Published online by Cambridge University Press: 09 October 2003
Two recognized strains of Schistosoma intercalatum, one from the Democratic Republic of Congo (DRC), formerly Zaire, and the other from Cameroon, have been investigated using DNA sequences from 3 mitochondrial genes, cytochrome oxidase subunit 1 (cox1), NADH dehydrogenase subunit 6 (nad6) and the small ribosomal RNA gene (rrnS). In addition, partial DNA sequences from the nuclear large subunit ribosomal RNA gene (lsrDNA) were included within the study. Although partial lsrDNA alone reveals little taxonomic information, phylogenetic analysis of the mitochondrial data demonstrates a clear dichotomy between the 2 purported strains and it is proposed that they should be treated as distinct taxa. The ‘original’ S. intercalatum now falls relatively basal in the S. haematobium group, while the proposed new species is more derived and sister taxon to S. bovis and S. curassoni.
Two strains of Schistosoma intercalatum have previously been recognized; one from Lower Guinea (Cameroon, Equatorial Guinea, Gabon, Nigeria and São Tomé; Southgate, Rollinson & Kaukas (1994); Wright, Southgate & Knowles (1972)) and the other from the Democratic Republic of Congo (DRC: Upper Zaire River and Kinshasa; Fisher (1934); Tchuem Tchuenté, Southgate & Vercruysse (1997)). The molluscan intermediate host for the former is Bulinus forskalii whilst for the latter it is B. globosus (Wright et al. 1972; Frandsen, 1978). The parasite strains differ from each other in a number of features including: pre-patent periods in the intermediate and definitive hosts (Wright et al. 1972; Bjørneboe & Frandsen, 1979), egg morphology (Wright et al. 1972; Frandsen, 1978), intermediate host–parasite relationships (Wright et al. 1972), and characteristics for certain isoenzyme systems (Wright, Southgate & Ross, 1979; Brown et al. 1984). The features distinguishing the 2 geographically isolated strains of S. intercalatum have traditionally been regarded as the result of divergence between 2 allopatric populations rather than the existence of 2 cryptic species.
Given that the strains are supposedly the same species, it might be anticipated that resultant crosses should remain viable over a significant number of generations. However, Frandsen (1978) conducted hybridization experiments between the 2 strains of S. intercalatum and found that it was only possible to obtain F2 cercariae. More recently, Pagès et al. (2002), using different isolates from those employed by Frandsen (1978), have demonstrated that the intraspecific cross between the 2 strains, is only viable until the F3 adult generation but with low cercarial productivity. F4 cercariae proved non-infective to mice.
Pagès et al. (2001 a) examined the RAPD profiles from males and females of both strains and found that the 2 isolates could be differentiated unambiguously. Taken in conjunction with the other differing biological characters between the latter, Pagès et al. (2001 a) support the concept that they are 2 distinct species rather than strains of the same species.
The current study seeks to contribute to the molecular data previously obtained by Pagès et al. (2001 a) in order to resolve the phylogenetic position of these S. intercalatum ‘strains’ with respect to each other and also to additional species within the genus Schistosoma. This was achieved by sequencing 3 mitochondrial genes from each isolate used. In order to best utilize the results from a recent work on the phylogeny of the Schistosomatidae (Lockyer et al. 2003) a large section of the cytochrome oxidase subunit 1 (cox1) gene was sequenced along with NADH dehydrogenase subunit 6 (nad6), the small ribosomal RNA gene (rrnS) and also the nuclear large subunit ribosomal RNA gene (lsrDNA).
Two isolates of S. intercalatum from Kinshasa, DRC (DRC-A and DRC-C) and 2 from Edea, Cameroon (CAM-B and CAM-D) were selected for study, together with a further S. intercalatum isolate from San Antonio, São Tomé (ST-E). For comparative analysis, 4 other species from the S. haematobium group, S. haematobium, S. curassoni, S. mattheei and S. bovis were chosen as well as S. mansoni (see Table 1). DNA from individual adult, male and female worms was extracted according to the method outlined by Walker, Rollinson & Simpson (1986) with minor modification. Worms were digested in 10 μl of DNA extraction buffer (50 mM Tris–HCl (pH 8·0), 50 mM EDTA, 100 mM NaCl), with 1% lauryl sulphate, sodium salt (SDS) and 4 μl of proteinase K at a concentration of 20 mg/ml. Following digestion for 2 h at 37 °C, the digest was purified with 2 phenol/chloroform (50[ratio ]50) and 1 chloroform extraction. Subsequent to precipitation in absolute ethanol and a wash in 70% ethanol, the DNA pellet was dried for 10 min at 94 °C and re-suspended in 20 μl of deionized water.
Table 1. Details of isolates and species used in the study (Perp is used to indicate that the isolates are held in the collections at Perpignan.)

Amplifications were performed using Amersham Pharmacia Biotech ‘Ready-To-Go’ PCR beads containing 1·5 units of Taq DNA polymerase, 10 mM Tris–HCl (pH 9·0), 50 mM KCl, 1·5 mM MgCl2, 200 μM of each dNTP and stabilizers, including BSA. Each PCR reaction contained approximately 80 ng of template DNA and 25–50 pmol of oligonucleotide, depending on the primer used (see Tables 2 and 3). Amplification of the cox1 fragment was achieved using the following cycling parameters, 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C for 1 min/52 °C for 1 min/72 °C for 2 min and a final cycle at 72 °C for 10 min. The nad6 was amplified using 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C for 1 min/50 °C for 1 min/72 °C for 1, 2 or 3 min, depending on the expected size of the fragment and a final cycle of 72 °C for 10 min. The rrnS fragments were produced using 1 cycle of 5 min at 94 °C, 30 cycles at 94 °C for 1 min/50 °C for 1 min/72 °C for 2 or 3 min depending on the anticipated fragment size and a final cycle of 72 °C for 10 min. The lsrDNA fragments for all S. intercalatum isolates and S. haematobium were generated using cycling parameters of 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C for 1 min/50 °C for 1 min/72 °C for 2 min and 1 cycle at 72 °C for 10 min. The remaining species had the same cycling parameters except that annealing was accomplished at 58 °C with a 1 min extension phase. PCR products were purified using a Qiaquick PCR purification kit (Qiagen). The nad6, rrnS and lsrDNA fragments were all sequenced directly, whilst the cox1 fragments were cloned into pGEM-T Easy Vector (Promega) for subsequent sequencing. All sequencing reactions were performed using Fluorescent Dye Terminator Sequencing Kits (Applied Biosystems) and the sequencing reactions run on either an Applied Biosystems 377 or 373A ‘XL Stretch’ automated sequencer.
Table 2. Oligonucleotide primers used for the PCR amplification of DNA fragments in this study (sequences 5′-3′) (Lower case letters indicate polylinker sites for directional cloning.)


The gene, cox1 was chosen in order to utilize the results of a recent study on the phylogeny of the Schistosomatidae (Lockyer et al. 2003). The current data set contained sequences of 11 species of Schistosoma plus the 5 new and 1 published sequence of S. intercalatum, rooting the trees against S. mansoni and S. rodhaini. The data sets of nad6 and rrnS each contained 5 species of Schistosoma plus the 5 new sequences of S. intercalatum, rooting the trees against S. mansoni. Partial lsrDNA sequences of the D2 variable domain were also obtained for 5 isolates of S. intercalatum and compared with taxa where sequences for each of the 3 mitochondrial genes were available (Fig. 1B).

Fig. 1. (A) Phylogeny reconstructed from cox1 sequence data using maximum likelihood (ML). Numbers above each branch represent the bootstrap values for ML (n=100) while those below each branch are for maximum parsimony (MP) (n=1000). (B) Phylogeny reconstructed from combined cox1+nad6+rrnS sequence data; ML (n=100) and MP (n=1000) solutions are presented above and below branch lines respectively.
Protein coding regions cox1 and nad6 were readily alignable with reference to their open-reading frame and inferred amino acid sequences using MacClade ver. 4.03 (Maddison & Maddison, 2000), employing the rhabditophoran genetic code of Telford et al. (2000). ClustalX (Jeanmougin et al. 1998) was employed initially to align rrnS, using default parameters and then subsequently refined by eye, excluding ambiguously aligned positions. Individual gene alignments were concatenated in MacClade and data partitions defined.
Maximum parsimony (MP) and maximum likelihood (ML) analyses were performed using PAUP* ver. 4.0b10 (Swofford, 2002) and the resulting networks rooted with the outgroup taxon. Each gene was analysed both independently and combined using MP and ML. All genes were analysed only as nucleotides. For both protein-coding genes, third codon (synonymous) positions were removed prior to analysis as these sites were found to be saturated. The latter was demonstrated by plotting pair-wise sequence difference or p-dist (the proportion (p) of nucleotide sites at which the two sequences compared are different; see Kumar et al. (2001)) against the number of differences between isolates/species in terms of transitional or transversional substitutions.
For all ML analyses suitable nucleotide substitution models were estimated using Modeltest (Posada & Crandall, 1998). Subsequent analyses used a heuristic search strategy and tree-bisection-reconnection (TBR) branch-swapping options. Analyses by MP were performed using the branch-and-bound strategy ensuring that all tree space was sampled. All characters were run unordered and equally weighted. Gaps were treated as missing data. Nodal support was assessed by bootstrap resampling in MP (1000 replicates) and ML (100 replicates). In order to test whether there was significant conflict between the data partitions prior to combining them the criteria of conditional combination of independent data sets (Cunningham, 1997; Huelsenbeck, Bull & Cunningham, 1996) were examined using the incongruence length-difference (Farris et al. 1995) test as implemented in PAUP*. The test was performed with maximum parsimony, 10 heuristic searches (random sequence addition, TBR branch-swapping) each for 100 homogeneity-replicates on informative sites only (Lee, 2001).
Sequences have been submitted to the EMBL database with the following accession numbers: cox1, AJ519515–AJ519524, nad6, AJ416894–AJ416904, rrnS, AJ419779–AJ419789 and lsrDNA, AJ519525–AJ519529.
Regarding the full cox1 data set, including 22 taxa, there were 1134 unambiguously alignable sites, of which 635 were constant and 87 phylogenetically informative under parsimony. For the reduced data set, including only those taxa where all 3 genes had been sequenced (10 taxa), of the 1134 included sites, 656 were constant and 42 phylogenetically informative under parsimony.
For the reduced taxon set, nad6 (codon positions 1 and 2 only) provided a total of 280 unambiguously alignable sites, of which 151 were constant and 55 phylogenetically informative under parsimony. The rrnS data set provided a total of 757 unambiguously alignable sites, of which 613 were constant and only 39 phylogenetically informative under parsimony. The partition homogeneity test indicated that these independent data sets were compatible with one another (P=0·900), and may be combined under the principles of conditional combination.
As might be expected from data sets containing so few informative positions, trees obtained from analysing nad6 and rrnS were largely unresolved. However, both MP and ML for each gene demonstrated that isolates of S. intercalatum from the DRC grouped strongly to the exclusion of those from West Africa. The results of both data sets are shown in Fig. 1. The phylogeny estimated from the full cox1 data set includes 22 taxa (Fig. 1A) and the phylogeny estimated from the reduced taxon set has 10, but includes the combined data from each of the three genes (Fig. 1B).
ML and MP analyses for cox1 (Fig. 1A) resolved compatible tree topologies. MP resolved 4 equally parsimonious trees (length 211; CI=0·673; RI=0·773). Only the ML tree is shown, which was identical to one of the MP trees, with nodal support from each analysis. The ML model selected by Modeltest was K81+I+G; where base frequencies were A=0·2025, C=0·1391, G=0·2343, T=0·4241, the rate matrix (K81) was 14·7285 (A-G, C-T), 2·7364 (A-T, C-G) and 1·0000 (A-C, G-T), the proportion of invariable sites (I) was 0·7241, and the gamma distribution shape parameter (G) was 1·0925. Mitochondrial cox1 provides strong evidence for the separation of the West African and DRC isolates of S. intercalatum into 2 species. The phylogeny of Schistosoma follows closely that of Lockyer et al. (2003) apart from the S. intercalatum DRC strain which was not included in their study. With the African species S. mansoni and S. rodhaini used to root the topology, S. nasale, S. indicum and S. spindale form a monophyletic group at the base of the tree. The DRC strains of S. intercalatum form a clade that is the sister group to the remaining taxa. Although the grouping of certain taxa, such as S. mattheei and S. margrebowiei as sister groups, is equivocal there is strong nodal support for the S. haematobium group to the exclusion of the DRC isolates of S. intercalatum. Indeed, the West African isolates of S. intercalatum are strongly associated with S. bovis and S. curassoni. A fuller analysis, including nuclear genes, indicates that the West African isolates form the sister group to S. bovis+S. curassoni (Lockyer et al. 2003).
In order to test further whether the isolates of S. intercalatum formed 2 statistically distinct clades, a constraint analysis was performed under ML, constraining all the isolates of S. intercalatum as monophyletic. The constrained tree and the unconstrained tree were then subjected to a Shimodaira-Hasegawa test (Shimodaira & Hasegawa, 1999) as implemented in PAUP* with full optimization and 1000 bootstrap replicates. Results indicated that a tree with the S. intercalatum isolates held to be monophyletic (−lnL=2154·97) was significantly different (P=0·012) from the unconstrained solution shown (−lnL=2137·68) in Fig. 1A.
The partition homogeneity test indicated that the 3 genes cox1, nad6 and rrnS could be combined for a total evidence estimate of phylogeny (P=0·90). With the reduced taxon set employing all genes MP found 2 equally parsimonious trees (length 1245; CI=0·783; RI=0·668). One of these was identical to the ML solution shown. The ML model selected by Modeltest was HKY+I+G; where base frequencies were A=0·2640, C=0·1113, G=0·2234, T=0·4013, the transition/transversion ratio under the Hasegawa-Kishino-Yano (HKY) 2-parameter substitution model was 3·3793, the proportion of invariable sites (I) was 0·5487, and the gamma distribution shape parameter (G) was 0·6423. The relative placement of each taxon was identical to that shown for the cox1 analysis (Fig. 1A). High nodal support differentiated the DRC isolates of S. intercalatum as relatively basal taxa occupying a phylogenetic position between S. mattheei and the S. haematobium group taxa. Again, although the relative positions of S. bovis, S. curassoni and the West African isolates of S. intercalatum could not be resolved, it was clear that the 3 species form a strongly supported clade that is sister group to S. haematobium. As before, a constraint analysis on the full data set was run, employing a Shimodaira-Hasegawa test to see whether a tree holding the S. intercalatum isolates as monophyletic was significantly worse than the unconstrained solution. Again, the unconstrained solution, shown in Fig. 1B. (−lnL=4675·18) was significantly better (P=0·041) than the constrained solution (−lnL=4689·54).
In an attempt to mirror the work of Lockyer et al. (2003) partial lsrDNA sequences including the D2 variable domain were obtained for the 5 new isolates of S. intercalatum. The data set for partial lsrDNA provided very little phylogenetic information with only 11 parsimony informative positions within the alignment. Phylogenetic analysis of this region produced poorly supported trees, although they were in accord with the evidence from the mitochondrial genes.
The term ‘species’ within Schistosoma is sometimes difficult to define with any precision. Species ‘X’ may or may not hybridize with species ‘Y’ to produce offspring. The viability of the hybrid offspring in terms of fecundity, cercarial productivity or the number of generations through which infectivity can be maintained, may depend on the polarity of the cross i.e. male of species ‘X’ with female of species ‘Y’ or the converse. For example, S. haematobium, a major pathogen of man, will naturally hybridize in humans with S. mattheei, the latter usually infecting wild bovines, with both species of schistosome sharing the same intermediate host B. globosus in areas where these parasites are sympatric (Wright & Ross, 1980). Similarly, 2 schistosome species infecting man, S. haematobium and S. intercalatum readily hybridize in nature to produce viable hybrid offspring (Southgate, van Wijk & Wright, 1976). Mutani, Christensen & Frandsen (1985) crossed female S. intercalatum (Edea, Cameroon) with male S. haematobium (Dar es Salaam, Tanzania) to produce hybrids which stayed viable at least until the F7 generation, the reverse pairing being less successful. Pagès et al. (2001 b) demonstrated that matings between S. intercalatum (Cameroon) and S. intercalatum (DRC) occurred in a random manner, clearly indicating that there is no pre-zygotic isolation mechanism if the two strains were sympatric. Pagès et al. (2002) confirmed the existence of a hybrid breakdown between S. intercalatum (Cameroon) and S. intercalatum (DRC) characterized by an impaired viability of larval offspring from the F2 generation onwards.
As a consequence of all these factors there has been uncertainty over the precise relationship of the Lower Guinea and DRC strains of S. intercalatum to each other and to the S. haematobium group as a whole. Based on the previous available evidence, the characterization of the 2 strains as a single species does not appear to fit comfortably into the existing taxonomic framework. The presentation of the current molecular data set, in combination with the work of Pagès et al. (2001 a) is hoped to resolve this ambiguous position.
A recent and comprehensive phylogeny of the Schistosomatidae including 17 of the 20 known Schistosoma species has done much to detail the taxonomy of these digenean flukes (Lockyer et al. 2003). The S. intercalatum strain used was from São Tomé and thus corresponds to the West African/Cameroon isolates presented here. The clade formed by S. bovis, S. curassoni and the West African form of S. intercalatum in the phylogenetic trees of Lockyer et al. (2003) is reflected in the current mitochondrial data set. However, from the latter it is now clear that there is also a significant dichotomy between the DRC and West African forms of this parasite with the former occupying a position that is basal relative to the other purported strain and separated by all the remaining members of the S. haematobium group. From evidence available at the time, Wright et al. (1972) concluded that the divergence of the DRC and Cameroon S. intercalatum strains was not recent. The early nodal branching of the DRC isolate would tend to support this hypothesis.
Thus, in conclusion, when the differing biological characters apparent between the 2 purported strains of S. intercalatum are considered in combination with the results of mitochondrial molecular analysis and the RAPD study of Pagès et al. (2001 a), a convincing case can be made that these allopatric isolates should be considered as distinct taxa. This will necessitate a redescription and naming of S. intercalatum (Lower Guinea).
The authors wish to thank Dr R. Stothard of the Department of Zoology, The Natural History Museum, London for his helpful advice regarding the analysis of data. The study received financial support from the CNRS (Sciences de la Vie) and from the Royal Society-CNRS research project. D. T. J. L. and A. E. L. were funded by a Wellcome Trust Fellowship (043965) to D. T. J. L.

Table 1. Details of isolates and species used in the study

Table 2. Oligonucleotide primers used for the PCR amplification of DNA fragments in this study (sequences 5′-3′)

Table 3. Primer pairs used to amplify the genes
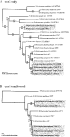
Fig. 1. (A) Phylogeny reconstructed from cox1 sequence data using maximum likelihood (ML). Numbers above each branch represent the bootstrap values for ML (n=100) while those below each branch are for maximum parsimony (MP) (n=1000). (B) Phylogeny reconstructed from combined cox1+nad6+rrnS sequence data; ML (n=100) and MP (n=1000) solutions are presented above and below branch lines respectively.