INTRODUCTION
Investigation of the relationship between parasite phylogeny and various biological traits can provide two important types of information. It serves as a background for understanding a parasite's evolutionary history, including evolution of the host-parasite interaction, and it indicates markers suitable for diagnostic and/or taxonomic purposes.
An immense amount of data and analyses were produced during the last few years for various groups of parasites (Page, Reference Page1996a; Barta et al. Reference Barta, Martin, Liberator, Dashkevicz, Anderson, Feighner, Elbrecht, Perkins-Barrow, Jenkins, Danforth, Ruff and Profous-Juchelka1997; Hoberg et al. Reference Hoberg, Brooks, Siegel-Causey, Clayton and Moore1997; Proctor, Reference Proctor1999; Jousson et al. Reference Jousson, Bartoli and Pawlowski2000; Paterson et al. Reference Paterson, Wallis, Wallis and Gray2000; Johnson and Clayton, Reference Johnson, Clayton and Page2001; Nieberding et al. Reference Nieberding, Morand, Libois and Michaux2004, Reference Nieberding, Libois, Douady, Morand and Michaux2005; Banks et al. Reference Banks, Palma and Paterson2006) including apicomplexan taxa, such as Adelina, Atoxoplasma, Cyclospora, Goussia, gregarines, etc. (Carreno et al. Reference Carreno, Martin and Barta1999, Eberhard et al. Reference Eberhard, Da Silva, Lilley and Pieniazek1999, Barta et al. Reference Barta, Martin, Carreno, Siddall, Profous-Juchelka, Hozza, Powles and Sundermann2001, Jirků et al. Reference Jirků, Modrý, Šlapeta, Koudela and Lukeš2002, Leander et al. 2006, Schrenzel et al. Reference Schrenzel, Maalouf, Gaffney, Tokarz, Keener, McClure, Griffey, McAloose and Rideout2005, Kopečná et al. Reference Kopečná, Jirků, Oborník, Tokarev, Lukeš and Modrý2006). Within coccidia, attention has been mainly given to the evolution of Sarcocystidae, the tissue cyst-forming coccidia with a typically heteroxenous life-cycle. A number of studies investigated the phylogenetic significance of specificity to intermediate and definitive hosts, or affinity to various tissues (Votýpka et al. Reference Votýpka, Hypša, Jirků, Flegr, Vávra and Lukeš1998; Doležel et al. Reference Doležel, Koudela, Jirků, Hypša, Oborník, Votýpka, Modrý, Šlapeta and Lukeš1999; Šlapeta et al. Reference Šlapeta, Modrý, Votýpka, Jirků, Lukeš and Koudela2003). For example, the species of the genus Sarcocystis were suggested to co-evolve with their final, rather than intermediate, hosts (Doležel et al. Reference Doležel, Koudela, Jirků, Hypša, Oborník, Votýpka, Modrý, Šlapeta and Lukeš1999; Holmdahl et al. Reference Holmdahl, Morrison, Ellis and Huong1999; Šlapeta et al. Reference Šlapeta, Modrý, Votýpka, Jirků, Lukeš and Koudela2003). Based on this finding, the final host is supposed to represent an ancestral type of the host within Sarcocystidae (Barta, Reference Barta1989; Tenter et al. Reference Tenter, Baverstock and Johnson1992).
Paradoxically, only few studies have been devoted to the largest group of coccidia, the monoxenous parasites of the genus Eimeria Schneider, 1875. Thus, evolution of various traits within this genus, such as morphology, host-specificity or pathogenicity, remains very unclear. Since Eimeria are typically parasites with extremely narrow host spectra, analysis of the dependence between phylogeny and host-specificity is relatively straightforward. The molecular data currently available show significant correlation between phylogenetic relationships and host-specificity; the genus splits into well-formed lineages, such as livestock-specific or fowl-specific species. This pattern, however, is not universal. For example, with an increasing number of available sequences, it became clear that rodent-associated fauna is composed of species from several lineages. While each of these lineages itself is monophyletic, their relationships are paraphyletic or even polyphyletic. Even single rodent species can harbour a set of parasites from different lineages. For example, certain species of the genera Peromyscus Gloger, 1841, Onychomys Baird, 1858 or Dipodomys Gray, 1841 were shown to host the parasite from at least 2 lineages (Zhao and Duszynski, Reference Zhao and Duszynski2001a, Reference Zhao and Duszynskib; Morrison et al. Reference Morrison, Bornstein, Thebo, Wernery, Kinne and Mattsson2004; Matsubayashi et al. Reference Matsubayashi, Takami, Niichiro, Kimata, Tani, Sasai and Baba2005).
An effort to put the morphological and biological features of Eimeria into a phylogenetic framework can be found in several studies; all of them focused on rodent species. First, Reduker et al. (Reference Reduker, Duszynski and Yates1987) recognized 2 different Eimeria lineages by cladistic and phenetic analyses of isozyme banding patterns, sporulated oocysts morphology and life-history traits. Similar results, based on phylogenetic analyses of molecular sequence and riboprinting data, were reported by Hnida and Duszynski (Reference Hnida and Duszynski1999a, Reference Hnida and Duszynskib). Another study by Zhao and Duszynski (Reference Zhao and Duszynski2001a, Reference Zhao and Duszynskib) determined the phylogenetic relationships among Eimeria species parasitizing 3 rodent families (Muridae, Heteromyidae and Cricetidae) based on plastid 23S rDNA, ORF470 and nuclear 18S rDNA sequences. Their results split the rodent Eimeria into 2 lineages according to the oocyst size/shape and the presence/absence of the oocyst residuum (OR).
The establishment of any concise evolutionary scenario across Eimeria is, however, limited by relatively scarce sampling of molecular data. Here, we focus on a so far neglected group of Eimeria, a group of rabbit-associated species. Of 65 Eimeria spp. described from 10 genera of lagomorphs (Duszynski and Upton, Reference Duszynski, Upton, Samuel, Pybus and Kocan2001), 11 parasitize the domestic rabbit, Oryctolagus cuniculus f. domesticus and also the wild Old World rabbit, Oryctolagus cuniculus (Coudert et al. Reference Coudert, Licois and Streun1979; Pakandl, Reference Pakandl1988; Coudert et al. Reference Coudert, Licois, Drouet-Viard, Eckert, Braun, Shirley and Coudert1995). The whole rabbit-specific fauna is relatively heterogeneous in respect to oocyst morphology, location within host or pathogenicity (Pellérdy, Reference Pellérdy1965; Coudert et al. Reference Coudert, Licois, Drouet-Viard, Eckert, Braun, Shirley and Coudert1995; Eckert et al. Reference Eckert, Braun, Shirley and Coudert1995). While 10 of the rabbit Eimeria species develop in the host intestine, E. stiedai parasitizes the epithelium of biliary ducts (Metzner, Reference Metzner1903; Minning, Reference Minning1936; Rutherford, Reference Rutherford1943). Some species are highly pathogenic, causing apathy, inapetency, severe growth depression, tympany, diarrhoea or even mortality (E. stiedai, E. flavescens, E. intestinalis); others express mild (E. magna, E. piriformis, E. irresidua, E. media) or low (E. coecicola, E. perforans, E. exigua, E. vejdovskyi) pathogenicity for their hosts (Pellérdy, Reference Pellérdy1965; Coudert et al. Reference Coudert, Licois and Streun1979; Licois and Mongin, Reference Licois and Mongin1980; Licois and Coudert, Reference Licois and Coudert1982; Vítovec and Pakandl, Reference Vítovec and Pakandl1989; Coudert et al. Reference Coudert, Licois, Drouet-Viard, Eckert, Braun, Shirley and Coudert1995; Pakandl and Coudert, Reference Pakandl and Coudert1999).
A number of studies have been published on rabbit coccidia. The majority of them were based on morphological and ultrastructural characters and focused on the descriptions of the life-cycle and selection of precocious lines of these parasites. The 3 molecular studies published thus far focused on diagnostics and inter- and intraspecific variation using RAPD techniques (Ceré et al. Reference Ceré, Licois and Humbert1995, Reference Ceré, Humbert, Licois, Corvione, Afanassieff and Chanteloup1996, Reference Ceré, Licois and Humbert1997). However, many questions concerning the rabbit-specific species can only be addressed with sufficient molecular data providing a clear phylogenetic signal. It has never been tested whether the rabbit-specific Eimeria form a monophyletic branch of species that has diversified on the rabbit hosts, or represent an assemblage of unrelated species colonizing the same host.
In this study, we present the first molecular data on rabbit-associated parasites to address the two following questions. Is the morphologically diverse group of rabbit-specific Eimeria a monophyletic branch or a polyphyletic assemblage of distant species? Is there a correlation between the phylogeny and any of the morphological/biological features of these species?
MATERIALS AND METHODS
Parasites
Eimerian oocysts were obtained from faeces of naturally infected domestic rabbits (Oryctolagus cuniculus f. domesticus) from various localities in the Czech Republic and France. Pure strains were prepared by a single-oocyst infection, or by a method previously described by Pakandl et al. (Reference Pakandl, Černík and Coudert2003). Briefly, about 20 individually collected oocysts examined by microscopy were administered into the oral cavity of a rabbit. The SPF (specific pathogen-free) rabbits originating from the Charles River Laboratories (Germany) and supplied by AnLab (Prague, Czech Republic) were used in the experiments.
Pure strains were obtained after 1 cycle of multiplication, and maintained in laboratory-reared rabbits. The purity of the strains was confirmed by microscopical examination. Oocysts were recovered and purified as described by Coudert et al. (Reference Coudert, Licois, Drouet-Viard, Eckert, Braun, Shirley and Coudert1995), and were sporulated in 2·5% potassium dichromate (K2Cr2O7) at room temperature on a shaker to ensure good aeration. Obtained oocysts were stored in potassium dichromate solution at ~4°C until used. All experiments comply with the current laws of the Czech Republic and were officially permitted.
DNA extraction, PCR and sequencing
Oocysts (approximately 3×105) were washed repeatedly by resuspension in sterile PBS, disrupted using a combination of glass-sand grinding and repetitive freeze-thaw cycles (liquid nitrogen – hot water) and incubated overnight on ice with lysis buffer (180 μl of NET 50, 24 μl of 30% sarcosin, 4 μl of pronase E). After digestion, total coccidian DNA was extracted using a standard phenol – chlorophorm protocol followed by ethanol precipitation.
Specific primers amplifying aproximately 1500 bp long region of 18S rDNA were designed according to published sequences of related Eimeria species. Their postion corresponded to the sites 62–86 (primer EF: 5′-GAAACTGCGAATGGCTCATT-3′) and 1568–1586 (primer ER: 5′-CTTGCGCCTACTAGGCATTC-3′) according to E. tenella sequence U67121. The PCR reaction was carried out in a 25 μl volume under standard conditions. The HotStarTaq DNA Polymerase (Qiagen) was used in all PCR reactions. The thermocycling protocol consisted of an initial inactivation-denaturation step at 95°C for 4 min, followed by 30 cycles of 92°C denaturation for 45 s, primer annealing at 53°C for 45 s, and elongation at 72°C for 90 s. A final primer extension of 10 min at 72°C completed the amplification process. Amplicons were directly cloned into the pGEM – T Easy Vector (Promega). Plasmids with DNA inserts of the expected size were isolated using a Plasmid Miniprep Spin Kit (Genomed). The sequencing of 5 clones from each sample was performed with the PCR primers and 4 internal primers using the Big Dye terminator cycle sequencing ready reaction kit (ABI Prism, Perkin Elmer) on an automatic sequencer ABI 3130xl. Obtained sequences were identified by BLAST analysis. The sequences were deposited in the GenBank database (NCBI) under the Accession numbers EF694007-EF694017.
Sequence alignment and phylogenetic analysis
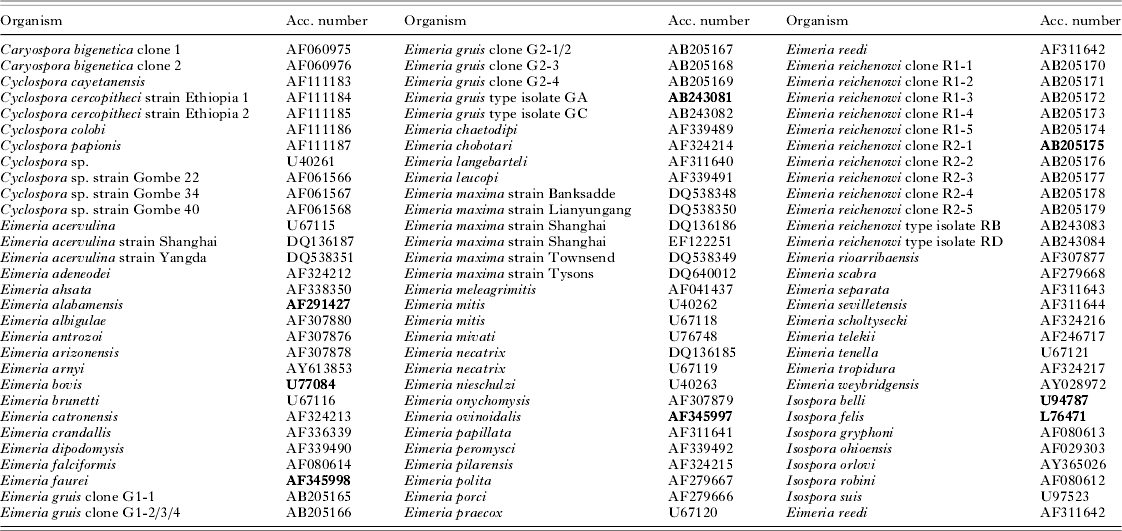
Sequences were assembled and edited using the DNASTAR program package (DNASTAR, Inc). The phylogenetic position and arrangement of the new sequences was investigated in 2 steps. First, we designed an Eimeria matrix to examine whether the rabbit-specific Eimeria formed a monophyletic branch. For this purpose, we downloaded Eimeria, Isospora, Caryospora and Cyclospora sequences of 18S rDNA currently available in GenBank (NCBI) (Table 1). The sequences were aligned in the MegAlign program (DNASTAR, Inc) using the ClustalW algorithm, and the matrix was analysed by fast neighbour-joining method (NJ) implemented in PAUP* version 4.0b10 (Swofford, Reference Swofford2001). Since several studies demonstrated that 18S rDNA is insufficient to establish deeper phylogenetic relationships among coccidian lineages (Olsen and Woese Reference Olsen and Woese1993; Mugridge et al. Reference Mugridge, Morrison, Jakel, Heckeroth, Tenter and Johnson2000; Tenter et al. Reference Tenter, Barta, Beveridge, Duszynski, Mehlhorn, Morrison, Thompson and Conrad2002), this step was only taken to verify that the rabbit-specific Eimeria form a monophyletic branch and can be further treated within taxonomically restricted context. After revealing the rabbit-specific Eimeria as a monophyletic cluster reliably characterized by long common branch, a Rabbit matrix was designed. It contained all sequences of rabbit-specific Eimeria, 6 eimerians from other hosts (E. alabamensis, E. bovis, E. faurei, E. gruis, E. ovinoidalis, E. reichenowi) and 2 outgroups, Isospora belli and I. felis (Table 1). This set of sequences was aligned in MegAlign program, manually adjusted in BioEdit (Hall, Reference Hall1999) and analysed by more precise methods than the first matrix. It resulted in alignment of 1518 positions, containing 164 parsimony informative sites. Maximum parsimony (MP) was carried out using an exhaustive branch-and-bound search mode. Analysis was repeated with transition/transversion ratios set to 1:1, 1:2 and 1:3. A consensus cladogram was calculated from all trees obtained under the different parameter settings. Bootstrap values were calculated by 1000 replicates of heuristic search with ts/tv ratio set to 1:1. A maximum likelihood (ML) tree was calculated in PHYML version 2.4.3 (Guindon and Gascuel, Reference Guindon and Gascuel2003) under the GTR+Γ+I model, with parameters of gamma distribution and proportion of invariable sites estimated from the data. The trees were visualized and exported using TreeView version 1.6.6. (Page, Reference Page1996b).
Table 1. Sequences included in the large Eimeria matrix
(Taxa used as outgroups for the Rabbit matrix are printed in bold.)

RESULTS
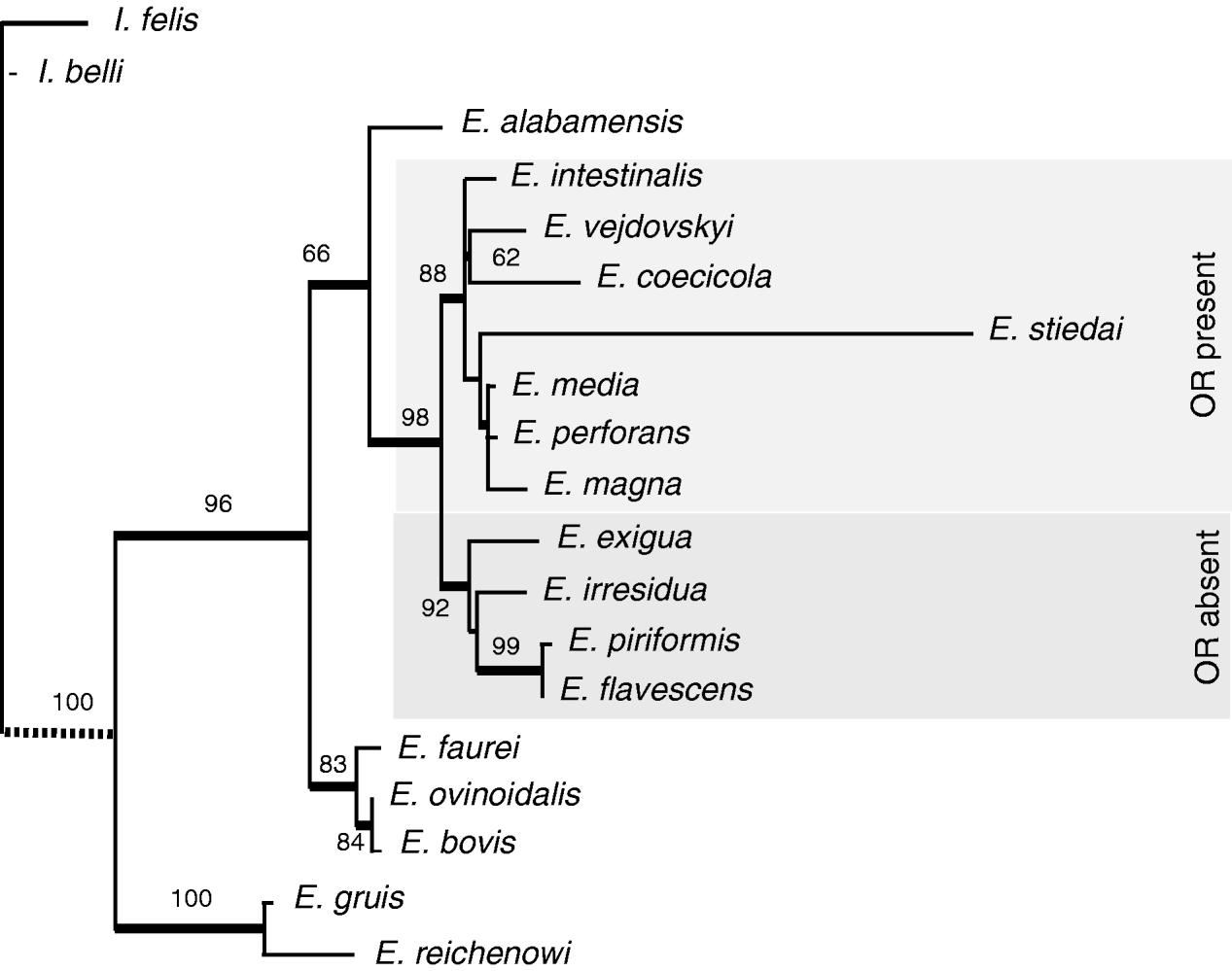
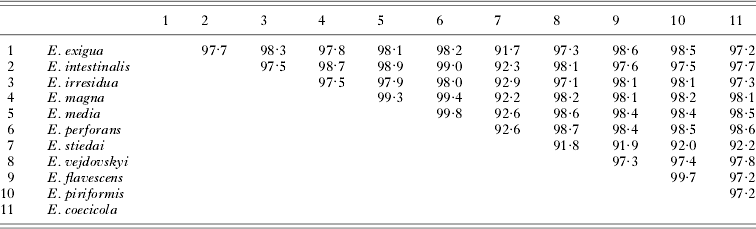
Partial sequences of 18S rDNA from 11 species of rabbit-infecting Eimeria were obtained. The sequences displayed a broad range of pairwise similarities, from 92% (e.g. E. stiedai vs E. flavescens) to more than 99% (E. piriformis vs E. flavescens) (Table 2). With 2 exceptions, the sequences were approximately of the same length (~1400 bp). The 2 exceptions were represented by E. coecicola, with a 20 bp long insertion, and particularly by E. stiedai, with an approximately 100 bp long deletion. When analysed in context of all Eimeria sequences available in GenBank, the rabbit-specific species formed a monophyletic cluster (result not shown; see the Materials and Methods section). This group was well-formed, possessing a relatively long common branch. The more restricted Rabbit matrix yielded several equally parsimonious trees; the number of the trees varied in relation to their ts/tv ratio (10, 26, 9). The trees exhibited high consistency indices (CI=0·79–0·81, RC=0·65–0·67) and their strict consensus produced a topology with several robust nodes supported by high bootstrap values (Fig. 1). The tree obtained by ML was entirely compatible with this topology. Of the morphological and biological characters (oocyst shape and size, presence/absence of oocyst inner structures, pathogenicity, site of infection, pre-patent period, patent period, sporulation time, number of asexual generations), mapped onto the topology, only the presence/absence of oocyst residuum strictly followed the phylogenetic division into 2 main branches (Fig. 1).

Fig. 1. A tree obtained by maximum parsimony analysis of the Rabbit matrix, with bootstrap values at the nodes. The branches preserved in the strict consensus of all MP trees and compatible with ML analysis are designated by bold lines. OR, oocyst residuum.
Table 2. Percentage similarity calculated for rabbit-associated species of Eimeria

DISCUSSION
Evolution of host specificity is one of the most intriguing questions of parasitological research. Amongst the coccidia, an important and diversified group of parasitic protists, Eimeria species are considered to exhibit high host specificity (Marquardt, Reference Marquardt, Hammond and Long1973; Joyner, Reference Joyner and Long1982; Long and Joyner, Reference Long and Joyner1984; Hnida and Duszynski, Reference Hnida and Duszynski1999a). Also, cyst-forming heteroxenous species (e.g. Sarcocystis) show significant host-specificity in respect to the intermediate and definitive host, as well as a strong site-specificity, e.g. affinity to the brain, muscles etc. (Votýpka et al. Reference Votýpka, Hypša, Jirků, Flegr, Vávra and Lukeš1998; Doležel et al. Reference Doležel, Koudela, Jirků, Hypša, Oborník, Votýpka, Modrý, Šlapeta and Lukeš1999; Šlapeta et al. Reference Šlapeta, Modrý, Votýpka, Jirků, Lukeš and Koudela2003).
The results of our phylogenetic analysis indicate that rabbit-specific Eimeria species form a monophyletic branch. This finding is in line with the ultimate phylogenetic significance of host-specificity within Eimeria in other studies (Morrison et al. Reference Morrison, Bornstein, Thebo, Wernery, Kinne and Mattsson2004; Matsubayashi et al. Reference Matsubayashi, Takami, Niichiro, Kimata, Tani, Sasai and Baba2005; Li et al. Reference Li, Xiao, Zhou, Li and Wadeh2007). It also implies that the 11 species with highly variable morphological characters diversified within a single host (or several closely related species at maximum). On the other hand, it should be recognized that in such a species-rich group as Eimeria, the study of its evolutionary history is usually highly compromised by the available samples. Thus, while more than 50 Eimeria sequences have so far been deposited into GenBank, this number represents only a small fraction of known species. The consequences of this problem can be easily demonstrated: for example, the fowl-infecting group of Eimeria is apparently monophyletic until the addition of Cyclospora species (Morrison et al. Reference Morrison, Bornstein, Thebo, Wernery, Kinne and Mattsson2004; Matsubayashi et al. Reference Matsubayashi, Takami, Niichiro, Kimata, Tani, Sasai and Baba2005).
An interesting aspect of the present study is a lack of congruence between phylogeny and most of the morphological and biological traits of the species. For example, the 2 most pathogenic species, E. intestinalis and E. flavescens, branch as very distant taxa; the latter particularly as one of the most derived offshots. At the same time, we have observed remarkable morphological and biological differences between genetically highly related species, supporting the ‘recent’ changes of some characteristics. Thus, while the 2 species with almost identical sequence of 18S rDNA, E. piriformis and E. flavescens, still share a unique location within the large intestine, they differ significantly in oocyst shape. Similarly, all of the 3 closely related species, E. media, E. magna and E. perforans, develop within the walls and tips of the small intestine villi, but only the first 2 possess a micropyle on their oocysts. Such a mosaic of phylogenetically conservative and versatile characteristics is likely to be a common phenomenon. An attempt to map biological and morphological traits on phylogeny within a monophyletic group of closely related eimerian species has previously been published for the fowl-specific group (Barta et al. Reference Barta, Martin, Liberator, Dashkevicz, Anderson, Feighner, Elbrecht, Perkins-Barrow, Jenkins, Danforth, Ruff and Profous-Juchelka1997). While authors of the study stress a generally high degree of phylogenetic congruence for various features, they also show that some characteristics may change rapidly. For instance, they report a shift of location for 3 related species, E. brunetti, E. maxima and E. praecox, and attribute it to a niche specialization process within the host. Within our set, the most striking example of uncoupled biology and phylogeny is the position of E. stiedai. Unlike all the other species inhabiting the host intestine, E. stiedai infects bile ducts (Pellérdy, Reference Pellérdy1965; Pellérdy and Dürr, Reference Pellérdy and Dürr1970; Eckert et al. Reference Eckert, Braun, Shirley and Coudert1995), and has also been reported in hares (Lepus europaeus Pallas, 1778) (Varga, Reference Varga1976; Entzeroth and Scholtyseck, Reference Entzeroth and Scholtyseck1977; Sugár et al. Reference Sugár, Murai and Mészáros1978; Scholtyseck et al. Reference Scholtyseck, Entzeroth and Pellérdy1979). Moreover, the structure of its OR is highly unusual, incompact, consisting of 1–7 or more granules situated centrally in the oocyst (Norton et al. Reference Norton, Catchpole and Rose1977; Eckert et al. Reference Eckert, Braun, Shirley and Coudert1995). This fact even lead Pellérdy (Reference Pellérdy1965) to claim that this species does not possess an OR at all. In our matrix, E. stiedai formed a relatively long branch due to a 100 bp deletion and many autapomorphic substitutions along the sequence. Despite the unique biological and molecular features, this species branched firmly within the OR+cluster of rabbit-associated eimeria in all analyses, regardless of the method, program and parameters.
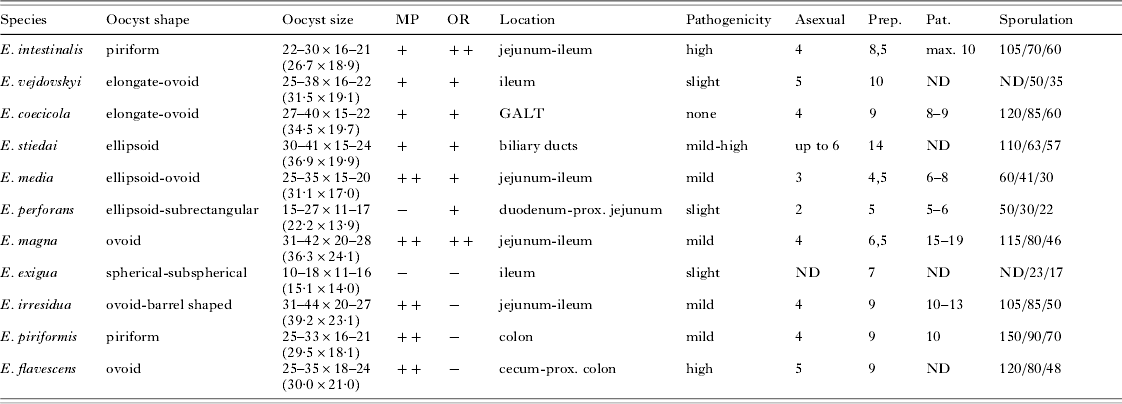
Interestingly, we identified a convincing correlation between the phylogenetic position of the species and presence/absence of an OR. This feature has previously been reported as the only phylogenetically consistent character differing in 2 lineages of rodent Eimeria (Zhao and Duszynski, Reference Zhao and Duszynski2001a, Reference Zhao and Duszynskib). In our analysis, the OR+ and OR- groups form 2 monophyletic sister lineages. It is therefore impossible to decide which state is plesiomorphic for rabbit Eimeria. Also, using light microscopy, we did not observe any further correlation between the inner phylogeny of the OR+ group and OR characteristics, such as size, shape or structure. Other morphological and biological characters (oocyst shape, size and presence/absence of oocyst inner structures, pathogenicity, pre-patent period, patent period, sporulation time, number of asexual generations) showed no influence on the phylogeny (Fig. 1; Table 3).
Table 3. Morphological and biological traits of the rabbit-associated species shown on Fig. 1
(The characteristics are adopted from the following descriptions: Eckert et al. Reference Eckert, Braun, Shirley and Coudert1995; Pakandl et al. 1996 a,b,c, Reference Pakandl, Černík and Coudert2003; Pakandl and Coudert, Reference Pakandl and Coudert1999; Pakandl and Jelínková, 2006. Oocyst size is provided in μm (average size in parentheses). MP, micropyle; OR, oocyst residuum; GALT, gut-associated lymphoid tissue (i.e. Peyer's patches, sacculus rotundus, vermiform appendix); ND, not determined; Asexual, number of asexual generations; Prep., pre-patent period (days); Pat., patent period (days); Sporulation time is provided as hours at 18°C/22°C/26°C.)

Since the presence/absence of OR seems to be an evolutionarily conserved feature, it would be interesting to determine the phylogenetic relationships of the rabbit-specific cluster to other OR+ groups. However, this was beyond the scope of the present study. In addition, resolving the OR issue at a higher phylogenetic level is problematic for 2 reasons. First, as demonstrated by conflicting results of several phylogenetic studies (Morrison et al. Reference Morrison, Bornstein, Thebo, Wernery, Kinne and Mattsson2004; Matsubayashi et al. Reference Matsubayashi, Takami, Niichiro, Kimata, Tani, Sasai and Baba2005; Li et al. Reference Li, Xiao, Zhou, Li and Wadeh2007), the 18S rDNA data currently available for analysis may be riddled with inconsistencies, or at least difficulties when extracting a phylogenetic signal. An overall sensitivity of the 18S rDNA dataset to selection of a particular type of analysis and evolutionary model has been demonstrated in a thorough study of Morrison et al. (Reference Morrison, Bornstein, Thebo, Wernery, Kinne and Mattsson2004). Second, and more importantly, as mentioned above, poor sampling across all known Eimeria species makes such an attempt naive. We are convinced that a much more complete data set, in terms of host spectrum and sequenced genes, has to be available before the evolution of various traits within Eimeria can be seriously addressed.
We thank Ryan Rego for useful advice on laboratory procedures and Hassan Hashimi for English correction. This work was supported by grants 524/03/H133 (Grant Agency of the Czech Republic), 46/2006/P-BF (Grant Agency of the University of South Bohemia), LC06073 and MSM 60076605801 (Ministry of Education, Czech Republic).