INTRODUCTION
The Bartonella genus includes fastidious haemotropic Gram-negative bacteria mainly transmitted by arthropod-vectors (Chomel et al. Reference Chomel, Boulouis, Breitschwerdt, Kasten, Vayssier-Taussat, Birtles, Koehler and Dehio2009). Over the last 20 years, the number of Bartonella species or subspecies identified from a wide range of mammals has increased considerably (Chomel et al. Reference Chomel, Boulouis, Breitschwerdt, Kasten, Vayssier-Taussat, Birtles, Koehler and Dehio2009). Among the species or subspecies known or suspected to be pathogenic for humans, three have the domestic cat as their natural reservoir, namely Bartonella henselae, Bartonella clarridgeiae (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005) and Bartonella koehlerae (Mogollon-Pasapera et al. Reference Mogollon-Pasapera, Otvos, Giordano and Cassone2009). Bartonella henselae and B. clarridgeiae are associated with cat-scratch disease (CSD) and other syndromes in humans and are the most commonly identified Bartonella species in cats, worldwide (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005; Breitschwerdt et al. Reference Breitschwerdt, Maggi, Chomel and Lappin2010b ). Bartonella koehlerae was previously reported as a cause of human endocarditis (Avidor et al. Reference Avidor, Graidy, Efrat, Leibowitz, Shapira, Schattner, Zimhony and Giladi2004; Chomel et al. Reference Chomel, Boulouis, Breitschwerdt, Kasten, Vayssier-Taussat, Birtles, Koehler and Dehio2009).
Considering that bacteria from the Bartonella genus are fastidious to grow in vitro, serological [Indirect Fluorescent Antibody Test (IFAT) and enzyme-linked immunosorbent assay (ELISA)] and molecular [conventional and real-time quantitative polymerase chain reaction (cPCR and qPCR), respectively] techniques are widely used for the diagnosis of Bartonella infection (Mogollon-Pasapera et al. Reference Mogollon-Pasapera, Otvos, Giordano and Cassone2009). Serological tests have limited specificity due to cross-reactions and inconsistent results (Maggi et al. Reference Maggi, Mascarelli, Pultorak, Hegarty, Bradley, Mozayeni and Breitschwerdt2011). Molecular techniques are sensitive and allow species identification (Fenollar and Raoult, Reference Fenollar and Raoult2004). cPCR assays have limited sensitivity compared with qPCR (André et al. Reference André, Dumler, Herrera, Goncalves, de Sousa, Scorpio, de Santis, Domingos, de Macedo and Machado2016). A pre-enrichment liquid culture medium (‘Bartonella Alpha Proteobacteria Growth Medium’, BAPGM) prior to PCR was suggested to improve the sensitivity of molecular techniques, mainly for detecting Bartonella species in biological samples from non-reservoir hosts, including humans (Maggi et al. Reference Maggi, Duncan and Breitschwerdt2005, Reference Maggi, Mascarelli, Pultorak, Hegarty, Bradley, Mozayeni and Breitschwerdt2011; Breitschwerdt et al. Reference Breitschwerdt, Maggi, Mozayeni, Hegarty, Bradley and Mascarelli2010a ; Pérez et al. Reference Pérez, Maggi, Diniz and Breitschwerdt2011).
The prevalence rates of Bartonella spp. detected by PCR vary considerably among cat populations, with an increase from cold (0% in Norway) (Bergh et al. Reference Bergh, Bevanger, Hanssen and Loseth2002) to warm and humid climates (61% in the Philippines) (Chomel et al. Reference Chomel, Carlos, Kasten, Yamamoto, Chang, Carlos, Abenes and Pajares1999). Although Bartonella spp. infection in cats can vary between areas, it is often associated with flea infestation (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005). Bartonella spp. infection is associated with extended, often subclinical (Kordick et al. Reference Kordick, Brown, Shin and Breitschwerdt1999), asymptomatic, with long-lasting intraerythrocytic bacteremia in domestic cats (Chomel et al. Reference Chomel, Boulouis, Breitschwerdt, Kasten, Vayssier-Taussat, Birtles, Koehler and Dehio2009). Understanding the potential associations between Bartonella spp. infection and clinical disease in cats is complicated. There is little information about haematological abnormalities in naturally infected cats (Breitschwerdt, Reference Breitschwerdt2008).
In Chile, few reports have assessed the prevalence of Bartonella in cats. An overall B. henselae seropositivity of 85% was found in cats sampled in three cities (Coquimbo, Santiago and Valdivia) (Ferrés et al. Reference Ferrés, Abarca, Godoy, García, Palavecino, Méndez, Valdés, Ernst, Thibaut, Koberg, Chanqueo and Vial2005). Another study in Valdivia, Southern Chile, described a B. henselae serosurvey of 71% (Zaror et al. Reference Zaror, Ernst, Navarrete, Ballesteros, Boroschek, Ferres and Thibaut2002). Additionally, 41·7% (25/60) of blood samples from cats in Santiago, Central Chile, were culture positive for Bartonella, and confirmed as B. henselae by 16S RNA gene sequencing (Ferrés et al. Reference Ferrés, Abarca, Godoy, García, Palavecino, Méndez, Valdés, Ernst, Thibaut, Koberg, Chanqueo and Vial2005). Nevertheless, the molecular prevalence of Bartonella spp. in cats from Southern Chile and its strain diversity are not yet known. Bartonella DNA was detected in 10·8% (4/37) of Ctenocephalides felis fleas collected from cats sampled in Chilean animal pounds (Pérez-Martínez et al. Reference Pérez-Martínez, Venzal, González-Acuña, Portillo, Blanco and Oteo2009). After PCR amplification and sequencing of rpoB, gltA genes and the 16–23S rRNA intergenic transcribed spacer, the species involved were identified as B. clarridgeiae and B. henselae (Pérez-Martínez et al. Reference Pérez-Martínez, Venzal, González-Acuña, Portillo, Blanco and Oteo2009). Increased exposure to cats, particularly kittens and cat-related trauma were associated with a higher prevalence of Bartonella-associated disease (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005; Breitschwerdt et al. Reference Breitschwerdt, Maggi, Mozayeni, Hegarty, Bradley and Mascarelli2010a ). In Chile, the disease is not of mandatory reporting. Nevertheless, more than 200 human cases of bartonellosis were diagnosed between 1997 and 2000 (Ferrés et al. Reference Ferrés, Abarca, Godoy, García, Palavecino, Méndez, Valdés, Ernst, Thibaut, Koberg, Chanqueo and Vial2005). According to previous studies, cats play a major role as B. henselae reservoirs in Chile; consequently, humans who have contact with those animals are at risk (Ferrés et al. Reference Ferrés, Abarca, Godoy, García, Palavecino, Méndez, Valdés, Ernst, Thibaut, Koberg, Chanqueo and Vial2005). The present study aimed at determining the prevalence, haematological findings and genetic diversity of Bartonella spp. in domestic cats from Valdivia, Southern Chile.
MATERIALS AND METHOD
Animals and area of study
The study was approved by the Universidad Austral de Chile (UACh) bioethics committee under the protocol number UACh 142/2013.
To accurately determine Bartonella spp. prevalence in Valdivia (39 48 30 S, 73 14 30 W), Southern Chile, the required sample size was estimated considering a prevalence of 50%, which fits the criteria when prevalence is unknown (Thrusfield, Reference Thrusfield2007), and corrected according to the cat population of Valdivia (Zuñiga, Reference Zuñiga2007), providing a sample of 370 cats. A 5% precision and 95% confidence interval were used (Thrusfield, Reference Thrusfield2007). Over a 15-month period (August 2013–November 2014), 370 client-owned cats had their blood sampled by a veterinary team. The cats came from all Valdivia city locations in order to acquire balanced and representative sampling. Samples were taken from: (1) cats during home visits to pet-owning households; and (2) cats admitted to the Veterinary Hospital of UACh, Valdivia (Fig. 1). Cats were sampled regardless of age, gender, health and reproductive status. Each owner signed a consent form before samples were taken.

Fig. 1. Map of Chile, showing Valdivia City located in the Los Ríos Region, where samples from cats were taken (MapInfo Professional 7·5 SCP).
Haematological analysis
Blood samples were collected aseptically by cephalic or jugular venipuncture, divided into two EDTA collecting plastic tubes (Vacutainer®), and sent to the UACh Veterinary Clinical Pathology Laboratory. One EDTA anticoagulated blood sample was stored at −20 °C until DNA extraction/purification. The other EDTA anticoagulated blood was used to perform a complete blood count (CBC). The following parameters were analysed: red blood cell, white blood cell (WBC) and platelet counts; haemoglobin concentration; packed red cell volume; mean corpuscular volume (MCV); and mean corpuscular haemoglobin concentration. An automated haematology analyser, KX-21N (Sysmex©, Japan), was used. The blood smears were stained with rapid staining (Hemacolor®, Merck) for a differential WBC count.
DNA extraction/purification
Frozen EDTA blood samples were thawed at room temperature and vortexed. DNA extraction from 100 µL of blood was performed using a DNeasy® Blood & Tissue Kit (QIAGEN®, Valencia, CA, USA) and was eluted with 100 µL of elution buffer, according to the manufacturer's instructions. Concentration and purity were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific©, USA). The absorbance ratio 260 and 280 nm (OD260/OD280) provided an estimate of sample purity, accepting a ratio of 1·8 ± 0·2 as ‘pure’.
Endogenous control real-time PCR
The 28S rDNA gene was used as an internal control for a PCR assay for feline genomic DNA (Helps et al. Reference Helps, Lait, Damhuis, Björnehammar, Bolta, Brovida, Chabanne, Egberink, Ferrand, Fontbonne, Pennisi, Gruffydd-Jones, Gunn-Moore, Hartmann, Lutz, Malandain, Möstl, Stengel, Harbour and Graat2005) using primers feline-28S rDNAFw (5′-AGCAGGAGGTGTTGGAAGAG-3′) and feline-28S rDNARv (5′-AGG GAGAGCCTAAATCAAAGG-3′) to discard the presence of PCR inhibitors. The reaction mixture was composed of 12·5 µL of Maxima®SYBR Green/Rox Master Mix (Thermo Scientific©, USA), 300 nm of the forward and reverse primers and 5 µL of DNA template, brought to a total volume of 25 µL with nuclease-free water (Thermo Scientific©, USA). The amplification conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Reactions were performed in a Stratagene Mx3000P™ (Agilent Technologies).
qPCR for Bartonella spp.
28S rDNA cPCR-positive samples were subsequently submitted to a previously described qPCR for Bartonella spp. targeting nuoG gene (André et al. Reference André, Dumler, Herrera, Goncalves, de Sousa, Scorpio, de Santis, Domingos, de Macedo and Machado2016). Amplification reactions were performed in duplicate using 10 µL of PCR mixtures containing 5 µL of Go Taq® Probe qPCR Master Mix, dTTP (Promega, Madison, WI, USA), 1,2 µ m of each primer [F-Bart (5′-CAATCTTCTTTTGCTTCACC-3′) and R-Bart (5′- TCAGGGCTTTATGTGAATAC-3′), hydrolysis probe [TexasRed-5′-TTYGTCATTTGAACACG-3′(BHQ2a-Q)3′] and 1 µL of the DNA sample. PCR amplifications were conducted in Low-Profile Multiplate™ Unskirted PCR Plates (BioRad©, Hercules, CA, USA) using a CFX96 Thermal Cycler (BioRad©). The amplification conditions were 95 °C for 3 min followed by 40 cycles of 95 °C for 10 min and 52·8 °C for 30 s. The qPCR was performed following the MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) (Bustin et al. Reference Bustin, Benes, Garson, Hellemans, Huggett, Kubista, Mueller, Nolan, Pfaffl and Shipley2009). Amplification efficiency (E) was calculated from the slope of the standard curve in each run using the following formula (E = 10–1/slope). Copy numbers were estimated using 10-fold serial dilutions of pIDTSMART plasmids (Integrated DNA Technologies, Coralville, IA, USA) encoding the nuoG B. henselae sequence (insert containing 83 bp). The number of plasmid copies was determined according to the formula [Xg µL−1 DNA/ (plasmid length in bp × 660)] × 6·022 × 1023 × plasmid copies µL−1. Bartonella henselae DNA obtained from a naturally infected cat (Miceli et al. Reference Miceli, Gavioli, Gonçalves, André, Souza, Marques de Sousa and Machado2013) was used as a positive control. All PCR runs were performed with nuclease-free water (Thermo Scientific©, USA) as a negative control. Replicates showing a Cq difference higher than 0·5 were retested.
cPCR for Bartonella spp.
For further molecular characterization and species differentiation, nuoG Bartonella qPCR-positive samples were tested using a previously described (Billeter et al. Reference Billeter, Gundi, Rood and Kosoy2011) cPCR targeting a 767-bp fragment of the citrate synthase gene (gltA), using primers CS443f (5′- GCTATGTCTGCATTCTATCA-3′) and CS1210r (5′-GATCYT CAATCATTTCTTTCCA-3′). Each DNA sample (5 µL) was used as a template in 25 µL reaction mixtures containing 10× PCR buffer, 1·5 mm MgCl2, 0·2 mm deoxynucleotide triphosphate (dNTPs) mixture, 0·625 U Platinum Taq DNA Polymerase (Invitrogen©, Carlsbad, CA, USA), and 0·5 µ m of each primer. cPCR amplification reactions were performed using a T100 BioRad termocycler (BioRad©) with the following cycling conditions: 94 °C for 2 min; 45 cycles of 94 °C for 30 s, 48 °C for 1 min and 72 °C for 1 min; and one cycle of 72 °C for 5 min. PCR products were separated by electrophoresis on a 1% agarose gel stained with ethidium bromide. To prevent PCR contamination, DNA extraction, reaction setup, PCR amplification and electrophoresis were performed in separate rooms. Gels were visualized under ultraviolet light using the Image Lab Software version 4.1 (BioRad©). The reaction products were purified using the Silica Bead DNA gel extraction kit (Fermentas©, São Paulo, SP, Brazil).
Sequencing and Phylogenetic analysis
Only gltA-cPCR-positive samples presenting strong band intensity were submitted for sequencing. Sanger sequencing was performed on purified amplified DNA fragments from positive samples in an automatic sequencer (ABI Prism 310 genetic analyser; Applied Biosystems©/Perkin-Elmer) for species identification and subsequent phylogenetic analysis. Consensus sequences were obtained through analysis of the sequenced products, from both the forward and the reverse oligonucleotides, using the CAP3 program (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py). Primer sequences were trimmed from the consensus sequences prior to Blastn analysis. Comparisons with sequences in GenBank were performed using the basic local alignment search tool (BLASTn). The sequences were aligned with sequences published in GenBank using Clustal/W and manually adjusted in Bioedit v. 7.0.5.3 (Carlsbad). Phylogenetic inference based on maximum-likelihood criterion (ML) was inferred with RAxML-HPC BlackBox 7.6.3 (Statamakis et al. Reference Statamakis, Hoover and Rougemont2008) through the CIPRES Science Gateway (Miller et al. Reference Miller, Pfeiffer and Schwartz2010) estimating the proportion of invariable sites by an evolutive model GAMMA GTR + I.
Nucleotide diversity
The alignment sequences of the gltA gene, amplified in the present study, were used to calculate the nucleotide diversity (π), polymorphic level [haplotype diversity (Hd)], number of variable sites (vs) and the average number of nucleotide differences (K) using the DnaSP v5.10 (Librado and Rozas, Reference Librado and Rozas2009).
Statistical analysis
To determine Bartonella spp. prevalence, qPCR-positive cats were divided by the total number of cats sampled and multiplied by 100. The observed prevalence rates were expressed in percentages and the 95% IC was calculated. Descriptive statistics were obtained for haematological parameters and the cats were divided into two groups according to their Bartonella spp. status based on the qPCR results: Bartonella spp. negative or Bartonella spp. positive. The normal distribution of data was evaluated by a Shapiro–Wilk's test. The non-normally distributed data were analysed using the Kruskal–Wallis test to determine if there were any significant differences between the haematological variables of the Bartonella spp. status groups. A P-value ⩽0·05 was considered statistically significant. Data were analysed using RStudio version 0.99.903 and were available for all 370 cats, except for platelet counts, which were available only for 171 cats.
RESULTS
Bartonella spp. qPCR results
All 370 DNA samples [median and standard deviation (s.d.) of DNA concentration = 26·5 ± 12·3 ng µL−1; mean and s.d. 260/280 ratio = 1·79 ± 0·07] were positive for the feline 28S rDNA endogenous gene. Molecular prevalence of Bartonella DNA in cats by qPCR (mean and s.d. efficiency of reactions: 96·1 ± 0·83%, r 2 = 0·998 ± 0·00046) was 18·1% (67/370) (95% CI 14·422·5%). Thirty-eight samples had a consistent Cq (mean and s.d. 30·21 ± 2·93) and quantification (mean 1·32 × 103; minimun–maximun 2·13 × 100–3·19 × 104 nuoG-copies µL−1) (Table 1). Twenty-nine cats had inconsistent Bartonella-qPCR quantification assays, due to that, their Cq and quantification results were not registered in the present work.
Table 1. Bartonella spp.-positive cat blood samples with their respective Cq (cycle of quantification) and quantification (nuoG-copies µL−1) mean values obtained by qPCR assays, and the Bartonella species identified by BLASTn analysis
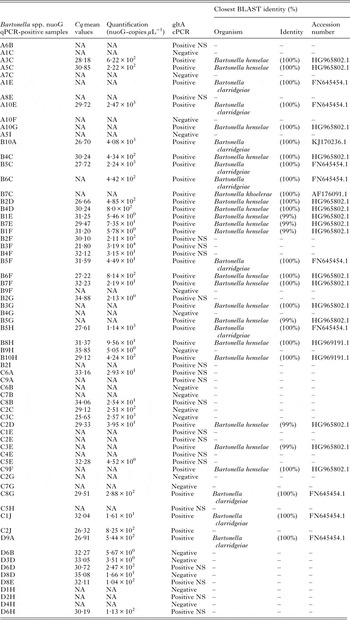
cPCR, conventional PCR; NS, cPCR-positive sample but with a weak band intensity, which precluded sequencing; NA, data not available due to inconsistent results.
cPCR results and phylogenetic analysis
Of 67 nuoG-qPCR-positive samples, 49 (73%) were gltA-cPCR-positive and 29 were sequenced. Twenty gltA-cPCR-positive samples presented weak band intensity, precluding sequencing. BLAST and phylogenetic analyses supported the identification of 62·0% (18/29) as B. henselae, 34·4% as B. clarridgeiae (10/29) and 3·4% (1/29) as B. koehlerae (Table 1).
Analysis of 14 sequenced products based on the gltA gene (GenBank accession numbers KX024499, KX024500, KX024503, KX024505, KX024509–KX024513, KX024515–KX024518, KX024520–KX024524) showed 99–100% identicalness with B. henselae (GenBank accession numbers HG965802; KJ170236). These fragments were positioned close to other B. henselae isolates, supported by high bootstrap values (96) in maximum-likelihood phylogeneitc analysis. Additionally, the analysis of six sequenced products based on the gltA region (GenBank accession numbers KX024501, KX024502, KX024504, KX024506, KX024507, KX024514, KX024519, KX024525–KX024527) showed 100% sequence identity with B. clarridgeiae (GenBank accession number FN645454 and KJ170236), and clustering with other B. clarridgeiae isolates. Finally, the analysis of one sequenced product based on the gltA region (GenBank accession number KX024508) showed 100% identicalness with B. koehlerae (GenBank accession number AF176091) and was positioned close to the American isolate (Fig. 2).
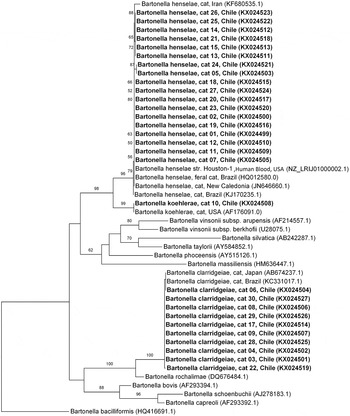
Fig. 2. Phylogenetic relationships within the Bartonella genus based on a 680 bp fragment of the gltA gene. The tree was inferred by using the ML method and evolutive model GAMMA GTR + I. The sequences detected in the present study are bold highlighted. The numbers at the nodes correspond to bootstrap values higher than 50% obtained with 1000 replicates. Bartonella bacilliformis was used as an outgroup.
Nucleotide diversity
Overall, the Bartonella species identified in the present study had a low genetic diversity. Of the 18 B. henselae gltA sequences analysed, only three different haplotypes were identified. The haplotype number #Bh1 with ten sequences was the most abundant haplotype, haplotypes # Bh2 and # Bh3 contained two and six sequences, respectively. Additionally, only two haplotypes were identified in the B. clarridgeiae sequences. While nine sequences formed the haplotype number #Bc1, only one sequence formed the haplotype number #Bc2. Both Bartonella species had a low number of variable sites, haplotypes and nucleotide diversity (Table 2).
Table 2. Polymorphism and genetic diversity of gltA Bartonella sequences detected in cats from Valdivia, Chile

N, number of sequences analysed; VS, number of variable sites; GC, G + C content; h, number of haplotypes; hd, haplotypes diversity; s.d., standard deviation; π, nucleotide diversity (per site = PI); K, average number of nucleotide difference.
Haematological analysis
All variables were non-normally distributed. The only haematological parameter that was significantly different was the MCV (P < 0·0001), which was significantly lower in Bartonella-positive cats (mean = 41·5 fL) than in Bartonella-negative ones (mean = 44 fL). There were no other significant differences between Bartonella-positive and -negative cats.
DISCUSSION
Cats are the major hosts of B. henselae, B. clarridgeiae and B. koehlerae (Mogollon-Pasapera et al. Reference Mogollon-Pasapera, Otvos, Giordano and Cassone2009). Since the pathogens can be transferred to humans through scratches or bites, public monitoring of its prevalence in cats is important (Breitschwerdt, Reference Breitschwerdt2008). This is the first study to investigate Bartonella spp. molecular prevalence in a population of domestic cats from southern Chile. Previous reports in Chile described high seroprevalence of B. henselae in Valdivia (Zaror et al. Reference Zaror, Ernst, Navarrete, Ballesteros, Boroschek, Ferres and Thibaut2002; Ferrés et al. Reference Ferrés, Abarca, Prado, Montecinos, Navarrete and Vial2006a ), Santiago and Coquimbo cities (Ferrés et al. Reference Ferrés, Abarca, Godoy, García, Palavecino, Méndez, Valdés, Ernst, Thibaut, Koberg, Chanqueo and Vial2005). Identification of B. henselae in solid cultures from cat (25/60) blood samples was performed in central Chile (Santiago City) (Ferrés et al. Reference Ferrés, Abarca, Godoy, García, Palavecino, Méndez, Valdés, Ernst, Thibaut, Koberg, Chanqueo and Vial2005).
Only few studies in South America have evaluated the molecular occurrence of Bartonella spp. in cats, mostly carried in Brazil (Staggemeier et al. Reference Staggemeier, Venker, Klein, Petry, Spilki and Cantarelli2010; Crissiuma et al. Reference Crissiuma, Favacho, Gershony, Mendes-de-Almeida, Gomes, Rozental, Barreira, Lemos and Labarthe2011; Braga et al. Reference Braga, de Paiva Diniz Diniz, André, de Bortoli and Machado2012; de Bortoli et al. Reference de Bortoli, André, Seki, Pinto, Machado and Machado2012; Miceli et al. Reference Miceli, Gavioli, Gonçalves, André, Souza, Marques de Sousa and Machado2013). Cats from Valdivia, Southern Chile, showed a similar prevalence (18·1%) to the one described in Buenos Aires, Argentina (17·8%) (Cicuttin et al. Reference Cicuttin, Brambati, De Gennaro, Carmona, Isturiz, Pujol, Belerenian and Gil2014) and Southern Brazil (17·0%) (Staggemeier et al. Reference Staggemeier, Venker, Klein, Petry, Spilki and Cantarelli2010), lower than that observed in cats from Galapagos Island, Ecuador (44·0%) (Levy et al. Reference Levy, Crawford, Lappin, Dubovi, Levy, Alleman, Tucker and Clifford2008) and higher than central-western (2·2%) (Miceli et al. Reference Miceli, Gavioli, Gonçalves, André, Souza, Marques de Sousa and Machado2013), northeastern (4·5%) (Braga et al. Reference Braga, de Paiva Diniz Diniz, André, de Bortoli and Machado2012) and southeastern (4·3%) (de Bortoli et al. Reference de Bortoli, André, Seki, Pinto, Machado and Machado2012) Brazil. However, comparison is difficult, because of the low number of cats and inclusion criteria used in some studies. In general, a higher prevalence was observed in stray cats (as high as 61·1%) (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005; Gutiérrez et al. Reference Gutiérrez, Morick, Gross, Winkler, Abdeen and Harrus2013), young adult cats living in shelters (36%) (Fleischman et al. Reference Fleischman, Chomel, Kasten, Stuckey, Scarlet, Liu, Boulouis, Haddad and Pedersen2015) and cats from spaying/neutering program (42·5%) (Crissiuma et al. Reference Crissiuma, Favacho, Gershony, Mendes-de-Almeida, Gomes, Rozental, Barreira, Lemos and Labarthe2011). Differences in prevalence may reflect variations in the groups of studied cats, or by geographical variations, such as climate and bloodsucking arthropod distribution, where the presence of fleas (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005) is a risk factor for Bartonella infection. Furthermore, a direct comparison between studies is difficult because of the differences in cPCR and qPCR diagnostic assays (André et al. Reference André, Dumler, Herrera, Goncalves, de Sousa, Scorpio, de Santis, Domingos, de Macedo and Machado2016).
As described worldwide (Chomel and Kasten, Reference Chomel and Kasten2010), B. henselae was the most prevalent species in Southern Chile, followed by B. clarridgeiae. The latter was less frequently isolated from domestic cats than B. henselae, as it appears to be difficult to isolate and is unevenly distributed in cat populations (Chomel et al. Reference Chomel, Boulouis and Breitschwerdt2004). Only one sampled cat was positive for B. koehlerae, which has rarely been detected in domestic cats worldwide (Avidor et al. Reference Avidor, Graidy, Efrat, Leibowitz, Shapira, Schattner, Zimhony and Giladi2004; Chomel and Kasten, Reference Chomel and Kasten2010; Fleischman et al. Reference Fleischman, Chomel, Kasten, Stuckey, Scarlet, Liu, Boulouis, Haddad and Pedersen2015). To the best of our knowledge, B. koehlerae is detected for the first time in cats from South America.
The low genetic diversity of Bartonella species identified in cats from Southern Chile is in accordance with the high intra species similarity between gltA gene sequences of various Bartonella spp. (99·80–100%) (Birtles and Raoult, Reference Birtles and Raoult1996). Since <1·00% genomic variety exists between various strains of B. henselae, a low diversity is a common finding (Guy et al. Reference Guy, Nystedt, Sun, Näslund, Berglund and Andersson2012). Due to the low genetic diversity, each Bartonella species from Chile clustered together and with other Bartonella spp. described in cats from Brazil, Iran and USA (Droz et al. Reference Droz, Chi, Horn, Steigerwalt, Whitney and Brenner1999; André et al. Reference André, Baccarim Denardi, Marques de Sousa, Gonçalves, Henrique, Grosse Rossi Ontivero, Lima Gonzalez, Cabral Nery, Fernandes Chagas, Monticelli, Alexandre de Santis and Machado2014; Fard et al. Reference Fard, Vahedi, Ashrafi, Alipour, Sharafi, Akbarein and Aldavood2016).
Bartonella henselae isolates clustered with both Houston-1 and Marseille strains, presenting a high similarity with these. Since Multi Locus Sequence Typing was not performed in the present study, it was not possible to determine which strains are circulating in Chilean cats. Most human cases of CSD are caused by B. henselae type Houston-1, suggesting that type Houston-1 strains could be more virulent to humans (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005) than Marseille (type II), which is more frequently identified in cats (Chomel et al. Reference Chomel, Boulouis and Breitschwerdt2004; Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005). Bartonella henselae-type Marseille is the dominant type in cat populations from Western Europe (France, Germany, Italy, The Netherlands and UK) and Australia (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005), whereas type Houston-1 is more frequently reported in human cases in the same regions (Arvand et al. Reference Arvand, Feil, Giladi, Boulouis and Viezens2007). Houston-1 is more frequent in cats from Asia (Japan and the Philippines) (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005). In North America (USA), type II is more prevalent in cats on the West Coast (California) (Chomel et al. Reference Chomel, Abbott, Kasten, Floyd-Hawkins, Kass, Glaser, Pedersen and Koehler1995; Chang et al. Reference Chang, Chomel, Kasten, Tappero, Sanchez and Koehler2002; Fleischman et al. Reference Fleischman, Chomel, Kasten, Stuckey, Scarlet, Liu, Boulouis, Haddad and Pedersen2015) but a 50–50% (types I and II) was described on the East Coast (North Carolina and Florida) (Guptil et al. Reference Guptill, Wu, Hogenesch, Slater, Glickman, Dunham, Syme and Glickman2004). Bartonella henselae isolates obtained from cats in Guatemala (Bai et al. Reference Bai, Rizzo, Alvarez, Moran, Peruski and Kosoy2015) and Argentina (Cicuttin et al. Reference Cicuttin, Brambati, De Gennaro, Carmona, Isturiz, Pujol, Belerenian and Gil2014) were Houston type I group, suggesting that it could be the major genotype in Central and South America. Nevertheless, more studies on B. henselae diversity in other countries, including Chile, are needed to prove this hypothesis.
The mean number of Bartonella spp. nuoG-copies µL−1 in cats from southern Chile was lower than that described in naturally infected cats from Brazil, using the same qPCR protocol (André et al. Reference André, Dumler, Herrera, Goncalves, de Sousa, Scorpio, de Santis, Domingos, de Macedo and Machado2016). Indeed, low initial DNA copies in some blood samples from Chilean cats could produce inconsistent quantification results in the Bartonella-qPCR assay, represented by the Monte Carlo effect (Bustin et al. Reference Bustin, Benes, Garson, Hellemans, Huggett, Kubista, Mueller, Nolan, Pfaffl and Shipley2009). The low number of nuoG-Bartonella copies in blood samples from cats in our study may be explained by the characteristics of Bartonella spp. infection. After infecting their hosts, Bartonella may cause a persistent bacteraemia in cats, which is undetectable (Breitschwerdt et al. Reference Breitschwerdt, Maggi, Mozayeni, Hegarty, Bradley and Mascarelli2010a ). Long-term intraerythrocytic bacteraemia in reservoir mammals is frequently described and represents a common strategy of Bartonella for achieving infection without producing organ damage, generating only chronic, asymptomatic, infection (Chomel et al. Reference Chomel, Boulouis, Breitschwerdt, Kasten, Vayssier-Taussat, Birtles, Koehler and Dehio2009).
A lower MCV, within the reference values, was the only haematological finding in Bartonella-positive cats and was not considered clinically relevant. As observed in Chilean cats, haematological abnormalities are rarely described in naturally infected cats that seem to be healthy carriers of the bacterium (Boulouis et al. Reference Boulouis, Chao-chin, Henn, Kasten and Chomel2005; Chomel et al. Reference Chomel, Boulouis, Breitschwerdt, Kasten, Vayssier-Taussat, Birtles, Koehler and Dehio2009). In neotropical felids haematological abnormalities were not associated with Bartonella spp. natural infection (Guimaraes et al. Reference Guimaraes, Brandão, Moraes, Kiihl, Santos, Filoni, Cubas, Robes, Marques, Neto, Yamaguti, Oliveira, Catão-Dias, Richtzenhain, Messick, Biondo and Timenetsky2010). On the contrary, eosinophilia (Kordick et al. Reference Kordick, Brown, Shin and Breitschwerdt1999) and neutrophilia (Guptill et al. Reference Guptill, Slater, Wu, Lin, Glickman, Welch and HogenEsch1997) were observed in experimentally infected cats. It is important to state that in experimentally infected cats, usually the inoculum dose was very high (Guptill et al. Reference Guptill, Slater, Wu, Lin, Glickman, Welch and HogenEsch1997). Furthermore, strain variability among B. henselae isolates may contribute to enhanced pathogenicity in experimentally infected cats (O'Reilly et al. Reference O'Reilly, Bauer, Freeland, Foil, Hughes, Rohde, Roy, Stout and Triche1999).
Circulation of the three Bartonella species in Valdivia cats strengthens the importance of the feline population as a source of zoonotic agents and represents a potential infection risk to humans. While most cats are asymptomatic after becoming infected with B. henselae, they serve as reservoirs of the agent and may transmit the infection to humans (Breitschwerdt et al. Reference Breitschwerdt, Maggi, Mozayeni, Hegarty, Bradley and Mascarelli2010a ). Data on CSD prevalence in Chile reports a 10·3% infection rate with B. henselae in children from Central Chile (Ferrés et al. Reference Ferrés, Abarca, Prado, Montecinos, Navarrete and Vial2006b ). Also, asymptomatic cat-owners from southern Chile showed serological exposure (18%) to B. henselae (Zaror et al. Reference Zaror, Ernst, Navarrete, Ballesteros, Boroschek, Ferres and Thibaut2002) and a high seroprevalence (60%) was observed in humans with an occupational risk in the Bío Bío region, Chile (Troncoso et al. Reference Troncoso, Fischer, Arteaga, Espinoza, Azócar and Abarca2016). The presence of B. clarridgeiae and B. koehlerae in cats suggests the need to consider these agents when testing clinical samples from suspected human cases in Chile, along with B. henselae.
Concluding remarks
The overall prevalence of Bartonella spp. in domestic cats from Valdivia, Southern Chile, is in accordance with that previously described in South America. Bartonella-positive cats had low DNA bacterial loads and their haematological parameters varied minimally. Low genetic diversity was reported among the B. henselae and B. clarridgeiae haplotypes in the present study. Three Bartonella species circulate in the studied cat population of Valdivia. Bartonella clarridgeiae is reported for the first time in cats from Chile and B. koehlerae in cats from South America.
ACKNOWLEDGEMENTS
We thank the veterinary team of the UACh Veterinary Hospital for their helpful contributions in collecting samples. We also wish to thank the team of the Inmunoparasitology Laboratory, Faculdade de Ciencias Agárias e Veterinarias of the Universidade Estadual Paulista, Jaboticabal, Brazil, for their technical support.
FINANCIAL SUPPORT
This work was supported by Beca Santander Iberoamérica Jovenes Investigadores, 2015, Chile.
CONFLICT OF INTEREST
The authors do not have any conflicts of interest to declare.







