Published online by Cambridge University Press: 16 April 2004
Previous studies have indicated that SAG1, the major surface molecule of the protozoan parasite Toxoplasma gondii, is an important attachment ligand for the host cell. However, the research data that supports this claim comes largely from studies investigating tachyzoite binding, and not SAG1 binding per se. In this study we successfully developed an in vitro attachment assay to directly evaluate the mechanism of SAG1-host cell binding. Competition experiments were then performed using SAG1 that had been pre-treated with the neoglycoprotein BSA-glucosamide or with antibody. Soluble BSA-glucosamide blocked SAG1 attachment to MDBK cells in a dose-dependent manner, implying that SAG1 binding is mediated, in part, via attachment to host cell surface glucosamine. Interestingly, pre-incubation of SAG1 in polyclonal sera from chronically infected mice failed to block binding. This challenges the assumption that anti-SAG1 antibodies block parasite attachment through the masking of SAG1 host cell binding domains. Taken together, this evidence presents new strategies for understanding SAG1-mediated attachment.
Toxoplasma gondii is a globally prevalent protozoan parasite affecting all warm-blooded animals (Miller, Frenkel & Dubey, 1972; Jackson & Hutchinson, 1989). It causes the disease toxoplasmosis which, although generally asymptomatic, can be life threatening in immunocompromised patients, such as those suffering from AIDS (Luft & Remington, 1992). The stage of the parasite responsible for dissemination of acute infection, termed the tachyzoite, is promiscuous in vitro, invading every mammalian cell line. While evidence that molecules secreted from the microneme organelles direct the process of host cell attachment is accumulating (Garcia-Réguet et al. 2000; Reiss et al. 2001; Brecht et al. 2001; Lourenco et al. 2001), the role of parasite surface molecules in this process remains equivocal.
The tachyzoite surface, which presumably represents the primary interface with the host cell surface, bears a number of glycophosphatidylinositol (GPI)-anchored surface proteins (Nagel & Boothroyd, 1989; Striepen et al. 1997), the most abundant of which is a 30 kDa protein named SAG1 (Kasper, Crabb & Pfefferkorn, 1983; Burg et al. 1988). Four lines of approach indirectly indicate that SAG1 binds host cell surfaces. In vitro antibody neutralization invasion assays demonstrate that a subset of anti-SAG1 MAbs block tachyzoite invasion, implying that SAG1 might function as a specific parasite attachment ligand (Grimwood & Smith, 1992, 1996; Mineo et al. 1993; Mineo & Kasper, 1994). Competitive invasion assays have shown that the neoglycoprotein (NG) BSA-glucosamide blocks the ability of wild-type parasites to bind host cells, whereas those expressing truncated SAG1 remain unaffected (Mineo et al. 1993). In more studies, solubilized SAG1 has been shown to attach directly to host cell surfaces (Fourmaux et al. 1996; Channon et al. 1999; Velge-Roussel et al. 2001). These studies have not, however, investigated the specificity of this binding. Finally, the solution of the structure of SAG1 reveals the presence of a basic groove at the interface, formed during SAG1 dimerization (He et al. 2002). This domain faces away from the parasite surface and is therefore perfectly positioned for interaction with host cell surface ligands.
Although anti-SAG1 MAbs and BSA-glucosamide inhibit tachyzoite attachment in vitro, it is not known whether their blocking effects are explicitly mediated through masking host cell binding domains on SAG1. For instance, antibody neutralization experiments were used to investigate the ability of anti-SAG1 MAbs to prevent tachyzoite binding, rather than SAG1 binding. To address this question we here investigate the influence of antibodies and sugar-conjugates directly on SAG1.
All reagents for mammalian tissue culture were purchased from GibcoBRL. HS27 Human foreskin fibroblast cells (ATCC, USA) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and incubated at 37 °C and 10% CO2 in air. MDBK cells (adult bovine kidney; Madin & Darby, 1958; Porton Down, UK) were grown in Ham's F-12 nutrient mixture containing L-glutamine supplemented with 10% FBS and incubated at 37 °C and 5% CO2 in air.
The production of MAb C1E3 has been previously described by Grimwood & Smith (1992). Polyclonal immune mouse serum (IMS) was taken by cardiac puncture from mice infected with the cystic strain ME49. Anti-mouse IgG-HRP conjugate was from Sigma–Aldrich, UK. Polyclonal IMS and MAb C1E3 were affinity-purified using a 1 ml Hi Trap Protein G column (Amersham) following manufacturer's instructions.
Tachyzoites of the T. gondii RH strain were maintained by twice-weekly intraperitoneal passage in Tucks number 1 mice. Exudates were washed 3 times by centrifugation at 3000 g for 10 min followed with resuspension in phosphate-buffered saline (PBS), pH 7·2. Parasite pellets were finally resuspended in 4 ml of solubilization buffer (50 mM NaHCO3, pH 9·0 containing 0·1% (v/v) NP40), incubated at 4 °C for 1 h then freeze-thawed 3 times. Material was sonicated on ice 5 times for 20 sec, centrifuged at 10000 g for 5 min at 4 °C and the supernatant containing solubilized tachyzoite lysate was used for SAG1 purification and binding assays.
SAG1 was purified in a one-step anion-exchange high performance liquid chromatography (HPLC) procedure. Four mg of 0·22 μm filtered tachyzoite lysate prepared in solubilization buffer was loaded onto an AP1 column (Waters; diameter=0·8 cm, height=7·5 cm) packed with 3·5 ml of DEAE fast-flow Sepharose (Amersham) and pre-incubated with the same buffer. Solubilization buffer (10 column volumes) was flushed through the column at a flow rate of 1 ml/min. Non-adherent protein, comprising pure SAG1, was collected manually in 1 ml fractions. Bound protein was eluted from the column using 2 M NaCl in solubilization buffer and dialysed against solubilization buffer at 4 °C overnight. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE) and immuno-blotting with anti-SAG1 mAb C1E3 were used to assess the protein profiles of fractions. Fractions containing purified SAG1 were pooled, flash-frozen in liquid N2 and stored in 100 μl aliquots at −20 °C.
SDS–PAGE was performed according to the method of Laemmli (1970) using 10% separating gels. Protein samples were not reduced. Following electrophoresis, samples were either silver-stained (Heukeshoven & Dernick, 1988), or transferred to PVDF membrane (Schleicher and Schuell, Dassel, Germany) as described by Towbin, Staehelin & Gordon (1978). Following blocking for 1 h with 2% (w/v) non-fat dry milk powder in PBST buffer (PBS, 0·1% (v/v) Tween 20, pH 7·4), membranes were incubated for 1 h at 37 °C in MAb C1E3 or polyclonal IMS diluted 1[ratio ]500 in PBST containing 5% (w/v) non-fat dry milk. After copious washes in PBST buffer, proteins were revealed using horseradish peroxidase (HRP)-conjugated anti-mouse IgG (diluted to 1[ratio ]1000) followed by 3,3-diaminobenzidine tetrahydrochloride substrate (DABS) staining (Sigma–Aldrich).
The in vitro protein attachment assay was based on a procedure by Fourmaux et al. (1996). Cells were grown on 6-well dishes (9·5 cm2 wells) until confluent. Medium was discarded and cells were blocked by 3×10 min washes with 10 ml of medium containing 2% (w/v) bovine serum albumin (BSA). After blocking, 0·5 mg of soluble tachyzoite lysate prepared in 5 ml of medium was incubated on the confluent monolayer for 30 min at 37 °C. Non-adherent lysate was discarded and following 6 washes in 10 ml of medium, cells were solubilized by the addition of 1 ml of 1× SDS–PAGE sample loading buffer (75 mM Tris–HCl, pH 6·8, 5% (w/v) SDS, 0·0025% (w/v) bromophenol blue) then detached using a cell scraper. Samples were immediately subjected to analysis by SDS–PAGE and immuno-blotting.
Anion-exchange chromatography separates molecules according to charge differences. Using the ExPASy Molecular Biology Server (http://www.ncbi.nlm.nih.gov), the theoretical pI of mature SAG1 was calculated to be 7·5. This moderately high pI was exploited to purify SAG1 by anion- exchange HPLC. Tachyzoite lysates were fractionated by anion-exchange HPLC in column buffer at pH 9·0. Silver staining and Western analysis (with MAb C1E3) did not detect SAG1 in the bound tachyzoite fraction (Fig. 1). At this pH SAG1 was the only parasite protein that eluted from the ion-exchange resin. Thus anion-exchange HPLC proved to be very effective in separating virtually all SAG1 from tachyzoite lysates. Interestingly, purification at pH 8·5 and 8·0 fractionated SAG1 from most, but not all, parasite proteins. From this, the actual pI of solubilized SAG1 was deduced as being above 9·0.

Fig. 1. Anion-exchange HPLC purification of SAG1. Tachyzoite lysates were fractionated by anion exchange at pH 9·0, 8·5 and 8·0. Flow-through (F and Fs) and bound (B and Bs) fractions were separated on a 12% (w/v) polyacrylamide gel. Lanes T and Ts are non-fractionated tachyzoite lysates. Lanes Ts, Fs and Bs were silver-stained, lanes T, F and B were analysed by immuno-blotting using anti-SAG1 MAb C1E3 as primary antibody. Cross-reacting bands were detected using anti-mouse IgG HRP conjugate followed by DABS staining. Values on the right side indicate molecular sizes of marker proteins in kDa.
The first step was to investigate the binding of proteins from tachyzoite lysates to the host cell surface. Immune mouse sera (IMS) identified major bands of approximately 28 kDa and 75 kDa in MDBK cell extracts, and 28 kDa and 48 kDa in HS27 cell extracts, following incubation of monolayers with tachyzoite lysate (Fig. 2A). Comparing these fractions to the protein profile of tachyzoite total cell lysate (Fig. 2B) revealed that many abundant tachyzoite proteins did not adhere to the host cell surfaces, indicating that the binding of the 28 kDa, 48 kDa and 75 kDa proteins was specific. Probing with MAb C1E3 confirmed the 28 kDa molecule as SAG1. No bands were present in control, unincubated cell extracts.

Fig. 2. Binding of tachyzoite molecules to host cell surfaces. Immuno-blot analysis of (A) tachyzoite proteins binding to MDBK and HS27 cells in the in vitro protein attachment assay, and (B) crude tachyzoite cell lysate. Host cells were incubated with (lanes 2 and 4) or without (lanes 1 and 3) soluble tachyzoite lysate (0·5 mg), then washed and host cell proteins separated by SDS–PAGE on a 12% (w/v) polyacrylamide gel. For immuno-blotting, proteins were analysed with IMS (A, lanes 1 and 2; and B) or MAb C1E3 (A, lanes 3 and 4) used as primary antibody. Cross-reacting bands were detected using anti-mouse IgG HRP conjugate followed by DABS staining. Values on the right side indicate molecular sizes of marker proteins in kDa.
To minimize the likelihood that tachyzoite proteins were present in the host cell fractions due to internalization or receptor recycling, the in vitro protein attachment assay was repeated at 4 °C to retard host cell endocytotic machinery. Comparing the results of attachment at 4 °C with attachment at 37 °C revealed no differences (data not shown), suggesting that the tachyzoite proteins in host cell fractions arose as a consequence of binding to the host cell surfaces.
We wanted to investigate the putative binding of SAG1 to host cell-surface glucosamine. Competition experiments were performed using SAG1 that had been pre-treated with serial dilutions of the neoglycoprotein BSA-glucosamide. Soluble BSA-glucosamide blocked SAG1 attachment to MDBK cells in a dose-dependent manner; virtually all SAG1 binding was prevented with 100 μM BSA-glucosamide (Fig. 3). The attachment blockade was shown to be specific for glucosamide since control incubations with BSA had no effect on SAG1 binding (Fig. 3, lane 8). Pre-incubation of host cells with BSA-glucosamide or BSA had no effect on their integrity or morphology (data not shown). Taken together these results illustrate that BSA-glucosamide competes with host cell surfaces for SAG1 binding.

Fig. 3. Inhibition of SAG1 attachment to MDBK cells using BSA-glucosamide. SAG1 was pre-incubated with the NG BSA-glucosamide prior to the in vitro protein attachment assay. Incubated cells were fractionated by SDS–PAGE on a 12% (w/v) polyacrylamide gel and proteins detected by silver staining. Lane 1 contained non-incubated MDBK cells, lane 2 is SAG1 and lane 3 contained MDBK cells incubated with 0·1 mg of SAG1. Lanes 4–7 contained MDBK cells incubated in SAG1 that had been pre-treated with serial dilutions of BSA-glucosamide (0·1 μM, 1 μM, 10 μM and 100 μM, respectively). Lane 8 is SAG1 pre-treated with BSA alone. Values on the right side indicate molecular sizes of marker proteins in kDa.
SAG1 was pre-incubated with MAb C1E3 or polyclonal IMS to determine whether these agents perturb the binding of SAG1 to host cell surface. Preliminary incubation of SAG1 with a 4-fold molar excess of MAb C1E3 did not inhibit attachment of SAG1 to MDBK cell surfaces, and so it appears that the MAb C1E3 epitope does not overlap with the SAG1 domain for host cell binding (Fig. 4). SAG1 could also bind host cell surfaces following pre-incubation with a 4-fold molar excess of IMS. These results demonstrate that immunodominant B cell epitopes on SAG1 do not directly bind to the surfaces of host cells. Interestingly, Western analysis with MAb C1E3 detected minor bands of approximately 55 kDa, 120 kDa and 180 kDa in MDBK cell extracts that had been incubated with the SAG1-antibody mixtures (Fig. 4, lanes 2). These molecules were resistant to reduction in 25 mM DTT, and were therefore not oligomers of IgG heavy chain (data not shown).

Fig. 4. The role of SAG1 epitopes in SAG1 binding. The binding of SAG1 epitopes to host cells was assessed by pre-incubating SAG1 with either MAb C1E3 or IMS. Incubated cells were fractionated by SDS–PAGE on a 12% (w/v) polyacrylamide gel and proteins were analysed by Western blotting using anti-SAG1 MAb C1E3 as primary antibody. Cross-reacting bands were detected using anti-mouse IgG HRP conjugate followed by DABS staining. Lane 1 contained MDBK cells incubated with 0·5 mg purified antibody. Lane 2 contained MDBK cells incubated with 0·1 mg SAG1 that had been pre-treated with 0·5 mg purified antibody. Values on the right side indicate molecular sizes of marker proteins in kDa. Arrows indicate the presence of higher molecular weight complexes of SAG1 detected by MAb C1E3.
We have shown that SAG1 specifically binds the surface of bovine kidney cells and human foreskin fibroblasts, thus adding to the collection of host cells to which SAG1 attaches, i.e. Vero cells (Fourmaux et al. 1996), monocytes (Channon et al. 1999) and epithelial cells (Velge-Roussel et al. 2001).
The finding that attachment of SAG1 to bovine kidney cells could be blocked by pre-incubation of SAG1 in BSA-glucosamide is in agreement with a previous study which found that the ability of BSA-glucosamide to block invasion was markedly reduced when SAG1-deficient mutants were used in place of wild-type parasites (Mineo et al. 1993). From this result it would be tempting to suggest that SAG1 binding to host cells is mediated through interaction with host cell-surface glucosamine. Even so, direct binding of SAG1 to glucosamide has not been confirmed, and further work is required to assess the specificity of interaction with other sugar-conjugates.
Interestingly, it has been established that attachment of T. gondii to host cells is mediated, in part, by the binding of parasite lectins to complex sugars such as glycosaminoglycans, e.g. heparin (Ortega-Barria & Boothroyd, 1999; Carruthers et al. 2000; Jacquet et al. 2001). Recently, recombinant SAG3 (rSAG3) was shown to recognize sulphated proteoglycans on the surface of CHO-K1 (Jacquet et al. 2001). Modelling the SAG3 structure reveals that the outside surface of the SAG3 groove is negatively charged, which may apparently function to direct specific binding of negatively charged heparin, a sulphated glycosaminoglycan, to the positively charged groove (He et al. 2002). It is tempting to speculate that the absence of this feature in rSAG1 allows the protein to ubiquitously attach to sulphated and non-sulphated cell surface gylcans. Even so, whilst Jacquet et al. (2001) demonstrated that rSAG3 binds with high affinity to heparin, these authors found no evidence for the binding of rSAG1 to heparin. However, host cell binding regions may not be correctly folded on rSAG1, and so the binding of native SAG1 to sulphated proteoglycans cannot be ruled out.
It is unlikely that all host cells will exhibit the same surface receptors or that a single parasite surface protein could mediate invasion of every cell type. Thus, it is likely that there is redundancy in these ligands. For example, in this paper we have shown that SAG1 can bind human foreskin fibroblasts, although Dzierszinski et al. (2000) have shown that SAG3 also binds this cell type. Disruption of the SAG3 gene reduces, but does not abolish invasion, and thus SAG1 could be responsible for mediating parasite attachment to this cell type. Even so, the presence of the diverse family of SAG1-related sequence (SRS) proteins may circumvent the need for both SAG1 and SAG3, making the parasite even more versatile. Interestingly, efforts to create a SAG1/SAG3 knockout have so far been unsuccessful.
One intriguing finding was the discovery that SAG1 forms high molecular weight complexes by self-association. These data corroborate the finding that rSAG1 forms dimers in solution (He et al. 2002), though the observations presented here also imply that SAG1 can also form heteromers and hexamers. The behaviour of SAG1 could be characterized in two ways. First, higher complexes were not present subsequent to purification, only appearing once SAG1 had been stored at −20 °C. Second, the absence of a band of approximately 80–90 kDa indicates that SAG1 does not form trimers, thus each complex is a multimeric association of SAG1 dimers.
In vitro invasion assays using anti-SAG1 MAb C1E3 have previously indicated that SAG1 has a role in host cell attachment (Grimwood & Smith, 1992, 1996). In these experiments, MAb C1E3 was used to perturb parasite invasion, and since only a subset of anti-SAG1 MAbs conveyed this effect it has been postulated that the blocking ability of MAb C1E3 was due to the masking of a specific epitope of SAG1. With the sole purpose of understanding the mechanism of anti-SAG1 MAb neutralization, the stoichiometry of these antibody--antigen interactions was addressed. Unexpectedly, the results presented here indicate that MAb C1E3 does not in fact block SAG1 binding to host cell surfaces. To comprehend this peculiar finding, the interaction between MAb C1E3 and SAG1 requires further investigation. Since MAb C1E3 binding appears to be implicitly linked to the process of host cell attachment, the identification of the epitope for this MAb on SAG1 could provide a valuable insight into the role of SAG1 epitopes in host cell attachment.
We thank Debra Evans and Sinead Moore for their expert technical assistance.

Fig. 1. Anion-exchange HPLC purification of SAG1. Tachyzoite lysates were fractionated by anion exchange at pH 9·0, 8·5 and 8·0. Flow-through (F and Fs) and bound (B and Bs) fractions were separated on a 12% (w/v) polyacrylamide gel. Lanes T and Ts are non-fractionated tachyzoite lysates. Lanes Ts, Fs and Bs were silver-stained, lanes T, F and B were analysed by immuno-blotting using anti-SAG1 MAb C1E3 as primary antibody. Cross-reacting bands were detected using anti-mouse IgG HRP conjugate followed by DABS staining. Values on the right side indicate molecular sizes of marker proteins in kDa.

Fig. 2. Binding of tachyzoite molecules to host cell surfaces. Immuno-blot analysis of (A) tachyzoite proteins binding to MDBK and HS27 cells in the in vitro protein attachment assay, and (B) crude tachyzoite cell lysate. Host cells were incubated with (lanes 2 and 4) or without (lanes 1 and 3) soluble tachyzoite lysate (0·5 mg), then washed and host cell proteins separated by SDS–PAGE on a 12% (w/v) polyacrylamide gel. For immuno-blotting, proteins were analysed with IMS (A, lanes 1 and 2; and B) or MAb C1E3 (A, lanes 3 and 4) used as primary antibody. Cross-reacting bands were detected using anti-mouse IgG HRP conjugate followed by DABS staining. Values on the right side indicate molecular sizes of marker proteins in kDa.

Fig. 3. Inhibition of SAG1 attachment to MDBK cells using BSA-glucosamide. SAG1 was pre-incubated with the NG BSA-glucosamide prior to the in vitro protein attachment assay. Incubated cells were fractionated by SDS–PAGE on a 12% (w/v) polyacrylamide gel and proteins detected by silver staining. Lane 1 contained non-incubated MDBK cells, lane 2 is SAG1 and lane 3 contained MDBK cells incubated with 0·1 mg of SAG1. Lanes 4–7 contained MDBK cells incubated in SAG1 that had been pre-treated with serial dilutions of BSA-glucosamide (0·1 μM, 1 μM, 10 μM and 100 μM, respectively). Lane 8 is SAG1 pre-treated with BSA alone. Values on the right side indicate molecular sizes of marker proteins in kDa.
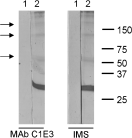
Fig. 4. The role of SAG1 epitopes in SAG1 binding. The binding of SAG1 epitopes to host cells was assessed by pre-incubating SAG1 with either MAb C1E3 or IMS. Incubated cells were fractionated by SDS–PAGE on a 12% (w/v) polyacrylamide gel and proteins were analysed by Western blotting using anti-SAG1 MAb C1E3 as primary antibody. Cross-reacting bands were detected using anti-mouse IgG HRP conjugate followed by DABS staining. Lane 1 contained MDBK cells incubated with 0·5 mg purified antibody. Lane 2 contained MDBK cells incubated with 0·1 mg SAG1 that had been pre-treated with 0·5 mg purified antibody. Values on the right side indicate molecular sizes of marker proteins in kDa. Arrows indicate the presence of higher molecular weight complexes of SAG1 detected by MAb C1E3.