Introduction
Cysticercosis caused by Cysticercus tenuicollis, larval stage (metacestode) of a canine tapeworm Taenia hydatigena, is endemic in many parts of the world (Boufana et al., Reference Boufana, Scala, Lahmar, Pointing, Craig, Dessì, Zidda, Pipia and Varcasia2015; Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019). A 2 distinct hosts relationship in the life cycle involves dogs and other wild canids as definitive hosts and small ruminants (sheep and goats) and pigs as intermediate hosts, resulting in domestic and sylvatic cycles of disease transmission (Abbas et al., Reference Abbas, El-Alfy, Janecek-Erfurth and Strube2021). Rarely, other animals such as cattle, deer, wild boars and other wild ungulates also act as intermediate hosts to the parasite (Cengiz et al., Reference Cengiz, Yucel Tenekeci and Bilgen2019; Filip et al., Reference Filip, Pyziel, Jeżewski, Myczka, Demiaszkiewicz and Laskowski2019; Sgroi et al., Reference Sgroi, Varcasia, Dessì, D'Alessio, Pacifico, Buono, Neola, Fusco, Santoro, Toscano, Fioretti and Veneziano2019). The intermediate hosts acquire the infection by ingesting the taeniid eggs from contaminated pastures, which further results in development of metacestodes (C. tenuicollis) (Cengiz et al., Reference Cengiz, Yucel Tenekeci and Bilgen2019). However, the definitive hosts get infected either by consuming the offal (mesenteries, omentum and less commonly the liver) of slaughtered intermediate hosts containing the cysticerci or by predation and hence, completing the domestic and sylvatic cycles, respectively (Sgroi et al., Reference Sgroi, Varcasia, Dessì, D'Alessio, Pacifico, Buono, Neola, Fusco, Santoro, Toscano, Fioretti and Veneziano2019). In infected animals, mortality occurs due to hepatitis cysticercosa, peritonitis and pneumonia (Scala et al., Reference Scala, Urrai, Varcasia, Nicolussi, Mulas, Goddi, Pipia, Sanna, Genchi and Bandino2016). The infection in the intermediate hosts also holds huge economic importance as it leads to mortality of heavily infected lambs and kids and also condemnation of heavily infected organs (especially liver) and carcasses (Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019).
In India, small ruminant (goat and sheep) farming/rearing plays a very important role in the livelihood of landless (pastoralists), small and marginal farmers (Chakraborty and Gupta, Reference Chakraborty and Gupta2017). It has been estimated that small ruminant farming contributes around 14, 4 and 15% of meat, milk and skin/hide production, respectively, and hence, adds approximately $314 014 671 per annum to the rural economy (Chakraborty and Gupta, Reference Chakraborty and Gupta2017). Despite being a potent benefactor to the underprivileged and poorest section of the society, the goat farming sector experiences many constraints including poor management practices leading to low productivity, less number of productive breeds, a very limited veterinary care especially to the animals reared under pastoralism and also issues related to marketing (Chakraborty and Gupta, Reference Chakraborty and Gupta2017).
In the past decade, only 2 abattoir-based studies demonstrated the occurrence of C. tenuicollis in small ruminants with 4.22% (Singh et al., Reference Singh, Sharma, Gill and Sharma2015) and 18.87% (Nimbalkar et al., Reference Nimbalkar, Shinde, Kamtikar and Muley2011) prevalence in north India and Maharashtra state, respectively. However, no such study has been carried out in definitive hosts. The transmission of various taeniid parasites becomes extremely successful in India as most ideal conditions aid in establishment, propagation and dissemination of these parasites (Moudgil et al., Reference Moudgil, Moudgil, Asrani and Agnihotri2019). The most common practices which result in easy transmission of taeniid parasites involve improper disposal of offal at unregulated abattoirs, easy access of dogs to the offal, absence of proper meat inspection and no or very limited public awareness about the various disease conditions (Singh et al., Reference Singh, Sharma, Ghatak, Sharma, Bal, Tuli and Gill2012).
Globally, a few reports targeting mitochondrial gene sequences (partial cox1 and nad1) had reported considerable levels of genetic diversity among T. hydatigena populations from different hosts and geographical regions (Kedra et al., Reference Kedra, Tkach, Swiderski and Pawłowski2001; Boufana et al., Reference Boufana, Scala, Lahmar, Pointing, Craig, Dessì, Zidda, Pipia and Varcasia2015; Braae et al., Reference Braae, Kabululu, Nørmark, Nejsum, Ngowi and Johansen2015; Rostami et al., Reference Rostami, Salavati, Beech, Babaei, Sharbatkhori, Baneshi, Hajialilo, Shad and Harandi2015; Omar et al., Reference Omar, Elmajdoub, Al-Aboody, Elsify, Elkhtam and Hussien2016; Adwan et al., Reference Adwan, Jayousi, Abuseir, Abbasi, Adwan and Jarrar2018; Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019; Sarvi et al., Reference Sarvi, Ebrahimi Behrestaghi, Alizadeh, Abdollah Hosseini, Gohardieh, Bastani, Yazdani Charati, Daryani, Amouei, Spotin and Gholami2020; Ohiolei et al., Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021).
In India, majority studies pertaining to incidence, pathology, biochemical and enzyme profiles/activities of C. tenuicollis and T. hydatigena started in 1980s (Pathak et al., Reference Pathak, Gaur and Sharma1982, Reference Pathak, Kumar and Gaur1984; Pathak and Gaur, Reference Pathak and Gaur1982). However, the disease condition in definitive and intermediate hosts is poorly documented from India. The existing few reports exhibit an infection rate ranging from 2.23 to 37.03% in small ruminants (Pathak and Gaur, Reference Pathak and Gaur1982; Nath et al., Reference Nath, Pal, Mandal and Parveen2010; Singh et al., Reference Singh, Sharma, Gill and Sharma2015). Since the data pertaining to basic research related to T. hydatigena are scant, a very little research regarding detailed studies related to molecular confirmation, phylogeny, molecular phylogeography and population-genetic structure of the parasite has been carried out in India. Hence, the present preliminary study was envisaged to perform phylogenetic analysis and to assess neutrality indices and haplotype/genetic diversity of C. tenuicollis isolates retrieved from slaughtered goats of north India by targeting partial cytochrome c oxidase subunit 1 (cox1) mitochondrial gene sequence.
Materials and methods
Study area
The present study targeted 3 geographically different locations of north India, Hisar, Palampur and Chandigarh, belonging to 2 different states (Haryana and Himachal Pradesh) and a union territory (Chandigarh), respectively. The study targeted the arid zone of Haryana state involving the district of Hisar (29.09°N, 75.42°E) having an elevation of 215 m above mean sea level and an average annual rainfall of 450 mm. Palampur (32.11°N, 76.54°E), located in the foothills of north-west Himalayas at an elevation of 1472–2350 m above mean sea level, receives an average annual rainfall of 1578 mm. Chandigarh (30.73°N, 76.78°E) possesses an elevation of 321 m above mean sea level and an average annual rainfall of 792 mm.
Study design, sample size estimation and collection of parasitic stages
Total goat population in Haryana, Himachal Pradesh and Chandigarh is 336 338, 1 108 413 and 998 units, respectively. The sample size was estimated to be 215 (Haryana = 73, Himachal Pradesh = 73 and Chandigarh = 69) by assuming the prevalence 5% [based on the study carried out in neighbouring state Punjab (Singh et al., Reference Singh, Sharma, Gill and Sharma2015)], confidence level 95% and desired absolute precision or margin of error 5% (Thrusfield, Reference Thrusfield2007). In the present cross-sectional study, a total of 613 goats (both male and female adult animals of all breeds) slaughtered at Hisar (n = 207), Palampur (n = 201) and Chandigarh (n = 205) were randomly selected and screened visually or palpated for the presence of C. tenuicollis infection during February to August 2021. Taenia hydatigena metacestodes (C. tenuicollis) were carefully collected/dissected out and morphologically identified as per Loos-Frank (Reference Loos-Frank2000). The cysticerci exhibiting calcification and caseation were avoided and only viable cysticerci were collected for the phylogenetic study. The cysticerci were cleaned with normal saline and transported to laboratory at 4°C (under cold chain) and further kept in 70% ethanol at −20°C until genomic DNA extraction. Before proceeding for the genomic DNA extraction, the excised portions (preferably scolices) were washed repeatedly (minimum 5 times) with phosphate-buffered saline (pH = 7.4) for the removal of ethanol (Kilinc et al., Reference Kilinc, Kesik and Simsek2019).
Genomic DNA extraction, PCR amplification and sequencing
Out of 59, 7 [Himachal Pradesh (n = 3; isolates 1–3), Haryana (n = 2; isolates 4–5) and Chandigarh (n = 2; isolates 6–7)] randomly selected representative isolates of C. tenuicollis were subjected to molecular analysis. Genomic DNA was extracted from the excised portions of the cysticerci; preferably scolex, using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. The retrieved DNA was stored at −20°C until further use. Mitochondrial cox1 gene was targeted for partial amplification of DNA sequences through PCR using primers; forward: 5′-TGCATTTAGCTGGTGCGTCAAGTA-3′ and reverse: 5′-ACAAACACGCCGGGGTAACC-3′ (Filip et al., Reference Filip, Pyziel, Jeżewski, Myczka, Demiaszkiewicz and Laskowski2019). The 25 μL PCR reaction mixture contained 12.5 μL of master mix (GoTaq Green Mater Mix, Promega, Madison, WI, USA), 0.75 μL of each primer (forward and reverse) (10 pmol), 1 μL of genomic DNA template and 10 μL of nuclease-free water. Amplification reactions were carried out in thermal cycler (Bio-Rad T100™ Thermal cycler, USA) using the conditions: initial denaturation (95°C for 1 min), denaturation (35 cycles of 95°C for 20 s), annealing (35 cycles of 56°C for 20 s), extension (35 cycles of 72°C for 40 s) and final extension (72°C for 5 min) (Filip et al., Reference Filip, Pyziel, Jeżewski, Myczka, Demiaszkiewicz and Laskowski2019). The amplified PCR products were electrophoresed on 1.25% agarose gel along with 100 bp marker (DNAmark™ 100 bp, G-Biosciences, Saint Louis, Missouri, USA) and were visualized under gel documentation system. The genomic DNA of T. hydatigena adult parasites and Echinococcus granulosus retrieved from definitive hosts (dogs) were used as positive and negative controls, respectively, for amplification reactions. The amplicons of approximately 830 bp obtained in the present study were subjected to custom sequencing (GeneBio Solutions, Dehradun, India). Sanger sequencing (by carrying out chain termination PCR) was performed on the amplicons in both the directions (forward and reverse) separately by using the aforementioned primers. Each isolate was sequenced in triplicate to eliminate any error in sequencing.
Data analysis
DNA sequences retrieved were analysed for misread sequences using BioEdit software and final alignment of resulting gene sequences was carried out (Hall, Reference Hall1999). The NCBI BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was employed for the identification and assessment of homology of the sequenced products by comparing them with reference sequences of different hosts archived in GenBank [goat (n = 2) – JN831297, KR337823; sheep (n = 1) – MT784879; small ruminants (n = 8) – MK945752, MK945753, MN175590–MN175595; swine (n = 2) – MF630923, MF630926; dog (n = 3) – MT784873, MT784890, MT784891]. The phylogenetic analysis was then performed using MEGA X (Molecular Evolutionary Genetic Analysis) software by constructing phylogenetic tree for representative isolates of T. hydatigena and other Taenia sp. [Taenia ovis (AB731675.1), Taenia serialis (AB731674.1) and Taenia multiceps (JX507220.1)] using maximum-likelihood method (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018). Bootstrap analyses were conducted using 1000 replicates and the distance scale was estimated at 0.05. The sequence of Moniezia expansa (AB099693.1) was included in the phylogenetic analysis as out-group species to root the tree. Population neutrality indices [Tajima's D (Tajima, Reference Tajima1989) and Fu and Li's D statistics values (Fu and Li, Reference Fu and Li1993)], nucleotide diversity (π) and haplotype diversity (Hd) were estimated using DnaSP v.5 software. Median-joining network (Bandelt et al., Reference Bandelt, Forster and Röhl1999) based haplotype diversity was inferred based on the sequences of mitochondrial cox1 using PopART (http://popart.otago.ac.nz). Fisher's exact test was used to determine the association between the prevalence of C. tenuicollis among different geographical locations (Haryana, Himachal Pradesh and Chandigarh). Significance was set at a 2-tailed P value <0.05.
Results
Prevalence studies
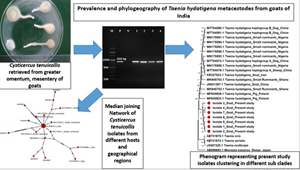
Out of 613 slaughtered goats screened, 59 (9.62%) showed the presence of C. tenuicollis (Fig. 1) attached to greater omentum, mesentery and even liver. Infection rates were highest in the slaughtered goats examined from Himachal Pradesh (12.43, 25/201, P = 0.11) followed by Haryana (8.69%, 18/207, P = 0.66) and Chandigarh (5.36%, 11/205, P = 0.013).

Fig. 1. Cysticercus tenuicollis (Taenia hydatigena metacestode) retrieved from slaughtered goats.
Molecular analysis
The partial amplification of cox1 gene and a PCR product size of approximately 830 bp confirmed the presence of C. tenuicollis (Fig. 2). An amplicon of 830 bp was also obtained for T. hydatigena adult parasites (positive control) whereas no amplification was observed for the DNA extracted from E. granulosus (negative control), validating the specificity of the primers.

Fig. 2. PCR amplification targeting the mitochondrial cox1 gene. M: 100 bp plus marker; P: positive (Taenia hydatigena) control; N: negative template (Echinococcus granulosus) control; 1–4: PCR products.
GenBank archived DNA sequences of T. hydatigena, retrieved from different intermediate (goats, sheep and pigs) and definitive hosts (dog) showed high similarity (>99%) with the present study isolates (Fig. 3). Phylogenetic tree analysis revealed that all the 7 isolates formed a major clade and grouped in a different cluster with T. hydatigena isolates retrieved from sheep, goats, pigs and dogs, originating from China, Iran, Nigeria, Ghana and Poland (Fig. 3). However, a single isolate (isolate 3) from Himachal Pradesh formed a subgroup within the clade.

Fig. 3. Phylogenetic tree of Cysticercus tenuicollis (Taenia hydatigena metacestode) isolates retrieved from goats in relation to different GenBank archived isolates of intermediate and definitive hosts based on mitochondrial cox1 gene. The phylogenetic tree was constructed by the maximum-likelihood method using MEGA X software. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. Moniezia expansa (AB099693.1) was used as out-group species to root the tree.
A total of 10 cox1 nucleotide GenBank archived sequences of T. hydatigena from China, Iran, Nigeria, Ghana and Poland were included with the present study isolates. The haplotype and nucleotide diversities (Hd and π) of the present study isolates were 0.99695 (0.95238–1.0000) and 0.49276, respectively, whereas the neutrality indices were negative with Tajima's D and Fu and Li's D statistics values of −1.26988 and −0.74556, respectively.
The haplotype network of T. hydatigena had all 17 (7 from present study and 10 retrieved from GenBank) haplotypes arranged within a star-like configuration (Fig. 4) with a main central haplotype (MT784873.1, retrieved from dog), separated from other haplotypes by 1–22 mutational steps. A total of 55 polymorphic regions containing 34.54% (19/55) parsimony informative sites were detected in 17 isolates analysed in this study. In Indian isolates, 27 polymorphic regions with 6 parsimony informative sites (68, 89, 235, 254, 365 and 487) and 21 singleton variable sites were recorded. The haplotype network containing Indian isolates of the present study also exhibited star-like configuration (Fig. 5) and the haplotypes were separated by 2–14 mutational steps. The 7 present study isolates subjected to haplotype network analysis represented 7 different haplotypes (Fig. 5).

Fig. 4. Median-joining network of Cysticercus tenuicollis isolates from different hosts and geographical regions using partial mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. Circle size is relative to haplotype frequency. Bars/hatch marks indicate the number of haplotype substitutions/mutations.

Fig. 5. Median-joining network of north Indian Cysticercus tenuicollis isolates retrieved from goats using partial mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. Circle size is relative to haplotype frequency. Bars/hatch marks indicate the number of haplotype substitutions/mutations.
Discussion
Taenia hydatigena is ubiquitous in distribution. Scanty information is available on prevalence and morphological characterization of the parasite in small ruminants in north India (Singh et al., Reference Singh, Sharma, Gill and Sharma2015). To the best of the authors’ knowledge, this is the first study from India assessing the genetic population structure relationships of C. tenuicollis from goats. The overall prevalence (9.62%) of cysticercosis recorded in this study was higher than reported in a previous study (4.83%, Singh et al., Reference Singh, Sharma, Gill and Sharma2015). These rising trends of the parasitic infections can be attributed to the improper disposal of offal from open space non-gazetted abattoirs, making offal easily accessible to the scavenger dogs (Singh et al., Reference Singh, Sharma, Ghatak, Sharma, Bal, Tuli and Gill2012). This leads to easy completion of life cycle of the parasite in dogs feeding on contaminated offal and its subsequent transmission to other healthy susceptible animals. High prevalence of C. tenuicollis infection in goats slaughtered in Himachal Pradesh can be attributed to this offal-based transmission since large migratory goat herds are usually guarded by dogs. These guard dogs are being fed with the offal from diseased goats (often infected with cysticercosis) by the goat herder. These infected dogs contaminate the pastures (feces containing eggs), thereby setting up the transmission cycle (Moudgil et al., Reference Moudgil, Moudgil, Asrani and Agnihotri2019).
Mitochondrial DNA genes are usually considered as most potent targets for genealogical studies (Celik et al., Reference Celik, Kilinc, Kesik, Ahemd and Simsek2021). Also, for phylogenetics and phylogeography of taeniid cestodes, mitochondrial cox1 gene is the most promising candidate to assess and establish intra- and interspecies variants (Bowles et al., Reference Bowles, Blair and McManus1992; Adwan et al., Reference Adwan, Jayousi, Abuseir, Abbasi, Adwan and Jarrar2018). This is due to relatively higher mutation and evolutionary rates, conserved structure, maternal inheritance, absence of recombination and high genetic divergence of mitochondrial DNA (Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019; Moudgil et al., Reference Moudgil, Nehra, Nehra, Sharma, Vohra and Moudgil2021). Mitochondrial DNA also holds high diagnostic values as it is present in more than 1 copy per cell (Celik et al., Reference Celik, Kilinc, Kesik, Ahemd and Simsek2021).
Phylogenetic analysis of cox1 DNA sequences revealed that all the isolates of the present study were accurately identified as T. hydatigena as they clustered with other GenBank archived isolates retrieved from different regions of the world. The present study T. hydatigena isolates also exhibited relative distance from other Taenia species (T. ovis, T. serialis and T. multiceps), in concordance to previous study (Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019). The neutrality tests (Tajima's D, Fu and Li's D statistics) based on selective neutrality of nucleotide variability exhibited negative values, indicating deviations from neutrality and both propounded excess of polymorphic sites and recent population expansion or purifying selection (Adwan et al., Reference Adwan, Jayousi, Abuseir, Abbasi, Adwan and Jarrar2018; Cengiz et al., Reference Cengiz, Yucel Tenekeci and Bilgen2019). The neutrality and diversity indices revealed high values of Hd and low π, which was indicative of demographic expansion and low gene flow, suggesting that Indian T. hydatigena isolates were not genetically differentiated (Adwan et al., Reference Adwan, Jayousi, Abuseir, Abbasi, Adwan and Jarrar2018). We observed 7 different haplotypes corresponding to 7 different isolates of north India, which is an indication of cross-fertilization driven gene flow in mature T. hydatigena parasites (Kilinc et al., Reference Kilinc, Kesik and Simsek2019). The prevailing conditions will definitely affect T. hydatigena host adaption and life cycle in future as new strain and genotype emergence would take place due to intra-species variation due to incessant nucleotide changes as a result of cross-fertilization (Kilinc et al., Reference Kilinc, Kesik and Simsek2019).
The consumption of meat infected with C. tenuicollis does not cause any harm to human beings, however it remains a threat to livestock production (Ohiolei et al., Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021). The genetic diversity of T. hydatigena/C. tenuicollis within populations is poorly understood due to limited studies (Ohiolei et al., Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021). A proper understanding of the genetic diversity is required for epidemiological investigation, anthelmintic treatment, vaccine/drug development and control (Ohiolei et al., Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021). Recent studies had reported a high variation between T. hydatigena populations, establishing its 2 haplogroups (Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019, Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021). However, in the present study, we had observed a low genetic diversity of T. hydatigena across north India.
While analysing the haplotype diversity through median-joining network, we observed a central haplotype in a star-like population structure considering the different isolates from different regions of the world. Our finding was in agreement with Boufana et al. (Reference Boufana, Scala, Lahmar, Pointing, Craig, Dessì, Zidda, Pipia and Varcasia2015). However, while considering the 7 isolates retrieved in the present study, we did not find any central haplotype. Similar findings were also reported by Ohiolei et al. (Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021), which indicated uncertainty about the existence of a central or widespread haplotype of T. hydatigena/C. tenuicollis (Kilinc et al., Reference Kilinc, Kesik and Simsek2019; Ohiolei et al., Reference Ohiolei, Luka, Zhu, Yan, Li, Magaji, Alvi, Wu, Li, Fu and Jia2019; Ohiolei et al., Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021). Extensive DNA sequence analysis data targeting different mitochondrial genes from a wide range of hosts and different geographical locations are requisite to resolve the issues of existence of central or widespread haplotype of T. hydatigena (Ohiolei et al., Reference Ohiolei, Yan, Li, Li, Wu, Alvi, Zhang, Fu, Wang and Jia2021).
Conclusion
The results of the present pilot study on phylogeographical analysis of C. tenuicollis from goats highlighted a low genetic diversity of T. hydatigena metacestodes across north India. Considering the present study as first step, future detailed studies involving different mitochondrial genes, vast host range and different geographical locations of India are warranted for understanding the molecular ecology and population genetics of T. hydatigena prevailing in India. This will also help in designing and implementation of effective control strategies.
Acknowledgements
The authors are thankful to the Dean, College of Veterinary Sciences and Director of Research, Lala Lajpat Rai University of Veterinary and Animal Sciences for providing the necessary facilities to carry out the research.
Author contributions
A. D. M. was involved in conceptualization, sample collection, investigation, methodology and writing. A. K. N. was involved in sample collection, investigation and laboratory work. S. V. was involved in supervision and editing. S. D. T. drafted, reviewed and edited the manuscript. D. S. was involved in sample collection and investigation. All the authors contributed to the review.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
None.
Ethical standards
No studies involving laboratory animals or invasive techniques were conducted. The samples were collected from slaughtered animals at abattoirs.