing were taken from a heterologous individual, these authors collected parasites and sera from 5 Mf+ patients. None of the patients’ sera reacted to their own (autologous) Mf, although one showed fluorescent binding to heterologous Mf. When a panel of unrelated patient sera was tested against these 5 isolates of Mf, all elephantiasis sera reacted to all isolates. Most significantly, of sera from 15 additional Mf+ patients, no fewer than 7 recognized one or more of the 5 test isolates from unrelated patients. These results suggest that Mf can express a variety of surface antigens, and that those that survive in the human bloodstream are those which the host antibody response fails to recognize.
The target antigens of this reaction have yet to be defined. The Mf sheath is composed of a tightly cross-linked set of repeat-rich proteins, together with some carbohydrate structures (Hirzmann et al. 1995; Zahner, Hobom & Stirm, 1995). The key proteins (SHP-1, -2, -3 and variants thereof) have all been defined, but the accessibility of antibodies to each protein on acetone-fixed Mf has not been established. Interestingly, Ravindran and colleagues found that antibody binding was not sensitive to protease treatment of the Mf. This does not necessarily indicate a carbohydrate target, as the dense cross-linking of sheath proteins is likely to protect them from proteolysis.
Few systematic analyses have yet been carried out at the molecular level for variation of any filarial antigen, but interesting data have already emerged as a result of studies executed for other purposes. For example, the Filarial Genome Project has provided partial cDNA sequences from some 22,000 cDNA clones from B. malayi (The Filarial Genome Project, 1999). All these clones are derived from the TRS strain, and yet minor sequence variants representing putative alleles can be observed within this one dataset. These presumably correspond to polymorphisms which have survived repeated passage through laboratory animals. Another window available is to compare the pattern of evolutionary change in homologous molecules from related species (B. malayi/B. pahangi and B. malayi/W. bancrofti). We review below our current information drawn from these sources on selected filarial antigens; these antigens have been reviewed in more detail elsewhere (Maizels, Blaxter & Scott, 2001).
The major surface protein in the cuticle of adult filarial parasite is a 29 kDa glycoprotein (Maizels et al. 1989) shown both by sequence analysis (Cookson, Blaxter & Selkirk, 1992) and functional characterization (Tang et al. 1996) to be the anti-oxidant enzyme glutathione peroxidase. The GPX-1 antigen is also strongly recognized by human antibodies, indicating that it could be under immune selection. By sequencing genomic copies of this gene from B. pahangi, B. malayi and W. bancrofti (Cookson, Tang & Selkirk, 1993), the analysis presented in Table 3 emerged. Remarkably, GPX-1 from the two Brugia species has identical amino acid sequence, with only 7/669 (1.0%) synonymous nucleotide differences. The rate of nucleotide substitution in the introns (9.3%) is nearly ten times higher than in the exons, in comparisons of the same two species. The complete conservation of coding sequence is more compatible with a strict functional constraint than with a model of immune evasion by antigen variation. Even when differences between B. malayi and W. bancrofti are considered, and coding changes become evident (3.1%), the numbers of non-synonymous and synonymous nucleotide substitutions (7 and 10 respectively) do not support the hypothesis that variability of this antigen is driven by the host immune response.

Another prominent surface and secreted product from adult filarial worms is a 15 kDa member of the cysteine protease inhibitor (cystatin) family (Gregory and Maizels, unpublished). Represented frequently in the EST database, sequences for CPI-2 display a dimorphism: approximately equal numbers have either lysine or asparagine at residue 89. No functional difference is apparent in terms of protease inhibition, and the residue is not involved in either of the two protease-reactive sites. However, the asparagine-encoding form provides a potential N-glycosylation site. Most interestingly, the same dimorphism is observed in the Onchocerca volvulus EST dataset.
A most intriguing set of L3-associated antigens in filarial nematodes is the Abundant Larval Transcript (ALT) family (Gregory et al. 2000; Maizels et al. 2001), which represent 1–10% of cDNAs in various filarial species at the L3 stage, but are barely found at other points in the life cycle. They are the front-running candidates for vaccine antigens. So far, the family has been characterized by numerous related genes rather than variants at any one locus, and coding polymorphisms have yet to be identified. However, we have established that within the TRS laboratory strain, one of the ALT family (ALT-2) exhibits variant forms of intron 3. Intron 3 is itself unusual in consisting of a repeat sequence, and two forms of differing repeat numbers are found (Fig. 1). Individual worms possess either or both these forms, consistent with the notion that they are Mendelian alleles (W. F. Gregory & N. Gomez-Escobar, unpublished).

Fig. 1. Intron polymorphism in B. malayi abundant larval transcript-2 gene. Gene structures of B. malayi alt-1 and alt-2, as described by Gregory et al. (2000) with further data on the 1st intron from unpublished data of Dr Natalia Gomez-Escobar (personal communication). Alleles shown are both represented in the TRS laboratory strain. Boxes represent exons, thin lines are introns with nucleotide lengths given. Not to scale.
Another ALT family member is the dominant product from larvae of Onchocerca species (Bianco, Wu & Jenkins, 1995; Joseph, Huima & Lustigman, 1998). In a striking experiment, individual L3 of O. lienalis were labelled by injecting infected blackflies with radioactive methionine. The resulting analysis showed a size polymorphism in this protein taken from 11 individual parasites (Blanco et al. 1990). Thus, there are appear to be coding variants as well as intronic variants of this important gene family.
The significance of the intron dimorphism remains to be defined, but intronic alleles are known to affect levels of expression in Drosophila (Laurie & Stam, 1994; Stam & Laurie, 1996) as well as in higher animals. Generally, greater effects are attributed to substitutions in the first intron, where enhancer sequences are often positioned, but in other cases polymorphisms in repeat motifs of downstream introns can regulate levels of gene expression (Shimogiri et al. 1998; MacKenzie & Quinn, 1999). Allelic diversity in other nematode intron sequences has been described, both for Haemonchus contortus β-tubulin (Beech, Prichard & Scott, 1994) and for a number of Ascaris genes (Anderson & Jaenike, 1997). These findings were based on restriction site polymorphisms rather than primary sequence, and so cannot at this stage be directly compared to our observations on filarial intron variation.
Because morphological and behavioural variation appears most prominently in the microfilarial stage, Mf proteins from B. malayi are of particular interest with respect to their variability. The sheath of microfilariae is formed from the vestiges of the eggshell, and is found in some genera (Brugia, Loa, Litomosoides, Wuchereria) but not others (Onchocerca, Acanthocheilonema, Dirofilaria, Mansonella). In sheathed Mf, this structure represents the physical interface between host and parasite, and is therefore likely to be subject to immune pressure. This hypothesis can be tested by investigating whether sheath proteins shows higher sequence diversity than other filarial products.
We have recently been studying the SHP-1 antigen, originally named MF-22 after its molecular weight (Selkirk et al. 1991). Comparison of full-length sequences from B. pahangi (Selkirk et al. 1991) and B. malayi (Zahner et al. 1995) show extensive coding changes, including one-codon insertions/deletions (indels) in the repeat region of the protein. The majority of nucleotide substitutions are, in this case, non-synonymous. Overall, the two species show 10% amino acid sequence divergence (Table 3), the highest yet reported for filarial genes known to be orthologues. Moreover, the TRS strain EST dataset reveals that some copies of this gene from B. malayi have the B. pahangi-like indel pattern.
We have also examined the SHP-1 sequence by PCR amplification from genomic DNA of several isolates from Indonesia. These isolates have previously been characterized from the point of view of non-coding DNA markers (Underwood et al. 2000) but our work is the first to study protein coding sequences. We have found not only that there is more extensive sequence variation in natural B. malayi populations, including additional indels (Fig. 2), but that there is a previously unknown gene which shares sufficient sequence identity with SHP-1 to be amplified with the same primers.

Fig. 2. Polymorphism of microfilarial sheath protein genes SHP-1 and SHP-5 from Brugia malayi. Allelic forms of two sheath protein genes, SHP-1 (also known as MF-22) and SHP-5 (also termed Shp-1a by J. Hirzmann and H. Zahner, Univ. Giessen, personal communication). Boxes represent exons, thin lines are introns with nucleotide lengths given. Vertical bars denote coding changes in sequence; synonymous substitutions are not shown. Alleles shown are from the TRS laboratory strain and isolates from Indonesia (Kurniawan-Atmadja et al. unpublished). Not to scale.
This novel gene has been named SHP-5. Remarkably, its 5′ and 3′ ends are very similar to SHP-1, but the central domain is very different and the repeat sequence shows little relationship. Most importantly, SHP-5 sequences from different isolates show variation in amino acids and contain indels (Fig. 2), which with the preliminary data currently to hand appears to be no less extensive than that observed among SHP-1 sequences.
An additional microfilarial protein of importance is chitinase (Fuhrman, 1995), which is expressed as 2 distinct forms in B. malayi (p70 and p75), and 3 forms in B. pahangi (Fuhrman, Lee & Dalamagas, 1995). The chitinase proteins are easily surface-labelled (Maizels et al. 1983) and monoclonal antibody p70/p75 (antibody MF1) confers passive protection on recipient animals (Fuhrman et al. 1992). There is a substantial level of sequence heterogeneity in these proteins, although the existence of multiple genes has so far prevented assignation of alleles and splice variants to any one locus. Thus, RT-PCR from B. malayi cDNA revealed two forms differing by 48-nt encoding a serine/threonine-rich repeat motif. Similar amplification from B. pahangi yielded a 42-nt (14aa) difference for a similar sequence motif (Arnold et al. 1996). Analysis of the multiple B. malayi ESTs for chitinase show a relatively high level of variants represented, indicating that a more extensive study of this gene family would be well worthwhile.
Although the focus of this review is on lymphatic filariae, it is important to include Onchocerca volvulus, another major filarial parasite the adults of which occupy a subcutaneous niche in humans. Microfilariae infest the skin, from where a blackfly vector uptakes parasites to continue transmission. Primarily found in Africa, two biological types have been described: a less malignant, non-blinding forest form, and a more pathogenic, blinding savanna biotype (Duke, 1980; McMahon et al. 1988). Latin American O. volvulus is thought to have been imported from Africa in recent centuries, although it displays distinct preferences for local Simulium species over the African vector (Romeo De Leon & Duke, 1966). In the Upper Orinoco region of Venezuela, it has been suggested that a distinct biotype exists, defined by microfilarial morphology, isozyme pattern, and a high level of blood microfilaraemia (Botto, Arango & Yarzábal, 1984). Perhaps this instance should remind us that only a small fraction of parasite biodiversity has been sampled, even for a geographically restricted species such as O. volvulus.
Most molecular studies have concentrated on comparisons of the two African biotypes of O. volvulus, as this distinction is important in disease control. Isoenzyme analyses revealed a number of polymorphic loci, but no alleles unique to either type. Allele frequencies differ between forest and savanna isolates, but this cannot be used to distinguish individual worms (Flockhart et al. 1986). At the molecular level, a DNA probe for a repeat sequence has been described which hybridises only to the forest form (Erttmann et al. 1987), although success depends on conditions which discriminate between different copy numbers of similar repeat sequences in the two strains (Harnett et al. 1989; Meredith et al. 1989).
In terms of coding sequence polymorphisms, Keddie looked at 4 antigens comparing savanna/blinding (Ghana and Mali) and forest/non-blinding (Côte d'Ivoire and Liberia) strains, namely calreticulin, protein disulphide isomerase, RAL-2 and aspartyl protease inhibitor (Ov-API-1 or Ov33-3). None had any coding differences (Table 1), and the rate of synonymous substitution varied from 0 to 2.4 per 1000 nucleotides (Keddie et al. 1999). These findings of only limited diversity have been used to argue that O. volvulus has emerged from a recent bottleneck (Unnasch & Williams, 2000). It would be interesting, therefore, to compare sequence diversity of the homologous genes in other species for which no such restriction is thought to have occurred.
It has previously been argued (Maizels & Lawrence, 1991) that the success of filarial parasites may depend on their inducing a form of immunological tolerance in their host. Such a state may place selective pressure on parasites to conform in antigenic terms, for to diversify may take them outside the protective shelter of tolerance and lead to immune recognition and destruction. Early evidence for such a purifying selection was drawn from examples such as GPX, discussed above, and appears to be supported by studies on several other filarial proteins which, like GPX, are associated with the established adult stage.
New data now emerging, however, highlights the opposite scenario. Microfilarial sheath proteins reveal a high level of polymorphism, although the allelic variants have yet to be shown to have any immunological distinction. Antibody staining studies on the microfilarial surface provide evidence that this stage displays some form of antigenic variation, although here the structural targets have yet to be identified. It is conceivable that while the adult worms are driven to uniformity, the microfilarial stage diversifies because it is under a very different form of selective pressure. Closer definition of the patterns of variation may allow us to consider exactly what these selective pressures may be.
Parasitologists have, with some honourable exceptions, been tardy in studying phenotypic polymorphism in helminths. Now that tools are available for sophisticated analyses at the molecular level, we anticipate major advances in this area. Data so far available are both scanty and patchy, revealing a spectrum of variability from total conservation to surprising levels of allelism. The latter instances (such as H. contortus P-glycoprotein and the B. malayi sheath proteins) should be studied with greater intensity if we are to probe the true extent of helminth genetic variation. More thought should be devoted to scanning the set of parasite-expressed proteins for those showing the highest level of diversity, so that variation is not sought only in those proteins which appear on our current understanding to be most likely candidates.
Once substantial datasets are in place, we may be in a position to address the key questions of what selective forces may be driving polymorphism, and what are the relative selective pressures exerted by host and vector (Read & Viney, 1996; Wakelin & Goyal, 1996; Anderson et al. 1998). Experimental genetic systems such as transgenesis for helminths are in their infancy, but if developed may allow variant forms of antigens to be functionally compared. Sophisticated manipulation of the host immune system is already possible, so that one can envisage at a future point being able to test not only whether particular variants are advantageous in immune evasion, but also which arm of the immune system they may have evolved to combat. The study of helminth polymorphisms on this broader stage, across the whole species in the wild, and driven by many possible forces from host, vector and environment, is sure to provide many such fascinating and insightful advances and make an essential contribution to the control and elimination of helminth diseases.
The authors thank the Wellcome Trust for support through a Programme Grant (RMM) and an International Research Development Award (AKA), and the European Commission (contract ICA4-2000-10195). We also thank Natalia Gomez-Escobar and Bill Gregory for permission to quote unpublished work on B. malayi genes.

Table 1. Sequence polymorphisms in coding genes of nematode parasites

Table 2. Morphological and ecological variants of lymphatic filarial nematodes

Table 3. Intron and exon variation in two filarial antigens
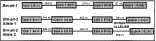
Fig. 1. Intron polymorphism in B. malayi abundant larval transcript-2 gene. Gene structures of B. malayi alt-1 and alt-2, as described by Gregory et al. (2000) with further data on the 1st intron from unpublished data of Dr Natalia Gomez-Escobar (personal communication). Alleles shown are both represented in the TRS laboratory strain. Boxes represent exons, thin lines are introns with nucleotide lengths given. Not to scale.

Fig. 2. Polymorphism of microfilarial sheath protein genes SHP-1 and SHP-5 from Brugia malayi. Allelic forms of two sheath protein genes, SHP-1 (also known as MF-22) and SHP-5 (also termed Shp-1a by J. Hirzmann and H. Zahner, Univ. Giessen, personal communication). Boxes represent exons, thin lines are introns with nucleotide lengths given. Vertical bars denote coding changes in sequence; synonymous substitutions are not shown. Alleles shown are from the TRS laboratory strain and isolates from Indonesia (Kurniawan-Atmadja et al. unpublished). Not to scale.