INTRODUCTION
The digenetic trematode genus Schistosoma Weinland, 1858, contains several species that are of medical or veterinary importance. These species cause schistosomiasis, a disease most common in Africa, Asia, and South America, afflicting at least 165 million domestic ruminants (De Bont and Vercruysse, Reference De Bont and Vercruysse1997), and as many as 207 million humans (Steinmann et al. Reference Steinmann, Keiser, Bos, Tanner and Utzinger2006). Worms within this genus form a monophyletic group with members of the genus Orientobilharzia (Farley, Reference Farley1971) based on molecular data (Snyder and Loker, Reference Snyder and Loker2000; Li et al. Reference Li, Yu, Zhu, Wang, Zhai and Zhao2008; Wang et al. Reference Wang, Li, Ni, Zhai, Chen, Chen and Zhu2009). All members of Schistosoma and Orientobilharzia inhabit the vascular system of mammalian definitive hosts.
Species of Schistosoma have historically been placed into 4 groups based on their egg morphology, intermediate and definitive hosts, and biogeography (Rollinson and Southgate, Reference Rollinson, Southgate, Rollinson and Simpson1987). Although molecular evidence has required modification of the 4 species group concept, (Snyder and Loker, Reference Snyder and Loker2000; Lockyer et al. Reference Lockyer, Olson, Ostergaard, Rollinson, Johnston, Attwood, Southgate, Horak, Snyder, Le, Agatsuma, McManus, Carmichael, Naem and Littlewood2003; Morgan et al. Reference Morgan, DeJong, Kazibwe, Mkoji and Loker2003; Webster et al. Reference Webster, Southgate and Littlewood2006), the group traditionally referred to as the Schistosoma haematobium group has been supported by molecular data as monophyletic. In these species, the ovary is in the posterior half of the body, males contain 4–7 testes, the uterus contains numerous eggs, and all but S. margrebowiei produce eggs with a terminal spine (Rollinson and Southgate, Reference Rollinson, Southgate, Rollinson and Simpson1987; Pagès et al. Reference Pagès, Jourdane, Southgate, Tchuem Tchuente, Combes and Jourdane2003). The S. haematobium group contains 8 species (Table 1), 4 of which can infect humans, and all of which use snails in the genus Bulinus as intermediate hosts (Brown, Reference Brown1994).
Table 1. Details of the Schistosoma haematobium group
(DRC, Democratic Republic of the Congo.)

* Experimental laboratory exposure.
§ Locality needs to be verified.
† Length/width ratio.
Due to the economic and health impact of schistosomiasis, representatives of Schistosoma have been intensely studied for nearly a century. Despite this research focus, our understanding of their diversity and taxonomy remains incomplete as evidenced by recent reports of new species (Attwood et al. Reference Attwood, Panasoponkul, Upatham, Meng and Southgate2002) or lineages (Morgan et al. Reference Morgan, DeJong, Kazibwe, Mkoji and Loker2003). During a parasitological survey of rodents in and around Kisumu, Kenya, located in the Lake Victoria Basin, several schistosomes were recovered, including worms, which produced small uniquely shaped eggs. Adult worm anatomy and egg morphology most closely resemble members of the S. haematobium group. In the present study, we describe these distinctive worms as a new schistosome species based on morphological and molecular characters.
MATERIALS AND METHODS
Parasites
Schistosomes were collected from rodent definitive hosts as follows. Sherman live traps were placed in and around Nyabera swamp in Kisumu, Kenya (0·10914°S, 34·775°E) from October 2006 to December 2008 (Fig. 1). Samples were taken monthly with the exception of a period between January and August 2009, a time of civil unrest in Kenya. This site is located along the Kisumu-Kericho highway on the eastern outskirts of the city, is bordered on all but one side by homes and businesses, and continues approximately 6·5 kilometers to the south to Winam Gulf, part of Lake Victoria. The swamp has been used previously for rice cultivation, but since 2004 has lain fallow and has mainly been used as a source for several macrophyte species used for roof-thatching materials. Human defecation is common in and around the waters within the swamp, which is also used as a communal bathing area. Schistosoma mansoni Sambon, 1907 has been reported from this site (Steinauer et al. Reference Steinauer, Mwangi, Maina, Kinuthia, Mutuku, Agola, Mungai, Mkoji and Loker2008a).

Fig. 1. Collection locality of Schistosoma kisumuensis n. sp. and the distribution of S. intercalatum. Inset map shows the area of detail. Circles represent reports for S. intercalatum: grey, confirmed infection foci; open, unconfirmed infection foci. Star represents collection locality of S. kisumuensis. Shaded area is the Congo River drainage Basin.
Sherman traps were baited with an even mixture of peanut butter and cupcakes (locally known as Queen Cakes). Traps were checked at dusk and dawn, and animals were returned live to the laboratory in Kisian, Kenya, 15 kilometers from Nyabera swamp. For all captured individuals, sex, weight, and several size measurements were taken. Animals were euthanized, injected with a mixture of heparin and sodium pentabarbitol, and worms were recovered by perfusion (Lewis, Reference Lewis, Coligan, Kruisbeek, Margulies, Shevach and Strober1998). A liver smear was made from each animal to confirm infection status and to view eggs passed from the female's body, which will be referred to as ‘expelled eggs’. Worms were placed in 100% ethanol, and stored at 4°C for 2–10 weeks. The identity and prevalence of all schistosomes recovered from these hosts will be presented in a separate survey paper. The following description focuses only on the rodents infected with S. kisumuensis n. sp.
Snails were collected at Nyabera swamp monthly throughout the study period. Snails were isolated in individual wells of tissue-culture plates in aged tap water for 6–12 h and examined for shedding of cercariae in a natural light cycle. Infected snails were given an individual identification number and their cercariae were (1) used to infect mice (Swiss Albino, male and female, 6–7 weeks old) or hamsters (males, 10 weeks old), or (2) preserved in 100% ethanol for subsequent genetic analysis.
Livers containing eggs were minced by passing through a 212 μm mesh metal sieve, and placed in water for miracidial hatching for 1–12 h. Snails, collected at Nyabera swamp, Bulinus globosus Morelet 1868, Bulinus forskalii (Ehrenburg, 1831), and previously screened numerous times for cercarial shedding and found to be negative, were exposed to water from a liver mixture and/or to water containing liver fragments. Exposed snails were maintained in the laboratory for up to 10 weeks, and periodically checked for cercarial shedding.
Morphological examinations
Eggs were measured fresh, either as intrauterine eggs or from liver smears using 400x magnification on an Olympus BX2 microscope (Olympus Corp., Center Valley, PA, USA) and an Nikon Coolpix 990 digital camera. Adult worms were photographed, alive or after placement in 100% ethanol. All body measurements were based on worms placed in ethanol. Therefore, the presented size data must be interpreted with caution, since the preservation in alcohol may shrink worms. Worm body length was measured, before the posterior 10–15% of each worm was removed, retained in 100% ethanol, and saved for DNA analysis. Worms were then stained with Semichon's acetocarmine (Pritchard and Kruse, Reference Pritchard and Kruse1982), counterstained with fast green and mounted in Canada balsam. Morphometric data were recorded (Table 2). Drawings were done by image overlay in Microsoft PowerPoint (Microsoft Corp., Seattle, WA, USA). All measurements are in micrometers (μm) unless otherwise stated.
Table 2. Measurements of adult Schistosoma intercalatum and Schistosoma kisumuensis n. sp.
(S. kisumuensis measurements are based on alcohol preserved specimens; S. intercalatum in both studies were recovered from experimentally infected rodents.)

Molecular analyses
Genomic DNA from whole adults, parts of adults, or single cercariae, was extracted using a modified HotShot method (Truett et al. Reference Truett, Heeger, Mynatt, Truett, Walker and Warman2000; Steinauer et al. Reference Steinauer, Agola, Mwangi, Mkoji and Loker2008b). A single Schistosoma bovis (Sonsino, 1876) worm recovered from the rodent Mastomys natalensis (Smith, 1834) was also included in the subsequent analysis. DNA was amplified using protocols and primers to mirror the work by Kane et al. (Reference Kane, Southgate, Rollinson, Littlewood, Lockyer, Pages, Tchuem Tchuente and Jourdane2003). Partial fragments of cox1, nad6, 12S and 28S were amplified using GO Taq (Promega, Madison, WI) and a standard PCR protocol using the following primers: Cox1 Schist 5′ and Cox1 Schist 3′ for cox1 (Lockyer et al. Reference Lockyer, Olson, Ostergaard, Rollinson, Johnston, Attwood, Southgate, Horak, Snyder, Le, Agatsuma, McManus, Carmichael, Naem and Littlewood2003), RKND6F and RKND6R3 for nad6, RK12SF and RK12SR2 for 12S (Kane et al. Reference Kane, Southgate, Rollinson, Littlewood, Lockyer, Pages, Tchuem Tchuente and Jourdane2003), and LSU5' and LSU3' for 28S (Littlewood and Johnston, Reference Littlewood and Johnston1995). Extracted DNA was stored at 4°C until PCR amplification. PCR reactions were analysed by agarose gel electrophoresis, with the use of 1·0% agarose gels, stained with 0·5% GelRed™ Nucleic Acid Gel Stain (Biotium, Hayward, California), and visualized on a UV transilluminator. Amplicons were purified by use of a Montage SEQ96 Cleanup Kit (Millipore, Billerica, Massachusetts). Purified DNA products were then sequenced with the use of the BigDye version 3.1 kit (Applied Biosystems, Foster City, California) in an ABI 3130x sequence analyser (Applied Biosystems). Both strands of the amplified DNA fragments were sequenced, edited in Sequencer version 4.7 (Gene Codes, Ann Arbor, Michigan), and manually corrected for ambiguous base calls. Sequences from other species of Schistosoma and outgroups were retrieved from GenBank (Table 3). Protein coding regions of cox1 and nad6 were aligned by eye. Alignments of 12S and 28S rRNA were done with ClustalX (Larkin et al. Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007), and refined by eye to minimize gap openings and to remove unalignable regions.
Table 3. List of taxa and sequences used in this study
(Taxa in bold type are previously unreported sequences.)

Two phylogenetics analyses were performed. First, we used combined cox1 and 28S sequence data to place the proposed new species within the larger context of the family Schistosomatidae (Snyder, Reference Snyder2004; Webster et al. Reference Webster, Southgate and Littlewood2006). The second analysis was performed with combined cox1-12S-nad6-28S regions to reconstruct relationships within the S. haematobium group, including our proposed new species (Table 3). Phylogenetic analyses using maximum parsimony (MP), maximum likelihood (ML), and minimum evolution (ME) were carried out using PAUP* ver 4.0b10 (Swofford, Reference Swofford2002) and Bayesian inference (BI) using MrBayes (Huelsenbeck et al. Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001; Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003). jModeltest (Posada, Reference Posada2008) was used to determine the best nucleotide substitution model for ML and ME analyses. In cases where the Bayesian Information Criteria (B.I.C) and Akaike Information Criteria (A.I.C) selected different models, both were used in analyses and in all cases, the tree topologies were the same. The model TIM+I+G, was selected for the cox1-28S data matrix, and GTR+I+G was selected for the cox1-12S-nad6-28S data matrix, for the ME and ML analyses. For BI, a mixed model approach was implemented to account for the potential differences in evolutionary model parameters between data partitions (both genes and codon positions). Parsimony trees were reconstructed using heuristic searches, random taxon-input order and tree-bisection and reconnection (TBR) branch swapping. Optimal ME and ML trees were determined from heuristic searches (500 replicates for MP and ME; 5 replicates for ML), random taxon-input order, and TBR. Nodal support was estimated by bootstrap (500 replicates) and was determined for the MP and ME trees (100 replicates for the ML bootstrap of the cox1-12S-nad6-28S matrix), using heuristic searches, each with random taxon-input order. In all BI the data were partitioned by codon position for cox1 and nad6, and by genes for 12S and 28S. For the BI of the cox1-28S data set, there were four partitions defined by cox1pos1, cox1pos2, cox1pos3 (Nst=6 rates-invgamma ngammacat=4), and 28S (Nst=6 rates=gamma ngammacat=4). For the BI of the cox1-12S-nad6-28S dataset, there were 8 partitions defined by first cox1pos1, cox1pos2, cox1pos3, nad6pos1, nad6pos2, nad6pos3 (Nst=6 rates-invgamma ngammacat=4) and 12S and 28S (Nst=6 rates=gamma ngammacat=4). All parameters were unlinked between partitions. For all analyses, 4 chains were run simultaneously for 1×105 generations, trees were sampled every 100 cycles, the first 10 000 trees with pre-asymptotic likelihood scores were discarded as burnin, and the retained trees were used to generate 50% majority-rule consensus trees and posterior probabilities. Genetic distances of the cox1 were calculated with MEGA 2.0 (Kumar et al. Reference Kumar, Tamura, Jakobsen and Nei2001) using the p-distance options.
The methods used in this project were approved by the University of New Mexico's Institutional Animal Care and Use Committee (IACUC), and the Scientific Steering Committee of the Kenya Medical Research Institute, and the KEMRI/National Ethics Review Board of Kenya. All investigators/assistants in this study attained animal use certification regarding the ethical treatment of animals.
RESULTS
Parasites
Over 70 trap days and nights, 276 small mammals were caught and perfused, including 247 rodents of 16 species and 29 insectivores of 4 species. Hosts were deposited in the National Museum of Kenya, Nairobi (Table 4). Of these, 11 rodents of 3 species were found to contain 15 unidentified schistosomes (Table 4). All were collected from rodents of the family Muridae, most often in Pelomys isseli (de Beaux, 1924), Issel's groove-toothed swamp rat. One specimen each was also recovered from Mastomys natalensis, the Natal multimammate mouse, and Dasymys incomtus (Sundevall, 1847), the African marsh rat. Of these 15 worms, DNA was extracted from 2 whole worms and from posterior ends of 13 worms. Four of these worms were immature; 1 female worm was destroyed during the staining procedure. Measurements used for the following analyses were thus based on the remaining mature males (n=6) and females (n=2). Eggs were collected from the livers of 4 hosts, and measurements are based on 12 intrauterine eggs and 22 expelled eggs. Although all hosts with patent infections, defined by the presence of eggs in the liver, contained at least 1 worm, both male and female worms were recovered together from only 1 host, and were collected in copula. The other female from this host was unpaired. The reason for not finding both sexes in all patently infected rodents could be loss during perfusion due to the small worm size, or death of worms after oviposition. However, in several cases where only a single worm was recovered by perfusion, the rodent was examined manually and in no cases were additional worms found in the vascular system or body cavity.
Table 4. Hosts collected with Schistosoma kisumuensis n. sp.
(Host No. – refers to the identification number in the National Museum of Kenya.)
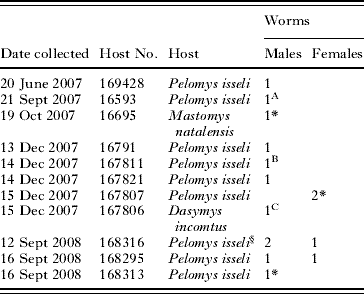
* Immature worm.
§ Male and 1 female worm collected in copula.
A–C, genotypes included in the phylogenetics analysis.
Snail collection at Nyabera swamp, based on shell morphology, consisted of the following gastropods: Biomphalaria sudanica (Martens, 1870), Bulinus globosus, Bulinus forskalii, Ceratophallus natalensis (Krauss, 1848), Lymnaea natalensis Krauss, 1848, and Melanoides tuberculata (Müller, 1774). Of 2066 B. sudanica snails collected at Nyabera swamp, 11 shed S. mansoni cercariae. Of 211 B. globosus, 3 shed mammalian cercariae, and of 194 B. forskalii, 1 shed mammalian cercariae. Genetic analysis of cercariae from these snails indicated that the infections were of S. bovis in B. forskalii and of Schistosoma haematobium (Bilharz, 1852) in B. globosus. The identity of these cercariae was confirmed by genetic analysis of adult worms retrieved from laboratory-infected mice and hamsters (data not shown). None of the other snail species shed mammalian schistosome cercariae (C. natalensis, n=190; L. natalensis, n=124; M. tuberculata, n=155). Snails shedding S. kisumuensis were not found and we hope that future work may identify the natural intermediate host.
Schistosoma kisumuensis eggs were noted in 8 livers. In each of these livers, only 1 egg type was observed. Five attempts were made to hatch miracidia from these livers. Unhatched eggs were observed within the homogenized liver tissue. Miracidia hatched only from a single sample (Fig. 2D). None of the snails exposed to miracidia or liver pieces shed cercariae within 10 weeks of exposure.
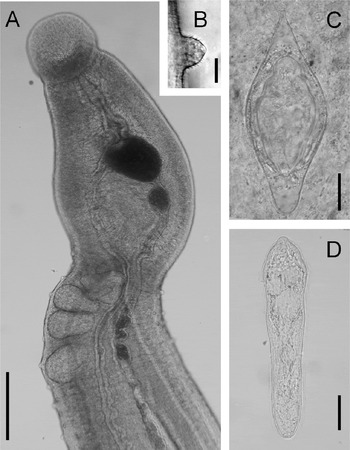
Fig. 2. Schistosoma kisumuensis n. sp. (A) Unfixed, live adult male, scale bar=100 μm (note that the ventral sucker is below the specimen in a separate plane of view). (B) Tubercle with spines, scale bar=5 μm. (C) Egg from liver, scale bar=25 μm. (D) Miracidium, scale bar=25 μm.
Morphology
Schistosoma kisumuensis n. sp.
Type-host: Pelomys isseli (de Beaux, 1924), Issel's groove-toothed swamp rat (Rodentia: Muridae).
Other hosts: Mastomys natalensis (Smith, 1834), the Natal multimammate mouse (Rodentia: Muridae), Dasymys incomtus (Sundevall, 1847), the African marsh rat (Rodentia: Muridae) deposited at the National Museum of Kenya (Table 1).
Intermediate host: unknown.
Type-locality: Nyabera swamp, Kisumu, Nyanza Province, Kenya, 0·10914°S, 34·775°E.
Type-specimens: Holotype (male) and allotype (female).
Site: Vascular system.
Type-material: The type series consists of 5 adult males, 1 adult female, 1 pair of adult worms in copula, 2 immature males and 1 immature female, all of which were preserved in ethanol. All types are deposited at the Parasitology Division, Museum of Southwestern Biology, University of New Mexico, New Mexico, USA, numbers MSBPARA164 – MSBPARA173. The holotype (male) is MSBPARA165 and the paratype (female) is MSBPARA172.
Etymology: The species is named for the collection locality, Kisumu, Kenya.
Description (Figs 2–5)
Male
Mature male is elongate, but relatively short, from 3·2–6·9 mm long by 0·18–0·32 mm wide (Table 2, Figs 2A and 3). Well-defined gynaecophoric canal extends from behind ventral sucker to tip of the posterior extremity. Posterior to ventral sucker, tegument is covered with tubercles (Fig. 2B), decreasing in size and number posteriorly. Tubercles covered with fine spines. Constriction present at the anterior end of body, just posterior to ventral sucker (Fig. 1A). Oral sucker, 87–163, is funnel shaped. Ventral sucker, 151–223, is large and does not appear to be supported by a fleshy peduncle. Oesophagus extends posteriorly to the ventral sucker where it bifurcates into the gut caecae. The 2 branches reunite near the posterior end of the body. The testes lie dorsally and their anterior extent is at the same position as the anterior end of the gynaecophoric canal. Testes from 4–6, not arranged in linear manner, and often overlap. Testes size tends to decrease anterior to posterior, oval and round to ovoid (nearly triangular) in shape. Seminal vesicle lies immediately anterior to testes, opens into anterior margin of gynaecophoric canal immediately posterior to ventral sucker. Immature males had small body size, lacked fully developed testes.
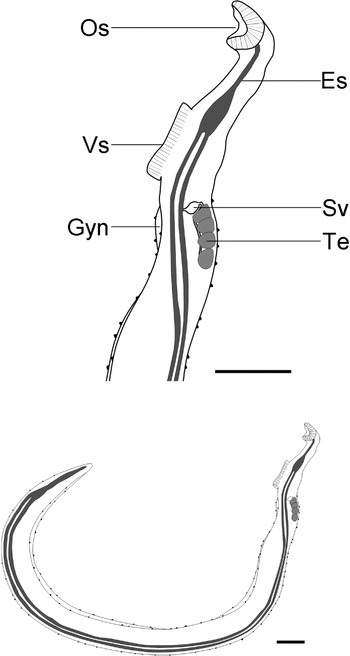
Fig. 3. Drawing summarizing the morphology of Schistosoma kisumuensis n. sp. adult male. Es, esophagus; Gyn, gynaecophoric canal; Os, oral sucker; Sv, seminal vesicle; Te, testes; Vs, ventral sucker. Scale bars=200 μm.
Female
Body filiform, longer than male, 5·2 and 7·6 mm long, 0·22 and 0·23 mm wide, posterior half of body expanded (Fig. 4). Posterior extremity tapered and rounded. Tegument smooth without tubercles or spines. Oral sucker similar to that of male, except smaller 36 and 58. Ventral sucker not as well developed and much smaller 27 and 33·5, than that of male, appearing rudimentary. Caecum branches in region of the ventral sucker reunites posterior to ovary, and terminates near posterior extremity of worm. Ovary, 257 and 412 long, 122 and 125 wide, situated at same region where the body expands, just anterior to the body's midpoint. Uterus straight tube, starts at ovary and terminates just posterior to ventral sucker where it opens through genital pore. Uterus has greater diameter posteriorly, and expands in anterior 25% only when containing egg. Vitellaria extensive, occupying roughly the posterior 50% of worm and extend further posteriorly than intestine.

Fig. 4. Drawing summarizing the morphology of Schistosoma kisumuensis n. sp. adult female. Gp, genital pore; Int, gut caecum; Iue, intrauterine egg; Os, oral sucker; Ov, ovary; Ovd, oviduct; Ut, uterus; Vg, vitelline gland; Vs, ventral sucker. Scale bars: bottom=250 μm; top=125 μm.
Eggs
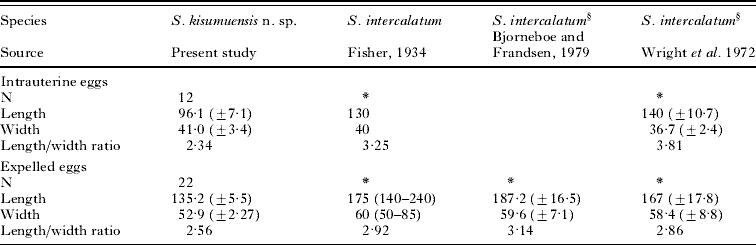
Females contain 5 and 10 intrauterine eggs, which are smaller and differently shaped than expelled eggs. Expelled eggs were obtained from the liver of the hosts (Figs 2C and 5; Table 5). Intrauterine eggs 41·0 (34·7–45·3) wide and 96·1 (81·3–106·7) long, ovoid shape, pointed end but lacking distinct spine. Expelled eggs, relatively small, compared with other S. haematobium group eggs, and diamond shaped, 52·9 (49·0–58·0) wide and 135·2 (123·0–142·7) long, similar in shape to eggs of S. intercalatum. As compared to intrauterine eggs, expelled eggs larger and longer and more diamond-shaped. Intrauterine eggs have a broader size range than expelled eggs.

Fig. 5. A comparison of egg morphology of (A) expelled and (B) intrauterine Schistosoma kisumuensis n. sp. eggs; (C) mature S. intercalatum eggs from the feces of rodents (redrawn from Wright et al. Reference Wright, Southgate and Knowles1972); and eggs from the original species description of S. intercalatum (D) expelled, and (E) intrauterine eggs (redrawn from Fisher, Reference Fisher1934). Scale bar=100 μm, but note that D and E were drawn without scale bars but eggs are sized to average measurements as given by Fisher (Reference Fisher1934).
Table 5. Measurements of eggs of Schistosoma intercalatum and Schistosoma kisumuensis n. sp.
(Data are mean followed by standard deviation or range; measurements are given in μm.)
* Unknown data.
§ Eggs were derived from experimentally infected rodents.
MOLECULAR ANALYSES
Fifteen worms of the proposed new species were sequenced for cox1 (1197 bp), 12S (779 bp), nad6 (610 bp), and 28S (1248 bp). The dataset used in the phylogenetic reconstructions was trimmed after alignment with the available sequence data in GenBank (Table 3) to include 1137 bp cox1, 729 bp 12S, 420 bp nad6, and 704 bp 28S. The combined 4 DNA regions of the 15 worms varied slightly between individuals (Genetic distances <0·01%); 3 of the more variable genotypes (Table 4) were included in the subsequent phylogenetic analyses, and are denoted as Schistosoma kisumuensis A, B, C (Figs 6 and 7).

Fig. 6. Maximum likelihood tree based on the combined partial cox1 and 28S sequence data. Schistosoma kisumuensis n. sp. isolates are indicated in bold. Numbers at branch nodes indicate MP and ME bootstrap values and Bayesian PP, respectively. * Indicates MP and ME bootstrap values of >65 and Bayesian PP of 100. – Indicates <55% bootstrap support.
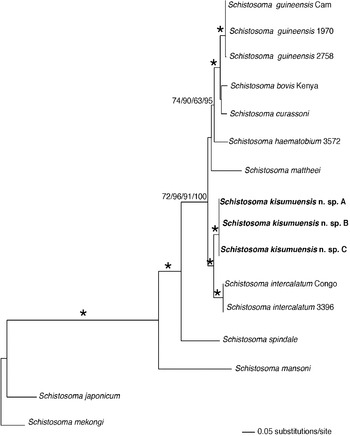
Fig. 7. Maximum likelihood tree based on the combined partial nad6, cox1, 12S, and 28S sequence data. Isolates of Schistosoma kisumuensis n. sp. are indicated in bold. Numbers at branch nodes indicate MP, ME, and ML bootstrap values and Bayesian PP, respectively. The ‘*’ indicates MP, ME, and ME bootstrap values of >90 and PP of 100.
Aligned cox1 and nad6 sequences appeared to be genuine mitochondrial sequence rather than nuclear copies, based on the lack of stop codons and the similarity of the translated protein sequences to those of close relatives. MP, ML, ME, and BI analyses each produced 1 tree per data matrix. Trees recovered from these analyses were largely congruent, and no conflicts received high support from bootstrapping or Bayesian posterior probabilities (Figs 6 and 7). Schistosoma kisumuensis was strongly supported as a clade. The cox1-28S tree supported the inclusion of S. kisumuensis within the family Schistosomatidae and within the clade containing species of Schistosoma and Orientobilhazia (Fig. 6). Schistosoma kisumuensis grouped within the clade defined by egg shape and by molecular data (Morgan et al. Reference Morgan, DeJong, Kazibwe, Mkoji and Loker2003; Kane et al. Reference Kane, Southgate, Rollinson, Littlewood, Lockyer, Pages, Tchuem Tchuente and Jourdane2003; Webster et al. Reference Webster, Southgate and Littlewood2006) as the S. haematobium group. For a more detailed analysis, a smaller data matrix was created that included all available gene regions for the S. haematobium group. The cox1-12S-nad6-28S tree (Fig. 7) shows strong node support for Schistosoma kisumuensis as sister to S. intercalatumFisher, Reference Fisher1934.
Genetic distances of cox1 among the 3 S. kisumuensis individuals were 0·1%, 0·2%, and 0·3%. The distance between S. kisumuensis and S. intercalatum is 6·3%, which falls within the pairwise distance values between other species of the S. haematobium group (Table 6).
Table 6. Mitochondrial (cox1) genetic distances of species within the Schistosoma haematobium group

DISCUSSION
Schistosoma kisumuensis n. sp. is member of the genus Schistosoma based on morphological and molecular characters. Genetic distance data indicate that S. kisumuensis is a distinct genetic lineage that merits recognition as a new species. Taken together, adult and egg morphology, and phylogenetic analyses firmly place this new species within the S. haematobium group, as the sister species to S. intercalatum.
Eight species comprise the S. haematobium group: S. bovis, S. curassoni Brumpt, 1931, S. haematobium, S. intercalatum, S. leiperi Le Roux, 1955, S. margrebowiei Le Roux, 1933, S. mattheei Veglia and Le Roux, 1929, and the most recently described S. guineensis Pagés, 2003. Previous studies have found that morphological features of adult flukes of the S. haematobium group vary and often overlap (Pitchford, Reference Pitchford1965; Pagès et al. Reference Pagès, Durand, Southgate, Tchuem Tchuente and Jourdane2001). Adult morphology of S. kisumuensis closely resembles that of members of the S. haematobium group, with the exception of the small adult size of S. kisumuensis, a trait likely affected by the preservation of worms we collected in alcohol. Available male and female worms are 2–3 times smaller than other species within the S. haematobium group (Loker, Reference Loker1983). It should be noted that adult worm length of schistosomes is variable and is not always a suitable diagnostic trait, since size variation can occur with host species (Wright et al. Reference Wright, Southgate and Knowles1972). Spined tubercles are shared by all members of the S. haematobium group, with the exception of S. bovis and S. mattheei, which possess tubercles without spines.
The eggs of S. kisumuensis are unique when considering their length to width ratio and size. Expelled eggs are about 75% the length of eggs of its sister species S. intercalatum. Similarly, intrauterine eggs are about 70% of the length of those of S. intercalatum. Expelled S. kisumuensis eggs are diamond-shaped, exhibiting blunt ‘shoulders’ on both ends and have a mean length to width ratio of 2·55, in contrast to mature S. intercalatum eggs, which are more elongate and oval, rarely contain shoulders, and have a mean length to width ratio of 2·9. The egg morphology of S. kisumuensis is more similar to some of the eggs of S. intercalatum drawn by Fisher (Reference Fisher1934) who based his drawings on eggs collected from humans (Fig. 5D), and less similar to those of S. intercalatum drawn by Wright et al. (Reference Wright, Southgate and Knowles1972), who based his drawings on eggs collected from experimentally infected mice (Fig. 5C). Additionally, unlike S. intercalatum, eggs of S. kisumuensis do not contain a true spine; rather the anterior end tapers to a spine-like point.
The intermediate host of S. kisumuensis is unknown. It has been well established that other members of the S. haematobium group primarily use snails of the genus Bulinus; Schistosoma intercalatum uses bulinid snails of the B. africanus group (Brown, Reference Brown1994). Thus, it is likely that S. kisumuensis also uses bulinid snails as an intermediate hosts, although the discovery of African members of Schistosoma in Ceratophallus snails (Morgan et al. Reference Morgan, DeJong, Kazibwe, Mkoji and Loker2003) suggests caution is warranted. Biomphalaria sudanica, B. globosus, B. forskalii, C. natalensis, L. natalensis, and M. tuberculata were all recovered from Nyabera, in proximity to where we found rodents infected with S. kisumuensis.
Schistosoma kisumuensis has only been found in murid rodents. Most of the species within the S. haematobium group are able to infect a relatively broad spectrum of definitive hosts, including rodents (Pitchford, Reference Pitchford1977). For example, the sister species of S. kisumuensis, S. intercalatum, naturally infects humans (Fisher, Reference Fisher1934), experimentally infects sheep and goats (Schwetz, Reference Schwetz1956b), and has been maintained in the laboratory in both hamsters (e.g. Wright et al. Reference Wright, Southgate and Ross1979) and mice (e.g. Frandsen, Reference Frandsen1977). In one additional report, S. intercalatum-like eggs were found in naturally infected rodents, Hybomys univittatus (Peters, 1876), near Kisangani, Democratic Republic of the Congo (DRC), near the parasite's type locality (Schwetz, Reference Schwetz1956a). Considering the fact that most species within the S. haematobium group can infect a relatively wide variety of hosts, it is conceivable that S. kisumuensis may also use hosts in addition to rodents, such as perissodactyls, artiodactyls, and primates.
Currently, S. kisumuensis has only been found in a single location in Kisumu, Kenya, located within the Lake Victoria Basin. Its sister species, S. intercalatum, appears to be confined to the adjacent Congo River Basin. Schistosoma intercalatum was first mentioned from a number of human intestinal schistosomiasis cases near Kisangani in what is now the Democratic Republic of the Congo (Chesterman, Reference Chesterman1923), but was not described as a distinct species until nearly 10 years later (Fisher, Reference Fisher1934). Fisher (Reference Fisher1934) deemed S. intercalatum as distinct from S. bovis and S. haematobium, based on egg shape and the observation that this schistosome produced terminal-spined eggs that were causing intestinal rather than urinary schistosomiasis in humans. Over the next decades, the range of S. intercalatum was extended to Lower Guinea forest regions, including Cameroon, Gabon, Equatorial Guinea, Nigeria and São Tomé (for review, see Tchuem Tchuente et al. Reference Tchuem Tchuente, Southgate, Jourdane, Webster and Vercruysse2003). However, all S. intercalatum-like worms collected outside of the DRC have been determined to be S. guineensis, a distinct and comparatively distantly related species (Webster et al. Reference Webster, Southgate and Littlewood2006). Since its original description from the infection focus within the upper reaches and tributaries of the Congo river, S. intercalatum has only been found in one other focus, Kinshasa city (De Clercq, Reference De Clercq1987; Tchuem Tchuente et al. Reference Tchuem Tchuente, Southgate, Vercruysse, Kaukas, Kane, Mulumba, Pages and Jourdane1997), located about 300 kilometers from the delta of the Congo River.
Due to the sporadic nature of human infections, it has been suggested that S. intercalatum is a zoonotic parasite, and that its natural host is a ‘forest-dwelling animal’ (Wright et al. Reference Wright, Southgate and Knowles1972). The distribution of many schistosomes is limited by that of their intermediate host species. However, S. intercalatum, remains highly restricted compared to the distribution of its intermediate hosts, members of the B. africanus group, found throughout large parts of Africa (Brown, Reference Brown1994). Therefore, the distribution of this parasite has been suggested to be limited by its normal definitive hosts, thought to include monkeys, such as mangabeys, and forest dwelling rodents (Wright et al. Reference Wright, Southgate and Knowles1972). Schistosoma intercalatum cercariae are shed during the day, concentrate at the water surface, congregate in large masses, and adhere easily to surfaces. Among mammalian schistosomes, this cercarial behaviour has been reported from Schistosomatium douthitti Cort, 1914), mainly a rodent parasite, S. japonicum Katsurada, 1904, which occurs in a large number of mammalian species, and S. margrebowiei, a parasite of antelopes. Thus, it is conceivable that S. intercalatum normally resides in a forest-dwelling mammal and only infects humans as a result of human incursions into pristine forests (Wright et al. Reference Wright, Southgate and Knowles1972).
Based on available data, S. intercalatum and its sister species S. kisumuensis are confined to adjacent hydrographic basins. One explanation for this distribution pattern is that these two species derived from a common ancestor, possibly with a rodent definitive host, and formed one lineage in the Congo Basin and another in the Lake Victoria Basin. This idea is supported by geological events that shaped east central Africa. From 8·0–2·3 MYA (million years ago) Lake Obweruka encompassed present day Lake Edward, Lake Albert, and the basin in between (Pickford et al. Reference Pickford, Senut and Hadoto1993). During this time, east-to-west rivers, such as the Kafu, Katonga, and Kagera rivers, are hypothesized to have drained much of the East Africa Dome into Lake Obweruka, the outlet of which flowed through the Beni Gap, into the Congo River and to the Atlantic (de Heinzelin, Reference de Heinzelin1962; Pickford et al. Reference Pickford, Senut and Hadoto1993). During the Pleistocene (1·8 MYA – 10 000 years ago) fossil and geological evidence suggests that the connection to the Congo Basin of Lake Obweruka was severed, largely separating the Congo Basin and what would become the Lake Victoria Basin (Van Damme and Pickford, Reference Van Damme and Pickford1999). Finally, about 12 000 years ago, fault warping along the Central Rift Valley caused a regional tilt to the north in the Albert-Edward area, causing the east-to-west rivers to reverse course, creating Lake Victoria, while the northwards tilting of the Lake Albert and Lake Victoria Basins resulted in flows into the Nile system (Pickford et al. Reference Pickford, Senut and Hadoto1993). Thus, it is conceivable that S. intercalatum and S. kisumuensis were separated as long as 1·8 MYA or as recently as 10 000 years ago.
It is interesting to note that S. kisumuensis falls in the middle of a clade of schistosomes infective to humans. Although, no reports exist from the Lake Victoria Basin of humans passing terminal-spined eggs in their stools, fitting the description of S. kisumuensis, an intriguing report of such an infection focus stems from the Nile drainage, one that is contiguous with the Lake Victoria Basin. In this report, 6 patients were passing terminal-spined eggs in their stools in Rhino Camp, located in northwestern Uganda, approximately 500 miles northwest of Kisumu, Kenya (Odongo-Aginya et al. Reference Odongo-Aginya, Mueller, Loroni-Lakwo, Ndugwa, Southgate, Schweigmann, Seitz and Doehring-Schwerdtfeger1994). It has been suggested that these eggs could represent non-viable eggs produced parthenogenetically by the heterospecific pairing of an S. mansoni male and a S. haematobium female, resulting in the production of terminal-spined eggs in the feces (Tchuem Tchuente et al. Reference Tchuem Tchuente, Southgate, Jourdane, Webster and Vercruysse2003). However, since S. haematobium has not been reported from this area of Uganda, this explanation may not hold. Although the eggs recovered in Rhino Camp resemble those of S. intercalatum, their size was relatively small, 156×59 μm, and the length to width ratio, 2·59, is much closer to that of S. kisumuensis (2·56) than to S. intercalatum (2·90). This raises the intriguing possibility that the infection focus described by Odongo-Aginya et al. (Reference Odongo-Aginya, Mueller, Loroni-Lakwo, Ndugwa, Southgate, Schweigmann, Seitz and Doehring-Schwerdtfeger1994) could have been caused by S. kisumuensis. Studies on eggs from this part of Uganda using molecular tools to identify and possibly reveal cryptic species are urgently needed.
Although there are currently no definitive reports of humans passing these relatively small terminal-spined schistosome eggs, we do not find it impossible to think that this species could infect humans, especially since Nyabera swamp, the type locality for S. kisumuensis, supports many human activities. In the future, testing the human population specifically for infection with this new species in both Kenya and elsewhere may reveal whether it contributes to the problem of schistosomiasis in Africa.
Primary funding was provided by NIH grant AI044913. We thank George Rosenberg and the Molecular Biology Facility at UNM, and NIH grant 1P20RR18754, IDeA Program of the National Center for Research Resources for providing access to microscope and sequencing facilities. We are indebted to Bernard Agwanda from the National Museum of Kenya, Nairobi, for field and rodent identification assistance. Finally, we would like to thank the Kenya Medical Research Institute for their support.















