INTRODUCTION
Plasmodium knowlesi is a non-human primate malaria from the Plasmodium vivax clade, that has split from P. vivax between 18 and 34 million years ago (Silva et al. Reference Silva, Egan, Arze, Spouge and Harris2015). The parasite can infect a wide range of non-human primates (Coatney et al. Reference Coatney, Collins, Warren and Contacos1971), among which are the natural host, Macaca fascicularis and its close relative, the rhesus monkey (Macaca mulatta). The disease progression is quite different between the natural and experimental host, suggesting different parasite–host interaction mechanisms, which may be relevant to the variable disease outcomes observed in falciparum malaria in humans. Over the last decade it has become clear that P. knowlesi is not only a non-human primate malaria, but also an important zoonosis in South-East Asia, sometimes with fatal disease outcome (Singh & Daneshvar, Reference Singh and Daneshvar2013). Plasmodium knowlesi is now the most common cause of malaria in East and Peninsular Malaysia, both in travellers (Singh & Daneshvar, Reference Singh and Daneshvar2013) and in residents (Yusof et al. Reference Yusof, Lau, Mahmud, Fong, Jelip, Ngian, Mustakim, Hussin, Marzuki and Mohd Ali2014). Thus, historical and current studies on P. knowlesi are relevant for human disease, with the parasite not only being a model malaria parasite, but importantly also a human pathogen. The classification of P. knowlesi as the fifth human malaria parasite has given research interest in this parasite a significant boost. Of the 1065 publications found in PubMed using ‘knowlesi’ as keyword (query date October 31, 2016), 251 were published in the last 5 years (24%, first publication in 1935). In this review we will focus on the use of the animal and in vitro models, rather than on the human infections. An extensive overview of reported human P. knowlesi infections can be found in Siregar (Siregar et al. Reference Siregar, Faust, Murdiyarso, Rosmanah, Saepuloh, Dobson and Iskandriati2015).
Plasmodium knowlesi offers a versatile experimental system, as illustrated in Fig. 1. Rhesus monkeys form the most commonly used experimental host for in vivo studies (blue shaded area in Fig. 1) and can be infected by mosquito bite, isolated sporozoites, or blood stage parasites. Transmission, both to macaques and humans, occurs through mosquitoes from the leucosphyrus group of forest-dwelling Anopheline mosquitoes, including Anopheles dirus (Singh & Daneshvar, Reference Singh and Daneshvar2013; Moyes et al. Reference Moyes, Shearer, Huang, Wiebe, Gibson, Nijman, Mohd-Azlan, Brodie, Malaivijitnond, Linkie, Samejima, O'Brien, Trainor, Hamada, Giordano, Kinnaird, Elyazar, Sinka, Vythilingam, Bangs, Pigott, Weiss, Golding and Hay2016). Natural infections in wild macaques (Siregar et al. Reference Siregar, Faust, Murdiyarso, Rosmanah, Saepuloh, Dobson and Iskandriati2015) and P. knowlesi transmission dynamics from the natural host to humans (Vythilingam et al. Reference Vythilingam, Tan, Asmad, Chan, Lee and Singh2006) have been the subject of extensive studies since macaques have been recognized as transmission reservoirs for human P. knowlesi malaria.
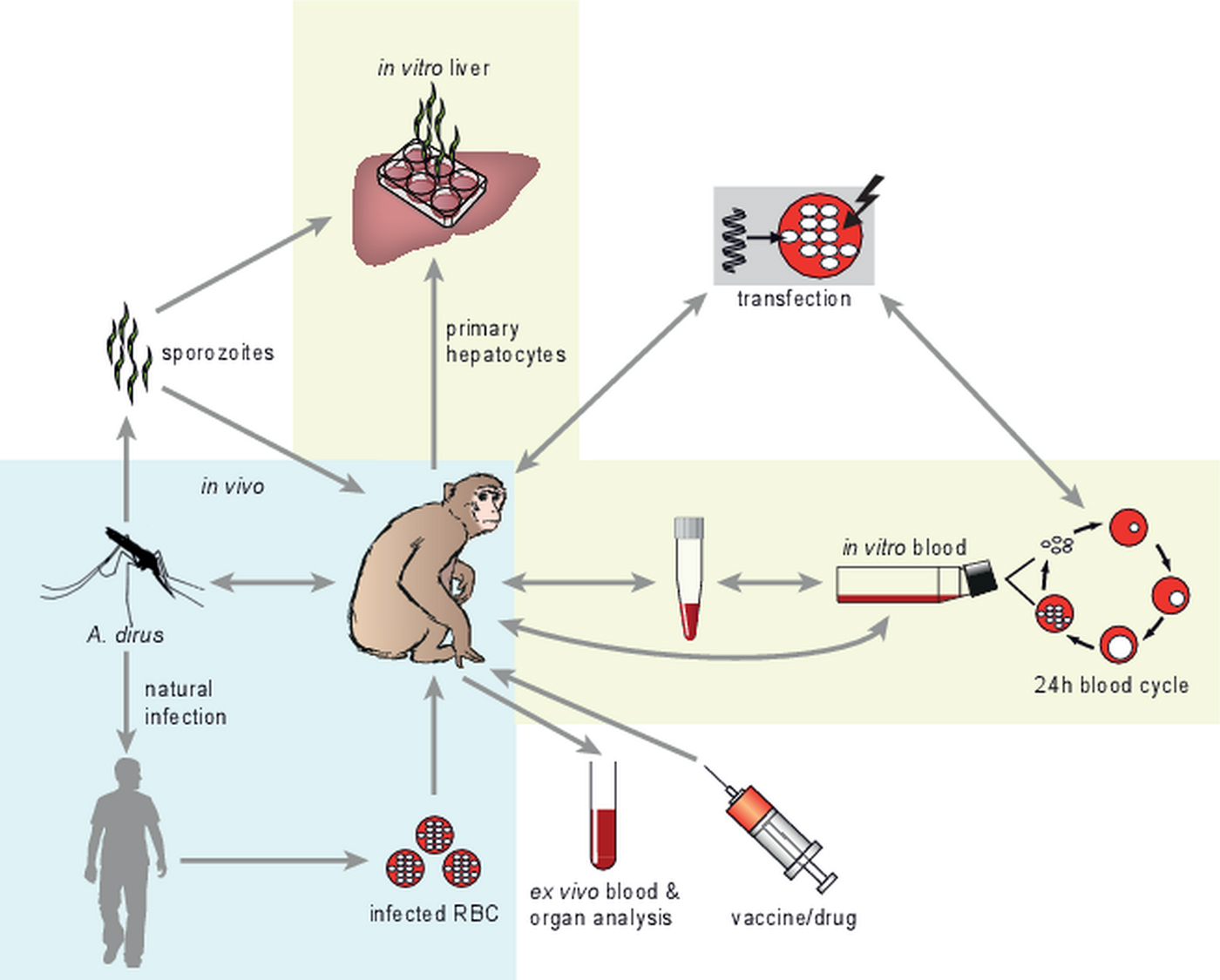
Fig. 1. The versatile P. knowlesi model. The blue-shaded area depicts the in vivo part of P. knowlesi work, with the monkey being the natural/experimental host, the mosquito providing transmission and the human as the zoonotic host. Different infection routes are shown, where parasite-infected human erythrocytes can be used to infect monkeys. Invasive sampling of organs and vaccine/drug application is also shown. The yellow-shaded areas depict in vitro work with P. knowlesi. Isolated sporozoites can be used to initiate in vitro liver stage cultures in primary macaque hepatocytes. Importantly, blood stage parasites have been adapted to continuous blood stage culture, in macaque or human erythrocytes. Transfection technology has been developed for in vivo- as well as in vitro derived parasites. Application of the various parts of this versatile model system is reviewed in detail in the text.
While in the wild infections exclusively occur through mosquito bites, in experimental conditions both mosquito bites and intravenous parasite injection can be used. Sporozoite infections by intravenous injection of 100 isolated sporozoites or by the bite of as little as 1 infected mosquito yield 100% infection in rhesus monkeys (Murphy et al. Reference Murphy, Weiss, Fryauff, Dowler, Savransky, Stoyanov, Muratova, Lambert, Orr-Gonzalez, Zeleski, Hinderer, Fay, Joshi, Gwadz, Richie, Villasante, Richardson, Duffy and Chen2014). Experimental blood stage infections in vaccine challenge studies are generally performed with 10 000 blood stage parasites (Mahdi Abdel Hamid et al. Reference Mahdi Abdel Hamid, Remarque, van Duivenvoorde, van der Werff, Walraven, Faber, Kocken and Thomas2011). Infected monkeys are used for parasite–host interaction studies and in vaccine- and drug efficacy studies. Importantly, invasive sampling of immunologically important organs is possible, either by needle biopsy during the study or by sampling larger parts of organs during necropsy at the end of vaccine efficacy studies. This allows the detailed study of immunological events that may lead to the discovery of correlates of protection (Epstein et al. Reference Epstein, Tewari, Lyke, Sim, Billingsley, Laurens, Gunasekera, Chakravarty, James, Sedegah, Richman, Velmurugan, Reyes, Li, Tucker, Ahumada, Ruben, Li, Stafford, Eappen, Tamminga, Bennett, Ockenhouse, Murphy, Komisar, Thomas, Loyevsky, Birkett, Plowe, Loucq, Edelman, Richie, Seder and Hoffman2011; Ishizuka et al. Reference Ishizuka, Lyke, DeZure, Berry, Richie, Mendoza, Enama, Gordon, Chang, Sarwar, Zephir, Holman, James, Billingsley, Gunasekera, Chakravarty, Manoj, Li, Ruben, Li, Eappen, Stafford, K, Murshedkar, DeCederfelt, Plummer, Hendel, Novik, Costner, Saunders, Laurens, Plowe, Flynn, Whalen, Todd, Noor, Rao, Sierra-Davidson, Lynn, Epstein, Kemp, Fahle, Mikolajczak, Fishbaugher, Sack, Kappe, Davidson, Garver, Bjorkstrom, Nason, Graham, Roederer, Sim, Hoffman, Ledgerwood and Seder2016). Such immunological studies are less relevant in rodent malaria models because of the large differences between human and rodent parasites and immune systems, and are not feasible in humans, where sampling is generally restricted to peripheral blood. Interestingly, to complete the arsenal of in vivo studies with P. knowlesi, a recent study has shown that parasites isolated from a human infection can be easily adapted to grow in monkeys, opening new possibilities to study human infection-adapted parasites e.g. at the level of red blood cell invasion characteristics (Amir et al. Reference Amir, Russell, Liew, Moon, Fong, Vythilingam, Subramaniam, Snounou and Lau2016).
Apart from in vivo/ex vivo studies, the P. knowlesi experimental model offers important in vitro possibilities, especially for liver and blood stage research (yellow shaded areas in Fig. 1). Isolated P. knowlesi sporozoites can be used for in vitro infection of primary rhesus hepatocytes, following similar protocols as described for the primate malaria Plasmodium cynomolgi (Zeeman et al. Reference Zeeman, van Amsterdam, McNamara, Voorberg-van der Wel, Klooster, van den Berg, Remarque, Plouffe, van Gemert, Luty, Sauerwein, Gagaring, Borboa, Chen, Kuhen, Glynne, Chatterjee, Nagle, Roland, Winzeler, Leroy, Campo, Diagana, Yeung, Thomas and Kocken2014). In vitro liver stage cultures provide the opportunity to study liver stage biology (Millet et al. Reference Millet, Collins, Aikawa, Cochrane and Nguyen-Dinh1990) and to perform immunological (sporozoite inhibition) and drug sensitivity studies (Fisk et al. Reference Fisk, Millet, Collins and Nguyen-Dinh1989).
Plasmodium knowlesi is the only parasite, apart from P. falciparum, for which continuous long-term in vitro blood stage cultures have been developed, initially in rhesus monkey red blood cells (Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002) and more recently also in human red blood cells (Moon et al. Reference Moon, Hall, Rangkuti, Ho, Almond, Mitchell, Pain, Holder and Blackman2013). These advances provide easy access to blood stage parasite material for in vitro drug- and (inhibition of) invasion studies (Mahdi Abdel Hamid et al. Reference Mahdi Abdel Hamid, Remarque, van Duivenvoorde, van der Werff, Walraven, Faber, Kocken and Thomas2011; Fatih et al. Reference Fatih, Staines, Siner, Ahmed, Woon, Pasini, Kocken, Singh, Cox-Singh and Krishna2013), -omics studies facilitated by the availability of the annotated genome sequences of both parasite (Pain et al. Reference Pain, Bohme, Berry, Mungall, Finn, Jackson, Mourier, Mistry, Pasini, Aslett, Balasubrammaniam, Borgwardt, Brooks, Carret, Carver, Cherevach, Chillingworth, Clark, Galinski, Hall, Harper, Harris, Hauser, Ivens, Janssen, Keane, Larke, Lapp, Marti, Moule, Meyer, Ormond, Peters, Sanders, Sanders, Sargeant, Simmonds, Smith, Squares, Thurston, Tivey, Walker, White, Zuiderwijk, Churcher, Quail, Cowman, Turner, Rajandream, Kocken, Thomas, Newbold, Barrell and Berriman2008) and experimental rhesus monkey host (Zimin et al. Reference Zimin, Cornish, Maudhoo, Gibbs, Zhang, Pandey, Meehan, Wipfler, Bosinger, Johnson, Tharp, Marcais, Roberts, Ferguson, Fox, Treangen, Salzberg, Yorke and Norgren2014), and for in vivo and in vitro transfection studies (Fig. 1). In this review we will further illustrate the versatility of the P. knowlesi experimental model by highlighting advances in parasite–host interaction studies, vaccine development and technological developments in P. knowlesi studies.
PLASMODIUM KNOWLESI AS A MODEL FOR PARASITE–HOST INTERACTION STUDIES
After the discovery of P. knowlesi in the early 1930s (Sinton and Mulligan Reference Sinton and Mulligan1932), experimental P. knowlesi infections in various hosts (including human subjects (Knowles, Reference Knowles1935; Chin et al. Reference Chin, Contacos, Coatney and Kimball1965)) have extensively been used to model malaria host–parasite interactions. This has been useful to underpin some of the major discoveries in the fields of parasite invasion (Bannister et al. Reference Bannister, Butcher and Mitchell1977), antigenic variation (Barnwell et al. Reference Barnwell, Howard and Miller1982, Reference Barnwell, Howard, Coon and Miller1983; Howard et al. Reference Howard, Barnwell and Kao1983; Howard & Barnwell, Reference Howard and Barnwell1985; al-Khedery et al. Reference al-Khedery, Barnwell and Galinski1999; Barnwell, Reference Barnwell1999; Corredor et al. Reference Corredor, Meyer, Lapp, Corredor-Medina, Huber, Evans, Barnwell and Galinski2004; Lapp et al. Reference Lapp, Mok, Zhu, Wu, Preiser, Bozdech and Galinski2015) immunity and vaccine development (reviewed in the next section).
Plasmodium knowlesi infections in experimental and natural hosts
Plasmodium knowlesi infects a wide range of Old- and New World monkeys, with variable disease outcome (Table 1). In the natural host, M. fascicularis, parasitaemia levels are relatively low (1–3%) and short-lived, and disease is mild (reviewed in Butcher, Reference Butcher1996). However, the genetic background and origin of the monkey play an important role in disease outcome (Schmidt et al. Reference Schmidt, Fradkin, Harrison and Rossan1977; Collins et al. Reference Collins, Skinner, Broderson, Filipski, Morris, Stanfill and Warren1992; Flynn et al. Reference Flynn, Satkoski, Lerche, Kanthaswamy and Smith2009). While mild disease is observed in M. fascicularis from the Philippines, M. fascicularis from West Malaysia developed severe disease, leading to death. Disease in other natural hosts like Macaca nemestrina (pig tailed macaques), Presbytis (Malayan leaf monkeys) and Trachypithecus has not been studied in detail. Other macaque species (Macaca radiata, Macaca fuscata, Macaca mulatta; Coatney et al. Reference Coatney, Collins, Warren and Contacos1971) of which the rhesus monkey (M. mulatta) has been most widely used (Collins, Reference Collins2012), are experimental hosts for P. knowlesi. Infection with blood stage parasites generally results in fulminant parasitaemia leading to death if left untreated, but infection with sporozoites leads to controllable blood stage parasitaemia in 30% of the monkeys (reviewed in Collins, Reference Collins2012). The difference of disease outcome in M. fascicularis and M. mulatta is striking, given the fact that these two macaque species are closely related. A study with few animals showed that a primary infection in M. fascicularis leads to relatively fast development of parasite-inhibitory antibodies, as compared with rhesus monkeys (Butcher et al. Reference Butcher, Mitchell and Cohen2010). Also, spleen cells from infected M. fascicularis have been shown to have anti-parasitic activity in vitro (Langhorne et al. Reference Langhorne, Butcher, Mitchell, Cohen and Torrigiani1977). Both studies suggest different immunological reactivity upon infection in two closely related macaque species, but further studies with larger groups are needed to confirm and expand these data.
Table 1. Experimental P. knowlesi infections in natural and experimental hosts. Macaca radiata is the only experimental host that uniformly yields a chronic, non-fatal infection

a Macaca fascicularis is a natural host for P. knowlesi.
The olive baboon (Papio anubis) (Ozwara et al. Reference Ozwara, Langermans, Maamun, Farah, Yole, Mwenda, Weiler and Thomas2003b ) has been used to study severe disease, which will be detailed below. The New World monkeys Saimiri sciureus (Collins et al. Reference Collins, Contacos and Chin 1978 ), Callithrix jacchus (Langhorne & Cohen, Reference Langhorne and Cohen1979) and Aotus trivirgatus (Collins et al. Reference Collins, Contacos, Skinner, Stanfill and Richardson1981) are also experimental hosts with self-curing or lethal infections in all species. Plasmodium knowlesi infections in experimental hosts may have a severe outcome (Table 1). In past studies P. knowlesi–host interactions were investigated in the light of severe malaria modelling, focusing on the comparison of characteristics observed in P. falciparum-cerebral malaria pathology (Jerusalem et al. Reference Jerusalem, Polder, Wijers-Rouw, Heinen, Eling, Osunkoya and Trinh1983).
Severe malaria pathogenesis and animal models: a hotly debated topic
The latest WHO estimates suggest 214 million cases of malaria in 2015 and 438 000 deaths (World Health Organisation, 2015) mainly as a consequence of a series of complications termed severe malaria. Severe malaria is frequently identified as being synonymous with cerebral malaria but in reality this is only a subset of severe disease (Craig et al. Reference Craig, Grau, Janse, Kazura, Milner, Barnwell, Turner and Langhorne2012), which encompasses multi-organ failure (including kidney and liver), severe anaemia and metabolic derangement as well as pregnancy-associated malaria. While severe malaria has previously exclusively been ascribed to human infections with P. falciparum, recent investigations have highlighted similar disease in human infections with P. knowlesi (Daneshvar et al. Reference Daneshvar, Davis, Cox-Singh, Rafa'ee, Zakaria, Divis and Singh2009) and with lower frequency in Plasmodium vivax malaria (Price et al. Reference Price, Tjitra, Guerra, Yeung, White and Anstey2007). Patterns of disease in P. knowlesi patients show several similarities to those seen with P. falciparum, such as multi-organ failure, albeit with the exemption of coma (Cox-Singh et al. Reference Cox-Singh, Hiu, Lucas, Divis, Zulkarnaen, Chandran, Wong, Adem, Zaki, Singh and Krishna2010). In the context of disease, it is important to consider that P. knowlesi is unique among the primate and human malarias in that it has a 24 h erythrocytic cycle (reviewed in (Collins, Reference Collins2012)), a characteristic that is likely to accelerate the development of complications (Cox-Singh et al. Reference Cox-Singh, Davis, Lee, Shamsul, Matusop, Ratnam, Rahman, Conway and Singh2008) thus making accurate diagnosis and effective treatment an urgent priority. Until recently, human P. knowlesi infections were often misdiagnosed as P. malariae, which contributed to P. knowlesi-infected individuals not being treated appropriately (Rajahram et al. Reference Rajahram, Barber, William, Grigg, Menon, Yeo and Anstey2016).
The etiology of severe malaria is complex and involves an interplay between genetic background, immune response and parasite virulence, a hallmark of the latter being cytoadherence-mediated sequestration (the ability of parasite infected red blood cells to bind to surface markers of endothelial cells of the capillaries in critical organs e.g. brain, lungs). The pathogenesis of severe malaria is hotly debated: different phenomena are being considered as the overriding pathogenic mechanism, including cytoadherence-mediated sequestration (White et al. Reference White, Turner, Day and Dondorp2013), endothelial dysfunction (Kim et al. Reference Kim, Higgins, Liles and Kain2011) and inflammation (Clark & Alleva, Reference Clark and Alleva2009; Grau & Craig, Reference Grau and Craig2012). However, recently (Cunnington et al. Reference Cunnington, Riley and Walther2013) it has been postulated that a combination of the three proposed mechanisms with a different balance in each specific syndrome can best explain the variability in severe malaria pathogenesis observed across human cases.
Equally hotly debated are the animal models to be used for investigating the mechanisms behind the pathophysiological manifestations of severe disease (White et al. Reference White, Turner, Medana, Dondorp and Day2010; Langhorne et al. Reference Langhorne, Buffet, Galinski, Good, Harty, Leroy, Mota, Pasini, Renia, Riley, Stins and Duffy2011; Craig et al. Reference Craig, Grau, Janse, Kazura, Milner, Barnwell, Turner and Langhorne2012). It is well documented that many of the key features of human malaria can be replicated in a variety of non-human primate models. However, despite their great potential, non-human primate models remain very under-utilized (Langhorne et al. Reference Langhorne, Buffet, Galinski, Good, Harty, Leroy, Mota, Pasini, Renia, Riley, Stins and Duffy2011). While it may well be the case that no single animal model is able to reproduce all of the different features of severe malaria faithfully (Craig et al. Reference Craig, Grau, Janse, Kazura, Milner, Barnwell, Turner and Langhorne2012), it is important to bear in mind that severe malaria is highly heterogeneous (Cunnington et al. Reference Cunnington, Riley and Walther2013) as its pathophysiological manifestations vary between parasite (P. falciparum, P. knowlesi and P. vivax) infections and even between human hosts infected by the same parasite (P. falciparum). It is therefore reasonable to assume that the heterogeneity of the pathophysiological presentations in animal models, if properly documented, could provide complementary insights on specific severe disease mechanisms. As compared with proposed larger field studies in humans (Cunnington et al. Reference Cunnington, Riley and Walther2013), non-human primate studies on severe malaria offer the possibility to investigate severe disease in a controlled environment and to perform multivariate studies and invasive sampling that are not possible in human field studies. In terms of non-human primate models, different severe malaria models have been used, including P. coatneyi-M. mulatta/fuscata, P. fragile-M. mulatta (Craig et al. Reference Craig, Grau, Janse, Kazura, Milner, Barnwell, Turner and Langhorne2012) and P. knowlesi in a range of non-human primate hosts. The latter will be reviewed below in more detail.
Plasmodium knowlesi animal models for severe disease
New World monkeys, such as Callithrix jacchus and Saimiri sciureus, were shown to be differentially susceptible to disease caused by various P. knowlesi strains (Collins et al. Reference Collins, Contacos and Chin1978; Langhorne & Cohen, Reference Langhorne and Cohen1979). This characteristic makes them particularly interesting as a malaria model, as humans have also been shown to be differentially susceptible to P. falciparum (Andrade & Barral-Netto, Reference Andrade and Barral-Netto2011) and P. knowlesi disease, where – in the latter – a clear-cut correlation was established between parasitaemia levels and malaria disease patterns (Cox-Singh et al. Reference Cox-Singh, Hiu, Lucas, Divis, Zulkarnaen, Chandran, Wong, Adem, Zaki, Singh and Krishna2010). Unfortunately, the studies on infections with P. knowlesi in New World monkeys are very limited and more well-designed experiments with a statistically significant number of animals per group are required to properly establish and validate these models for wider use.
Differential susceptibility to disease has also been observed in the Papio anubis-P. knowlesi H strain infection model (Ozwara et al. Reference Ozwara, Langermans, Maamun, Farah, Yole, Mwenda, Weiler and Thomas2003b ). The course of infection has been shown to be either acute or chronic. Animals with acute infection developed multiple system organ failure and cerebral involvement, while chronically infected animals presented with moderately enlarged spleens. The parasitaemia profiles of both groups were initially comparable with those observed in rhesus monkeys. However, animals that were able to control the initial hyper-parasitaemia developed chronic infection. In both cases, P. anubis individuals initially developed clinical symptoms such as apathy, raised fur and lethargy with dyspnea, which later disappeared in animals that controlled the hyper-parasitaemia. This model has also been used to study pregnancy-associated malaria, as infiltration of parasitized erythrocytes and inflammatory cells in the placental intravillous space, a key feature of human pregnancy-associated malaria, is observed in these animals (Onditi et al. Reference Onditi, Nyamongo, Omwandho, Maina, Maloba, Farah, King, Moore and Ozwara2015). Placental plasma and serum samples, assayed by enzyme-linked immunosorbent assay for cytokine profiles, appear to confirm findings from human pregnancy-associated malaria (Barasa et al. Reference Barasa, Ng'ang'a, Sowayi, Okoth, Barasa, Namulanda, Kagasi, Gicheru and Ozwara2012), as elevated concentrations of tumour necrosis factor alpha (TNF-α) and interleukin 12 (IL-12) were found in both. A controlled study in the same P. anubis model addressed the question of whether co-infections with schistosoma enhance or attenuate severe malaria symptoms. This study clearly indicated that chronic Schistosoma mansoni infection attenuates the severity of P. knowlesi co-infection by mechanisms that may enhance innate immunity to malaria (Nyakundi et al. Reference Nyakundi, Nyamongo, Maamun, Akinyi, Mulei, Farah, Blankenship, Grimberg, Hau, Malhotra, Ozwara, King and Kariuki2016).
The rhesus monkey is uniformly susceptible to disease and infections are generally lethal, if left untreated. However, a limited set of data shows that sporozoite-induced infections can be milder (Richards et al. Reference Richards, Mitchell, Butcher and Cohen1977). While some papers have postulated that rhesus monkeys infected with P. knowlesi show no disease symptoms prior to death (Butcher et al. Reference Butcher, Mitchell and Cohen2010), our own experience (unpublished) and other data (Ibiwoye et al. Reference Ibiwoye, Howard, Sibbons, Hasan and van Velzen1993; Chen et al. Reference Chen, Li, Lu and Luo2001) show that monkeys can display apathy, loss of appetite, lethargy, dehydration, fever and in some cases, display raised fur. Some hallmarks of severe disease in rhesus monkeys are discussed below.
Cytoadherence-mediated sequestration
While a critical factor in P. falciparum severe malaria in humans is accepted to be cytoadherence-mediated sequestration (White et al. Reference White, Turner, Day and Dondorp2013), this may not necessarily be the case for P. knowlesi human severe malaria (Daneshvar et al. Reference Daneshvar, Davis, Cox-Singh, Rafa'ee, Zakaria, Divis and Singh2009; Cox-Singh et al. Reference Cox-Singh, Hiu, Lucas, Divis, Zulkarnaen, Chandran, Wong, Adem, Zaki, Singh and Krishna2010). A number of endothelial markers, such as ICAM-1, VCAM and CD36, have been linked to cytoadherence-mediated sequestration in human P. falciparum severe malaria studies (Helms et al. Reference Helms, Dasanna, Schwarz and Lanzer2016). In P. knowlesi severe malaria, infected erythrocytes from five human subjects have been shown to bind in a specific but variable manner to the inducible endothelial receptors ICAM-1 and VCAM (three isolates bound to ICAM-1 and VCAM, one isolate bound to VCAM and one isolate did not bind to ICAM-1 or VCAM), while binding to the constitutively expressed endothelial receptor CD36 was not detected (Fatih et al. Reference Fatih, Siner, Ahmed, Woon, Craig, Singh, Krishna and Cox-Singh2012).
In the P. knowlesi-rhesus monkey model, reports detailing the sequestration phenotype of infected erythrocytes in severe disease are limited and there has been debate on whether sequestration in this model is due to accumulation or cytoadherence. While all post-mortem light microscopy examinations documented in literature identified marked cerebral vascular congestion and widespread plugging of the brain capillaries and venules (micro-vessels) by heavily parasitized erythrocytes mixed with uninfected erythrocytes, light microscopy alone cannot properly document cytoadherence. Electron microscopy on rhesus monkey brains after infection with the P. knowlesi W1 strain showed major changes including adherence of large numbers of parasitized erythrocytes and macrophages to swollen microvasculature endothelial cells (Ibiwoye et al. Reference Ibiwoye, Howard, Sibbons, Hasan and van Velzen1993), increased number of fibroblasts and the deposition of collagen bundles in the extracellular matrix around damaged capillaries, parasite-packed micro-vessels and ischaemic hypoxia in several parts of the brain (Ibiwoye et al. Reference Ibiwoye, Sibbons, Hasan, Howard, Desalu, Singhal and van Velzen1995). Parasitic infiltration of all regions of the central nervous system and cytoadherence in some regions were further confirmed by an independent electron microscopy study (Mahdi & Ahmad, Reference Mahdi and Ahmad1991). The latter already postulated in 1991 that a triad of mechanisms is involved in the etiology of cerebral malaria pathogenesis, namely mechanical obstruction, biochemical events promoted by free radicals and immunological dysfunction mediated by activated macrophages, which increase lipid peroxidation.
Early studies attributed P. knowlesi sequestration and the obstruction of cerebral capillaries in non-human primates to decreased deformability of P. knowlesi-infected erythrocytes (Miller et al. Reference Miller, Usami and Chien1971). However, as reviewed in Galinski & Corredor (Reference Galinski and Corredor2004), studies with clonal populations of P. knowlesi expressing different variant surface antigens (SICAvar), indicate a role for some SICAvar antigens in sequestration. SICAvar genes in P. knowlesi and P. falciparum erythrocyte membrane protein 1 (Pfemp1) genes in P. falciparum malaria share a number of fundamental features relating to their role in antigenic variation mediated immune evasion. While the role of P. falciparum Pfemp1 genes in sequestration has been detailed in a number of studies, the role of SICAvar in sequestration has been much less clear. The newly emerging data, suggesting a definite involvement of some SICAvar in sequestration, has led Galinski & Corredor (Reference Galinski and Corredor2004) to conclude that, while antigenic variation mediated immune evasion is a primary function in common between the P. knowlesi and P. falciparum variant antigens, the cytoadherent properties of such variant antigens could be viewed as a secondary adaptation, which has evolved as a much stronger characteristic in P. falciparum than in P. knowlesi, where sequestration is only partial.
Concluding remarks on studying parasite–host interactions using P. knowlesi-non-human primate models
While it is well accepted that both host and parasite factors influence severe malaria pathogenesis (Cunnington et al. Reference Cunnington, Riley and Walther2013) and that the etiology of human severe malaria is dependent on the interplay between genetic background, immune response and parasite virulence, many P. knowlesi-rhesus monkey studies have been insufficiently documented to draw firm conclusions on the model. In a few cases the parasite strain is not mentioned and most often the origin of the rhesus monkeys is not well defined. Together with the parasite strain, the origin of the host represents one of the key factors in the characterization of a host–parasite interaction model. For example, the P. knowlesi H strain's sequestration profile in Indian rhesus monkeys is more pronounced than that of the Nuri strain (CHMK & AVW, unpublished observation). Similarly, P. knowlesi H and C strains infections in M. fascicularis of Malayan origin will result in death, while those of Philippine origin can control the parasitaemia (Schmidt et al. Reference Schmidt, Fradkin, Harrison and Rossan1977; Flynn et al. Reference Flynn, Satkoski, Lerche, Kanthaswamy and Smith2009), indicating that the genetic background of the monkeys is important for the course of infection. For rhesus monkeys, mitochondrial typing characterized different origins (Burmese, Chinese and Indian) (Doxiadis et al. Reference Doxiadis, Otting, de Groot, de Groot, Rouweler, Noort, Verschoor, Bontjer and Bontrop2003) and differential pathophysiological manifestations for other infectious diseases have been recorded in rhesus monkeys from various origins (Langermans et al. Reference Langermans, Andersen, van Soolingen, Vervenne, Frost, van der Laan, van Pinxteren, van den Hombergh, Kroon, Peekel, Florquin and Thomas2001). Such studies for P. knowlesi infections would be very informative in relation to the display of parasite–host interactions, including severe malaria symptoms. Similarly, some early non-human primate pilot studies, e.g. severe malaria studies in Callithrix jacchus (Langhorne & Cohen, Reference Langhorne and Cohen1979), are worth following up with sufficient numbers of animals to reach statistical significance in case of differential disease course, using state-of-the-art technology and taking the current knowledge on severe P. knowlesi malaria in humans into account (Cox-Singh et al. Reference Cox-Singh, Hiu, Lucas, Divis, Zulkarnaen, Chandran, Wong, Adem, Zaki, Singh and Krishna2010).
PLASMODIUM KNOWLESI AS A MODEL FOR VACCINE STUDIES
Gaining an understanding of immune responses against malaria using P. knowlesi
The P. knowlesi-rhesus monkey model has been extensively used in vaccine research. Besides testing vaccine concepts, it has also provided valuable information on the development of anti-malarial immune responses and parasite–host interactions following infection that formed the basis for human studies. In 1937, Coggeshall and Kumm showed that rhesus monkeys could be protected against a blood stage challenge with P. knowlesi upon passive immunization with serum from chronically P. knowlesi-infected rhesus monkeys (Coggeshall & Kumm, Reference Coggeshall and Kumm1937). This paved the way for the classical work of Cohen et al. (Reference Cohen, Mc and Carrington1961), who demonstrated that passive transfer of antibodies from hyperimmune people living in endemic areas protected young children against malaria. In hyperendemic areas, people develop immunity only upon prolonged repeated exposure to malaria parasites. In part this slow development of immunity has been attributed to the ability of malaria parasites to evade the host's immune response by undergoing repeated antigenic variation. This important phenomenon was first described in P. knowlesi. In a seminal study by Brown & Brown (Reference Brown and Brown1965), it was shown that antigens expressed on the surface of infected red blood cells changed during the course of infection in chronically infected rhesus monkeys. These antigens, later termed schizont infected cell agglutination (SICA) antigens (Howard et al. Reference Howard, Barnwell and Kao1983), also played a role in parasite sequestration, as described in the previous section. The identification and biochemical characterization of the P. knowlesi SICA antigens led to a similar line of experimentation to identify homologous antigens with similar characteristics in P. falciparum, which, in turn, led to the discovery of the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) variant protein family (reviewed in (Galinski & Corredor, Reference Galinski and Corredor2004)).
A non-human primate vaccine model relevant for humans
Rhesus monkeys have been widely used for vaccine studies, mainly because in line with their phylogenetic proximity to humans, they share many relevant immunological and biological features (Messaoudi et al. Reference Messaoudi, Estep, Robinson and Wong2011). In rhesus monkeys, P. knowlesi produces a fulminating fatal parasitaemia if left untreated (Table 1), representing a rigorous model for vaccine testing. Many immunological reagents developed for human studies cross-react with rhesus monkey cells and are available to characterize the immune responses induced in these animals in detail. In addition, the genome of the rhesus monkey has been sequenced allowing for in-depth genome-wide analyses of protective immunological pathways (Zimin et al. Reference Zimin, Cornish, Maudhoo, Gibbs, Zhang, Pandey, Meehan, Wipfler, Bosinger, Johnson, Tharp, Marcais, Roberts, Ferguson, Fox, Treangen, Salzberg, Yorke and Norgren2014). Together with the opportunity to dissect immunological processes in situ, by examination of relevant organs, this renders the P. knowlesi-rhesus monkey model as a unique model in the search for correlates of protection. This model may even be further expanded in the future with the development of in vivo imaging approaches for non-human primates infected with malaria parasites to allow real-time characterization of interactions between host immune cells and parasites in different parts of the body (Beignon et al. Reference Beignon, Le Grand and Chapon2014). As reviewed below, transfection technology for P. knowlesi is already well established, enabling the production of marker parasites for this purpose.
The availability of a primate-host combination with immunological features that are relevant for the human situation renders P. knowlesi in the rhesus monkey an attractive parasite for malaria vaccine studies.
Blood stage vaccination
Most of the early vaccination studies with P. knowlesi parasites were blood stage oriented. Taking advantage of the relatively large size and robustness of P. knowlesi merozoites, Dennis et al. (Reference Dennis, Mitchell, Butcher and Cohen1975) developed a sieving method to isolate free live merozoites. The availability of P. knowlesi merozoites enabled groundbreaking studies on the invasion process of the red blood cell (Bannister et al. Reference Bannister, Butcher and Mitchell1977; Aikawa et al. Reference Aikawa, Miller, Johnson and Rabbege1978). It also provided the opportunity for P. knowlesi vaccination studies using freshly isolated or freeze-dried merozoite preparations in Freund's adjuvant. Immunizations with these preparations elicited sterile protection against challenge with blood stage parasites (Mitchell et al. Reference Mitchell, Butcher and Cohen1975, Reference Mitchell, Butcher, Langhorne and Cohen1977) and this immunity was shown to involve merozoite blocking antibodies (Butcher et al. Reference Butcher, Mitchell and Cohen1978), eventually leading to the identification and characterization of one of today's major blood stage candidate vaccines, the conserved Apical Membrane Antigen-1 (Deans et al. Reference Deans, Alderson, Thomas, Mitchell, Lennox and Cohen1982; Peterson et al. Reference Peterson, Marshall, Smythe, Crewther, Lew, Silva, Anders and Kemp1989; Remarque et al. Reference Remarque, Faber, Kocken and Thomas2008). An early proof-of-concept vaccine study with PK66/AMA-1 affinity purified from P. knowlesi blood stage schizonts showed only marginal effects after challenge with blood stage P. knowlesi. However, following a booster vaccination, protective effects were observed after re-challenge with blood stage P. knowlesi (Deans et al. Reference Deans, Knight, Jean, Waters, Cohen and Mitchell1988). More recently, the PK66/AMA-1 antigen was expressed in yeast and vaccine efficacy of this antigen, formulated in a novel adjuvant co-vaccine HT, was assessed following P. knowlesi blood stage challenge (Mahdi Abdel Hamid et al. Reference Mahdi Abdel Hamid, Remarque, van Duivenvoorde, van der Werff, Walraven, Faber, Kocken and Thomas2011). Similar to the earlier study, this showed only modest protective effects (1 out of 6 animals controlled the parasitaemia; 4 out of 6 animals showed a small delay in onset of parasitaemia and 1 animal was not protected). A subsequent booster vaccination followed by a re-challenge showed significant improvement of protective effects: 4 out of 6 monkeys controlled P. knowlesi parasitaemia, 1 monkey had a delayed onset of parasitaemia and 1 animal was not protected. Furthermore, it was shown that in order to achieve protection, high levels of inhibitory antibodies were required and purified IgG isolated from the vaccinated monkeys showed in vitro parasite growth inhibition that correlated with protection (Mahdi Abdel Hamid et al. Reference Mahdi Abdel Hamid, Remarque, van Duivenvoorde, van der Werff, Walraven, Faber, Kocken and Thomas2011). This study showed that AMA-1, as a single subunit vaccine can confer protection against malaria in a highly stringent model, underlining the importance of AMA-1 as a malaria vaccine candidate, and thus supporting clinical development of this antigen from P. falciparum as P. falciparum vaccine component. However, in order to achieve sterile protection repeated infection and booster immunization were needed, indicating that a vaccination scheme that involves multiple immunizations is necessary. Alternatively, a combination vaccine involving multiple antigens, for example inclusion of another conserved vaccine candidate MSP119, may improve protective effects, a strategy that is currently investigated for P. falciparum (Faber et al. Reference Faber, Younis, Remarque, Rodriguez Garcia, Riasat, Walraven, van der Werff, van der Eijk, Cavanagh, Holder, Thomas and Kocken2013).
Multi-stage vaccination strategies
Over the past 15 years a number of P. knowlesi vaccine studies have been performed in which combinations of antigens were used for vaccination, using prime-boost approaches. This strategy not only involved multi-antigen combinations, but also antigens targeting different stages of the parasite life-cycle using a DNA vaccination strategy that offers the flexibility to easily express a combination of antigens. Building on success in murine models (Sedegah et al. Reference Sedegah, Hedstrom, Hobart and Hoffman1994, Reference Sedegah, Jones, Kaur, Hedstrom, Hobart, Tine and Hoffman1998, Reference Sedegah, Weiss, Sacci, Charoenvit, Hedstrom, Gowda, Majam, Tine, Kumar, Hobart and Hoffman2000), this strategy was also tested in the P. knowlesi-rhesus monkey model (Rogers et al. Reference Rogers, Baird, Kumar, Tine, Weiss, Aguiar, Gowda, Gwadz, Kumar, Gold and Hoffman2001). The vaccine strategy included three immunizations with a DNA vaccine containing two pre-erythrocytic stage antigens, the circumsporozoite protein (PkCSP) and sporozoite surface protein 2 (PkSSP2) and two blood stage antigens, apical membrane antigen 1 (PK66/AMA1) and merozoite surface protein 1 (PkMSP1p42), followed by a booster immunization with recombinant canarypox virus encoding the four antigens (ALVAC-4). This regimen did induce antibody and T-cell responses and provided partial protection against sporozoite challenge. One out of the 12 experimental monkeys was completely protected and the mean parasitaemia in the remaining monkeys was significantly reduced compared with control monkeys (Rogers et al. Reference Rogers, Baird, Kumar, Tine, Weiss, Aguiar, Gowda, Gwadz, Kumar, Gold and Hoffman2001). However, the levels of immune responses and protection were significantly lower than those observed in the Plasmodium yoelii mouse model (Sedegah et al. Reference Sedegah, Jones, Kaur, Hedstrom, Hobart, Tine and Hoffman1998, Reference Sedegah, Weiss, Sacci, Charoenvit, Hedstrom, Gowda, Majam, Tine, Kumar, Hobart and Hoffman2000). This could be the result of differences in the experimental set-up between the studies. In the murine model a single antigen was used, CSP, in contrast to the multistage cocktail that was used in the monkey study, potentially resulting in antigenic competition leading to lower protective effects. However, modest levels of immunity were also reported in human trials (Wang et al. Reference Wang, Epstein, Baraceros, Gorak, Charoenvit, Carucci, Hedstrom, Rahardjo, Gay, Hobart, Stout, Jones, Richie, Parker, Doolan, Norman and Hoffman2001; Epstein et al. Reference Epstein, Charoenvit, Kester, Wang, Newcomer, Fitzpatrick, Richie, Tornieporth, Heppner, Ockenhouse, Majam, Holland, Abot, Ganeshan, Berzins, Jones, Freydberg, Ng, Norman, Carucci, Cohen and Hoffman2004) in which, analogous to the murine study, CSP was used as the single antigen. Different levels in responsiveness to DNA vaccination in mice and (non-)human primates may be a factor in this, but it may also be the result of the different vaccination procedures. Boosting in the murine model was performed with an attenuated vaccinia virus, in the rhesus monkey model with recombinant canarypox virus and in the human studies with either DNA or protein. Taken together, these data provided a rationale for further work with the P. knowlesi-rhesus monkey model aiming at dissection of the immunological mechanisms and optimization of the vaccination regimen. In a later study (Rogers et al. Reference Rogers, Weiss, Kumar, Aguiar, Tine, Gwadz, Harre, Gowda, Rathore, Kumar and Hoffman2002), it was shown that the use of attenuated vaccinia virus rather than canarypox for boosting dramatically improved protection against challenge with P. knowlesi sporozoites. Two out of 11 monkeys showed sterile protection and 7 of the 11 monkeys spontaneously resolved their blood stage parasitaemia. In an attempt to further improve this, various vaccination strategies were tested in a head-to-head comparison (Jiang et al. Reference Jiang, Shi, Conteh, Richie, Banania, Geneshan, Valencia, Singh, Aguiar, Limbach, Kamrud, Rayner, Smith, Bruder, King, Tsuboi, Takeo, Endo, Doolan, Richie and Weiss2009). However, none of the new vaccine components, including viral replicon particles and recombinant Adenovirus-5 could prime immune responses as effectively as DNA plasmid priming. Strikingly however, high levels of sterile protection were observed in the group that received the DNA prime/pox virus boost regimen with increased time intervals between the vaccination dosages. In this group 3 out of 5 monkeys never developed parasitaemia after sporozoite challenge and 1 out of the 5 monkeys spontaneously resolved its blood stage parasitaemia. Cellular (Jiang et al. Reference Jiang, Shi, Conteh, Richie, Banania, Geneshan, Valencia, Singh, Aguiar, Limbach, Kamrud, Rayner, Smith, Bruder, King, Tsuboi, Takeo, Endo, Doolan, Richie and Weiss2009) and antibody responses (Hamid et al. Reference Hamid, Remarque, El Hassan, Hussain, Narum, Thomas, Kocken, Weiss and Faber2011) were observed, but correlates of protection could not be found. Notwithstanding the impressive protective effects induced in this highly stringent model, all P. knowlesi DNA prime/pox virus booster vaccine studies failed to demonstrate protection in a second challenge 4 months later, indicating that further optimization is needed to obtain long-term protection. Alternatively, live-attenuated parasite vaccine strategies may be more suited to accomplish long-term sterile protection.
Plasmodium knowlesi live attenuated whole-organism vaccination
Over the past few years, significant progress has been made in the field of whole-organism malaria vaccine approaches (Hoffman et al. Reference Hoffman, Billingsley, James, Richman, Loyevsky, Li, Chakravarty, Gunasekera, Chattopadhyay, Li, Stafford, Ahumada, Epstein, Sedegah, Reyes, Richie, Lyke, Edelman, Laurens, Plowe and Sim2010; Epstein et al. Reference Epstein, Tewari, Lyke, Sim, Billingsley, Laurens, Gunasekera, Chakravarty, James, Sedegah, Richman, Velmurugan, Reyes, Li, Tucker, Ahumada, Ruben, Li, Stafford, Eappen, Tamminga, Bennett, Ockenhouse, Murphy, Komisar, Thomas, Loyevsky, Birkett, Plowe, Loucq, Edelman, Richie, Seder and Hoffman2011; Roestenberg et al. Reference Roestenberg, Teirlinck, McCall, Teelen, Makamdop, Wiersma, Arens, Beckers, van Gemert, van de Vegte-Bolmer, van der Ven, Luty, Hermsen and Sauerwein2011) and some important hurdles in the development of a P. falciparum attenuated sporozoite vaccine for humans have been overcome (Richie et al. Reference Richie, Billingsley, Sim, James, Chakravarty, Epstein, Lyke, Mordmuller, Alonso, Duffy, Doumbo, Sauerwein, Tanner, Abdulla, Kremsner, Seder and Hoffman2015). In order to dissect the immune responses elicited by vaccination with attenuated whole-organisms in a non-human primate model, Weiss and Jiang (Weiss & Jiang, Reference Weiss and Jiang2012) immunized 9 rhesus monkeys with radiation attenuated P. knowlesi sporozoites. They found sterile protection in 5 out of 9 monkeys after challenge with P. knowlesi sporozoites. After treating 3 of these monkeys with a monoclonal antibody that removed CD8+ lymphocytes from the circulation, the monkeys all developed blood stage parasitaemia after re-challenge with sporozoites. After 4 months rest, CD8+ lymphocytes had re-appeared in the blood and upon a third challenge all monkeys were protected again. This indicates an important role for CD8+ T-cell responses in eliciting protection against malaria with live-attenuated pre-erythrocytic vaccine approaches. This type of response may be more prominent inside the liver as was recently shown by Ishizuka et al. (Reference Ishizuka, Lyke, DeZure, Berry, Richie, Mendoza, Enama, Gordon, Chang, Sarwar, Zephir, Holman, James, Billingsley, Gunasekera, Chakravarty, Manoj, Li, Ruben, Li, Eappen, Stafford, K, Murshedkar, DeCederfelt, Plummer, Hendel, Novik, Costner, Saunders, Laurens, Plowe, Flynn, Whalen, Todd, Noor, Rao, Sierra-Davidson, Lynn, Epstein, Kemp, Fahle, Mikolajczak, Fishbaugher, Sack, Kappe, Davidson, Garver, Bjorkstrom, Nason, Graham, Roederer, Sim, Hoffman, Ledgerwood and Seder2016) who demonstrated that in rhesus monkeys vaccinated with attenuated P. falciparum sporozoites, P. falciparum-specific interferon-γ-producing CD8+ T-cells were present at ±100-fold higher frequencies in liver than in blood. P. falciparum does not develop into blood stage parasites in rhesus monkeys, precluding challenge studies that can directly correlate immune responses in the liver to P. falciparum vaccine efficacy. The P. knowlesi-rhesus monkey model is well suited to fill this knowledge-gap, because in this model both protective effects induced by vacci-nation as well as underlying immunological mechanisms inside the liver can be studied. The recent establishment of a protocol for using infective mosquitoes to challenge rhesus monkeys with P. knowlesi (Murphy et al. Reference Murphy, Weiss, Fryauff, Dowler, Savransky, Stoyanov, Muratova, Lambert, Orr-Gonzalez, Zeleski, Hinderer, Fay, Joshi, Gwadz, Richie, Villasante, Richardson, Duffy and Chen2014) now allows for testing efficacy of P. knowlesi pre-erythrocytic vaccination strategies using the natural route of infection, thereby avoiding potential over/underestimation of vaccine effects due to artificial challenging.
The development of new technologies (such as transfection technologies and continuous long-term in vitro cultures) contributed to the use of P. knowlesi as a model parasite and dramatically increased the versatility of the P. knowlesi-non-human primate model for vaccine, drug and also more basic biological studies, as detailed below.
TECHNOLOGICAL DEVELOPMENTS IN PLASMODIUM KNOWLESI STUDIES
Long-term continuous in vitro cultures
Short-term in vitro cultures of P. knowlesi blood stage parasites in rhesus monkey red blood cells were first described by Ball et al. (Reference Ball, Anfinsen, Geiman, McKee and Ormsbee1945), but only in 2002 a P. knowlesi parasite line derived from P. knowlesi H strain (PkHcc), supporting continuous in vitro growth was developed (Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002). When cultured parasites were injected back into a monkey, the parasitaemia did not exceed 0·2% in the first acceptor monkey. The secondary recipient developed fulminant parasitaemia again (Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002), and these parasites could be grown in vitro without any adaptation period. This shows that this parasite can be shuttled from in vitro culture to in vivo infections with relative ease. The continuous blood stage culture reduced primate use for studying blood stage parasites, as rhesus monkey infections are not needed anymore as a source for blood stage parasites. However, during the adaptation to continuous in vitro culture, the PkHcc strain has lost its ability to form gametocytes, precluding transmission from in vitro cultures. PkHcc in monkeys did not regain gametocyte production, indicating that the gametocytogenesis pathway is irreversibly damaged in these parasites (Kocken et al. Reference Kocken, Zeeman, Voorberg-van der Wel and Thomas2009). Thus, for transmission and liver stage studies, monkey infections with gametocyte forming P. knowlesi strains are still necessary (Fig. 1).
Although the P. knowlesi H strain parasite that is most extensively used for research was originally isolated from a naturally infected person in 1965 (Chin et al. Reference Chin, Contacos, Coatney and Kimball1965), the adaptation of P. knowlesi to in vitro blood stage cultures in human red blood cells has been a challenge, and P. knowlesi research was restricted to facilities that have access to macaque blood.
Recently, Moon et al. (Reference Moon, Hall, Rangkuti, Ho, Almond, Mitchell, Pain, Holder and Blackman2013) have achieved a breakthrough by adapting the P. knowlesi A1 strain to growth in human red blood cells (RBC). The researchers started with a stock taken from a rhesus monkey and adapted the parasite to in vitro culture in 100% M. fascicularis RBC. After 2 months of culture, the blood was changed to 80% human and 20% M. fascicularis RBC and the parasite was cultured in this RBC-mix for 10 months. The mixture of RBCs was then replaced with 100% human RBC and 17 months after the start of the experiment, a clonal parasite line was generated, capable of growing both in M. fascicularis as well as in human red blood cells. This made in vitro culture of P. knowlesi accessible to laboratories without access to macaque blood. Unfortunately P. knowlesi A1-H had already lost the ability to form gametocytes before adaptation to growth in human RBC, so this line is not suited for transmission and liver stage research. In parallel to Moon, Lim et al. (Reference Lim, Hansen, DeSimone, Moreno, Junker, Bei, Brugnara, Buckee and Duraisingh2013) adapted P. knowlesi H strain to in vitro culture with human RBC. Before adaptation, this strain had a strong preference for young human RBC and cultures could only be maintained when 8% of the human RBC consisted of reticulocytes. Adaptation of P. knowlesi H to long-term in vitro culture in human RBC was achieved by culturing the parasite in a mixture of 90% human RBC and 10% rhesus monkey RBC for a month, followed by culturing in 100% human RBC for 4 months, after which a stable parasite line was achieved that was able to grow at normal rate in human RBC. The preference for reticulocytes disappeared during the adaptation process. Gametocyte production is not reported in Lim et al. (Reference Lim, Hansen, DeSimone, Moreno, Junker, Bei, Brugnara, Buckee and Duraisingh2013). It will be very interesting to compare the genomes of the two strains to find which adaptations were needed for the parasites to survive in human RBC cultures and whether they have adapted along similar paths. With the P. knowlesi in vitro culture technology available, scientists now have the opportunity to perform functional analysis of especially the genes that are selectively found in the P. vivax clade (±90 genes) (Frech & Chen, Reference Frech and Chen2011), to which P. knowlesi belongs. In that perspective, we will summarize the transfection technologies available for P. knowlesi.
Plasmodium knowlesi transfection technology
With the P. knowlesi genome fully sequenced (Pain et al. Reference Pain, Bohme, Berry, Mungall, Finn, Jackson, Mourier, Mistry, Pasini, Aslett, Balasubrammaniam, Borgwardt, Brooks, Carret, Carver, Cherevach, Chillingworth, Clark, Galinski, Hall, Harper, Harris, Hauser, Ivens, Janssen, Keane, Larke, Lapp, Marti, Moule, Meyer, Ormond, Peters, Sanders, Sanders, Sargeant, Simmonds, Smith, Squares, Thurston, Tivey, Walker, White, Zuiderwijk, Churcher, Quail, Cowman, Turner, Rajandream, Kocken, Thomas, Newbold, Barrell and Berriman2008) and the blood stage cultures in human RBC achieved, transfection technology is elemental to study the function of P. vivax clade-specific genes as well as the many hypothetical genes present in all malaria species including P. knowlesi.
The first Plasmodium transfection was performed in Plasmodium gallinaceum gametes and zygotes (Goonewardene et al. Reference Goonewardene, Daily, Kaslow, Sullivan, Duffy, Carter, Mendis and Wirth1993), followed by episomal transfection of P. falciparum (Wu et al. Reference Wu, Sifri, Lei, Su and Wellems1995) and stable transfection of Plasmodium berghei blood stages (van Dijk et al. Reference van Dijk, Waters and Janse1995). A versatile transfection system was also developed for P. knowlesi, initially in vivo (van der Wel et al. Reference van der Wel, Tomas, Kocken, Malhotra, Janse, Waters and Thomas1997) and subsequently using in vitro adapted parasites in rhesus monkey red blood cells (Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002).
The first P. knowlesi transfection (van der Wel et al. Reference van der Wel, Tomas, Kocken, Malhotra, Janse, Waters and Thomas1997) was performed with infected red blood cells taken from a rhesus monkey, which were injected back into a recipient monkey after the transfection procedure (using Biorad electroporation in incomplete Cytomix (Wu et al. Reference Wu, Sifri, Lei, Su and Wellems1995)) (Fig. 1). Five years later, a long-term P. knowlesi in vitro culture was genetically modified side by side with in vivo parasites using double crossover recombination to target the CSP gene. Double crossover integration is preferred over single crossover integration, as a more stable genotype can be achieved because the insert can no longer loop out. The resulting transgenic parasite strains were analysed in vitro (Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002) except for the transmission phenotype, which, due to lack of gametocyte production, could not be tested in the in vitro adapted strain. The transmission phenotype (lack of sporozoite production in mosquito midgut oocysts) confirmed the earlier observed csp knockout phenotype from rodent malaria (Menard et al. Reference Menard, Sultan, Cortes, Altszuler, van Dijk, Janse, Waters, Nussenzweig and Nussenzweig1997; Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002).One of the major advantages of in vitro cultures is the possibility to clone the parasite after transfection, to verify that any phenotype fits the genotype. To date only few genes have been genetically modified in P. knowlesi, either through knockout, overexpression or tagging. Table 2 provides a comprehensive overview, including observed phenotypes, if any. Limited use of the P. knowlesi system is most likely due to the fact that up to recently the in vitro culture was restricted to facilities that had access to macaque blood. Together with the adaptation of P. knowlesi to human red blood cells (Moon et al. Reference Moon, Hall, Rangkuti, Ho, Almond, Mitchell, Pain, Holder and Blackman2013), transfection methodologies for P. knowlesi have been refined and optimised to obtain high efficiency, both for episomal and genome integration transfections (Moon et al. Reference Moon, Hall, Rangkuti, Ho, Almond, Mitchell, Pain, Holder and Blackman2013). The transfection efficiency of P. knowlesi achieved by Moon is outperforming the transfection efficiency of P. berghei, which was by far the most efficient malaria transfection system available (Janse et al. Reference Janse, Franke-Fayard, Mair, Ramesar, Thiel, Engelmann, Matuschewski, Gemert, Sauerwein and Waters2006). For an excellent recent review covering the available molecular genetic systems in malaria see de Koning-Ward et al. (Reference de Koning-Ward, Gilson and Crabb2015). In conclusion, the P. knowlesi transfection system is versatile, in terms of vitro-vivo shuttling, which can be essential for the detection of certain phenotypes that are not displayed in vitro (e.g. overexpression of IFN-γ (Ozwara et al. Reference Ozwara, Langermans, Kocken, van der Wel, van der Meide, Vervenne, Mwenda and Thomas2003a ) could have impact on host immune response), and it is highly efficient and suitable for both episomal transfection as well as single and double crossover integration into the genome.
Table 2. Chronological overview of genetic modifications in P. knowlesi

Although the tools are available, only few genetic modifications have been reported.
a Performed in P. knowlesi adapted to culture in human red blood cells.
b Parasite modification is briefly mentioned in the section Methods, but no experiments described using the transgenic parasite.
TgDHFR, T. gondii-mutant dhfr gene conferring resistance to pyrimethamine; CSP, circumsporozoite protein; KO, knockout; CO, crossover; IFN-γ, interferon gamma; GFP, green fluorescent protein; rRNA-ssu D-type ribosomal RNA small subunit; hDHFR human mutant DHFR gene conferring resistance to pyrimethamine and WR99210; Bsd, Blasticydin S deaminase, confers resistance against blasticydin; Neo, neomycin phsphotransferase II, confers resistance against neomycin; TK, thymidine kinase, negative selection marker which sensitises for ganciclovir; DBP, duffy binding protein; hRBC, human red blood cells; CDP-DAG, cytidine diphosphate diacyglycerol synthase; Pf, Plasmodium falciparum; MyoB, myosin B; MLC-B, myosin light chain-B.
Although current transfection efficiencies for P. knowlesi parasites are relatively high, further improvements are expected to ensue from the application of the CRISPR/Cas tool for gene-editing of human cells that was developed by Doudna and Charpentier in 2012 (Jinek et al. Reference Jinek, Chylinski, Fonfara, Hauer, Doudna and Charpentier2012) and reviewed in Doudna & Charpentier (Reference Doudna and Charpentier2014). Up to very recently, double crossover integration into the P. falciparum genome could only be achieved by positive and negative drug selection (Duraisingh et al. Reference Duraisingh, Triglia and Cowman2002), while currently, P. falciparum (Ghorbal et al. Reference Ghorbal, Gorman, Macpherson, Martins, Scherf and Lopez-Rubio2014; Wagner et al. Reference Wagner, Platt, Goldfless, Zhang and Niles2014) and P. yoelii (Zhang et al. Reference Zhang, Xiao, Jiang, Zhao, Li, Gao, Ling, Wei, Li, Lu, Su, Cui and Yuan2014) mutants can easily be generated using the CRISPR/Cas tool. Applying CRISPR/Cas technology in a high-throughput like fashion to P. knowlesi parasites could rapidly give insight into the ±90 genes that seem to be specific for vivax-type parasites (Frech & Chen, Reference Frech and Chen2011).
Concluding remarks
In this review the P. knowlesi-rhesus monkey model was shown to be a valuable tool for the evaluation of vaccine approaches for P. falciparum. Plasmodium knowlesi is increasingly recognized as causative agent for disease in humans in South East Asia. Its transmission in humans has been reported in all countries in Southeast Asia except Laos. In fact, P. knowlesi is responsible for the majority of malaria cases in Malaysia (Singh & Daneshvar, Reference Singh and Daneshvar2013). The increasing incidence of P. knowlesi infections in humans demands measures aimed at reducing this burden. It should be possible to translate the obtained knowledge from animal models directly to human P. knowlesi vaccine development, speeding up this process.
The possibilities offered by the combination of what was already a powerful model system supplemented with innovative technologies (including the emerging CRISPR/Cas transfection technology), provide for the continuous attractiveness of the P. knowlesi-non-human primate/in vitro model. Well-designed and controlled studies will increase our knowledge on the biology of specific malaria parasite features, parasite–host interactions and on immunological mechanisms behind successful vaccination strategies. The exploitation of genomic (Pain et al. Reference Pain, Bohme, Berry, Mungall, Finn, Jackson, Mourier, Mistry, Pasini, Aslett, Balasubrammaniam, Borgwardt, Brooks, Carret, Carver, Cherevach, Chillingworth, Clark, Galinski, Hall, Harper, Harris, Hauser, Ivens, Janssen, Keane, Larke, Lapp, Marti, Moule, Meyer, Ormond, Peters, Sanders, Sanders, Sargeant, Simmonds, Smith, Squares, Thurston, Tivey, Walker, White, Zuiderwijk, Churcher, Quail, Cowman, Turner, Rajandream, Kocken, Thomas, Newbold, Barrell and Berriman2008; Zimin et al. Reference Zimin, Cornish, Maudhoo, Gibbs, Zhang, Pandey, Meehan, Wipfler, Bosinger, Johnson, Tharp, Marcais, Roberts, Ferguson, Fox, Treangen, Salzberg, Yorke and Norgren2014), proteomic (Pasini et al. Reference Pasini, Kirkegaard, Mortensen, Mann and Thomas2010) and metabolomic (Salinas et al. Reference Salinas, Kissinger, Jones and Galinski2014) methods as well as bio-imaging are key to further our understanding of malaria parasite–host interactions in general and the severe malaria syndrome in all its facets in particular (Cunnington et al. Reference Cunnington, Riley and Walther2013).
ACKNOWLEDGEMENTS
We thank Francisca van Hassel for preparing figure 1.
FINANCIAL SUPPORT
Some of the work presented in this review was supported by the following funding: EC EVIMalaR contract 242095; EC EMVDA contract LSPH-CT-2007-037506; EC SysMalVac contract 305869 and NWO Computational Life Sciences grant number 600·635·100·08N28 (RUMPHI).






