Introduction
The Socotra Archipelago, located in the northwest Indian Ocean, is notable for its unique flora and fauna. It comprises four islands of continental origin, Socotra, Abd al Kuri, Samha and Darsa, with a total area of 3976 km2 (Fig. 1). The islands detached from Gondwana (more specifically from present-day Southern Oman) as the Arabian plate moved northward into Eurasia about 20 Mya (Autin et al., Reference Autin, Bellahsen, Leroy, Husson, Beslier and d'Acremont2013), and have been isolated from the mainland since then. Although less than 100 km away from the Horn of Africa, this archipelago is a governorate of Yemen. The ancient continental origin and geographical position of Socotra (in between Africa and Arabia), together with its climatic conditions and varied topography, have given rise to a remarkable endemic biodiversity (Van Damme, Reference Van Damme, Gillespie and Clague2009). Due to these high levels of endemism, Socotra was designated a Natural World Heritage site by UNESCO in 2008, and was included in the Horn of Africa biodiversity hotspot, one of the most threatened in the world (Mittermeier et al., Reference Mittermeier, Turner, Larsen, Brooks, Gascon, Zachos and Habel2011). Among vertebrates, Socotra has no amphibians and the endemic fauna includes probably one mammal species, 11 birds and 29 reptiles (Razzetti et al., Reference Razzetti, Sindaco, Grieco, Pella, Ziliani, Pupin, Riservato, Pellitteri-Rosa, Butikofer and Suleiman2011; Sindaco et al., Reference Sindaco, Metallinou, Pupin, Fasola and Carranza2012; Vasconcelos and Carranza, Reference Vasconcelos and Carranza2014). In fact, all native reptiles, with the exception of two recently introduced geckos, Hemidactylus robustus and Hemidactylus flaviviridis (Razzetti et al., Reference Razzetti, Sindaco, Grieco, Pella, Ziliani, Pupin, Riservato, Pellitteri-Rosa, Butikofer and Suleiman2011), are endemic to the archipelago. Reptiles therefore constitute one of its most relevant examples of endemism, with many species being the result of in situ radiation (Gómez-Díaz et al., Reference Gómez-Díaz, Sindaco, Pupin, Fasola and Carranza2012; Tamar et al., Reference Tamar, Simó-Riudalbas, Garcia-Porta, Santos, Llorente, Vasconcelos and Carranza2019), and five out of 12 genera of reptiles being endemic.

Fig. 1. Geographic location of the Socotra Archipelago and distribution of the reptiles screened for parasites for this study, including infected and non-infected hosts. Three groups of parasites were detected: haemogregarines (squares), sarcocystids (diamonds) and eimeriids (triangles). Colours indicate the parasite haplotypes identified based on the 18S rRNA gene. It should be noted that some points might overlap, please check Supplementary Table S1 for further information on the location of haplotypes.
While a recent study indicates that reptile diversity in the Socotra Archipelago might be under-estimated by 13.8–54.4% (Vasconcelos et al., Reference Vasconcelos, Montero-Mendieta, Simó-Riudalbas, Sindaco, Santos, Fasola, Llorente, Razzetti and Carranza2016), the situation is certainly worse for the microorganisms that inhabit them, such as blood and intestinal parasites. In particular, parasites of the phylum Apicomplexa are one of the most poorly studied organisms (Morrison, Reference Morrison2009). They are among the most frequently reported parasites of reptiles (Jacobson, Reference Jacobson2007). Previous genetic studies have revealed high levels of apicomplexan diversity in reptiles, especially in biodiversity hot spots and endemic communities (e.g. Harris et al., Reference Harris, Maia and Perera2012; Tomé et al., Reference Tomé, Maia and Harris2013, Reference Tomé, Rato, Perera and Harris2016, Reference Tomé, Pereira, Jorge, Carretero, Harris and Perera2018; Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016; Megía-Palma et al., Reference Megía-Palma, Martínez, Nasri, Cuervo, Martín, Acevedo, Belliure, Ortega, García-Roa and Selmi2016). Due to their isolation from the mainland and other characteristics, islands frequently have unique apicomplexan lineages (Harris et al., Reference Harris, Maia and Perera2011; Maia et al., Reference Maia, Crottini and Harris2014; Tomé et al., Reference Tomé, Pereira, Jorge, Carretero, Harris and Perera2018). However, interest in parasite communities goes beyond descriptive purposes. Parasites are also ideal models for studying biogeographic patterns and diversification of their insular hosts, and the effect of colonization and host population expansion on parasite traits, such as host-switching and specificity (Tomé et al., Reference Tomé, Pereira, Jorge, Carretero, Harris and Perera2018). For instance, when a parasite first arrives in a new habitat, it may remain restricted to its original host or adapt to use the new hosts it encounters, if it manages to arrive and successfully establish in the first place. In the long term, this may promote a diversity decrease, higher prevalence values and a larger niche (reflected in a lower host-specificity and more frequent host-switching events) than mainland species (collectively called as the ‘parasite island syndrome’; Nieberding et al., Reference Nieberding, Morand, Libois and Michaux2006). Understanding such processes is important to gain a better knowledge on inter-species interactions and the role of parasites in wild communities, but also on disease transmission dynamics.
In the current study, we screened tissue samples from 461 Socotran reptiles for the presence of apicomplexan parasites, namely haemogregarines, sarcocystids and eimeriids, which are frequently found infecting reptiles (e.g. Harris et al., Reference Harris, Maia and Perera2012; Tomé et al., Reference Tomé, Maia and Harris2013; Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016). Haemogregarines are blood parasites with heteroxenous life-cycles with a definitive invertebrate host, and an intermediate vertebrate host, such as reptiles (Telford, Reference Telford2009; Karadjian et al., Reference Karadjian, Chavatte and Landau2015). Sarcocystidae is a large family of parasites, which includes most notably the genus Sarcocystis. These parasites include two types of vertebrate hosts, often a predator and a prey (Duszynski and Upton, Reference Duszynski and Upton2009). Eimeriorina are intestinal parasites that mainly infect only one type of host (Upton, Reference Upton, Lee, Leedale and Bradbury2000). As exceptions, the reptile-infecting Lankesterella and Schellakia are transmitted by various hematophagous invertebrates, which act solely as paratenic hosts (Telford, Reference Telford2009). Traditionally, these genera have been grouped in the same family, Lankesterellidae, but recent phylogenetic studies have shown these to be polyphyletic (Megía-Palma et al., Reference Megía-Palma, Martínez, Nasri, Cuervo, Martín, Acevedo, Belliure, Ortega, García-Roa and Selmi2016). Even though the taxonomy of these groups is still under review, it is now accepted that Schellackia forms its own family, while Lankesterella is more closely related to some species of Isospora and Eimeria, members of the family Eimeriidae (Megía-Palma et al., Reference Megía-Palma, Martínez, Nasri, Cuervo, Martín, Acevedo, Belliure, Ortega, García-Roa and Selmi2016). The objective of our study was to assess the diversity and host associations of apicomplexan parasites to ascertain if, like their hosts, Socotran parasites presented endemic diversity and how the conditions on the archipelago affected their ecology.
Materials and methods
Sample collection
Between 2007 and 2014, a total of 461 reptiles were sampled across 200 localities in the four islands of the Socotra Archipelago (see Fig. 1 and Table 1). For each individual, a small tail tip was collected and preserved in 96% ethanol for subsequent molecular analysis. After processing, all individuals were released at the site of capture, which was registered using a GPS device (see Supplementary Table S1 for coordinates and screening information per locality). Sampling included 26 of the 31 reptile species occurring in the archipelago of nine families from the following reptile groups: snakes (n = 23 samples), geckos (n = 328), skinks (n = 41), lacertids (n = 59) and chamaeleonids (n = 10).
Table 1. List of reptile taxa screened for the three groups of apicomplexan parasites in the Socotra Archipelago

Their distribution (So, Socotra; Ab, Abd al Kuri; Sa, Samha; Da, Darsa; Ot, other localities in Africa, Arabia and Asia outside Socotra), sample sizes (n), number of locations sampled (nl), prevalence (P, number of infected individuals and %, percentage) and 18S rRNA haplotypes detected (h) are also given. The asterisk indicates the two introduced host species.
Molecular screening of parasites
DNA was extracted from tail-tip tissues using the Speedtools tissue DNA extraction kit (Biotools, Madrid, Spain) or the standard saline protocol (Sambrook et al., Reference Sambrook, Fritsch and Maniatis1989; Maia et al., Reference Maia, Crottini and Harris2014). Conventional PCR amplification was made using the primers HepF300 (5′-GTTTCTGACCTATCAGCTTTCGACG-3′) and HepR900 (5′-CAAATCTAAGAATTTC ACCTCTGAC-3′) (Ujvari et al., Reference Ujvari, Madsen and Olsson2004), targeting part of the 18S rRNA gene region. For each PCR, 20 μL reaction mixture was prepared containing 1 U of GoTaq® DNA Polymerase (5μ μL−1), 1.5 mm MgCl2 (25 mm), 0.125 mm of each nucleotide, 1X GoTaq® Flexi Buffer, 0.6 mm of each primer and 2 μL of DNA extraction. The reaction mix was then heated to 94 °C for 3 min, and amplification was performed through 35 cycles at 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min, followed by a final 10 min extension at 72 °C. Negative (ultrapure water) and positive controls (selected from KX453558–KX453648 for haemogregarines, KX453649–KX453660 for Lankesterella and KX453661–KX453662 for Sarcocystis) were included in each run. Lastly, the positive PCR products were purified and sequenced by a commercial sequencing facility (Macrogen Europe, Amsterdam, The Netherlands). Sanger sequencing was performed in both directions in all cases. Retrieved sequences were then manually inspected in Geneious 6.1.6 (www.geneious.com). We performed a similarity analysis with our sequences using the Basic Local Alignment Search Tool (BLAST) to find the best match against published sequences in GenBank (blast.ncbi.nlm.nih.gov/Blast.cgi). Heterozygous positions were identified in some individuals and the corresponding IUPAC codes were used. In the impossibility to clone the amplicons to investigate the intra-individual diversity of haplotypes, only good quality sequences with clear double peaks matching polymorphic positions previously identified in single infection haplotypes were considered as mixed infections. DnaSP v5 (Librado and Rozas, Reference Librado and Rozas2009) was used to estimate the number of haplotypes for non-heterozygous individuals.
All 18s samples matching sarcocystids were amplified with two additional markers: the nuclear gene 28S rRNA and the mitochondrial gene COI. The 28S rRNA gene was amplified using the primers CR1 (5′-CTGAAATTGCTGAAAAGGAA-3′) and CR2 (5′-CCAGCTACTAGATGGTTCGA-3′) (Zhu et al., Reference Zhu, Hartigan, Reppas, Higgins, Canfield and Šlapeta2009). Amplification of the COI gene was tested using a combination of primers SF1 (5′-ATGGCGTACAACAATCATAAAGAA-3′) and SR5 (5′-TAGGTATCATGTAACGCAATATCCAT-3′), and SF1 and COIRm (5′-CCCAGAGATAATACAAAATGGAA-3′) (Gjerde, Reference Gjerde2013). The PCR reaction protocols were the same as for the 18S rRNA, except for the annealing temperatures and times (55 °C for 30 s in the case of 28S rRNA, and 57 °C for 45 s in the case of COI). Amplification of the 28S rRNA worked for eight of the nine 18S rRNA sarcocystids positives, but for the COI, only four samples were successfully sequenced. Due to the low sequencing success rate and the small number of COI and 28S rRNA sequences available in GenBank for these parasite groups, only the 18S rRNA was used in subsequent phylogenetic analyses. However, we did compare the number of haplotypes and the genetic distances among the three genes (Supplementary Table S2). The new sequences were deposited in the GenBank database under the accession numbers MW076441–MW076453 for the 18S rRNA, MW076437–MW076440 for the 28S rRNA and MW074128–MW074129 for the COI, so they can be available for future phylogenetic studies.
Estimation of phylogenetic relationships and evolutionary divergences
Phylogenetic analyses were conducted separately for each apicomplexan group – haemogregarines, sarcocystids and eimeriids – following the same protocol. The obtained sequences were aligned with published GenBank sequences using MAFFT v7 algorithm (Katoh and Standley, Reference Katoh and Standley2013) with the Q-INS-i iterative method and applying the default parameters. The haemogregarine alignment was 579 base pairs (bp) long and included 197 sequences, seven of which were representative of the 35 from this study (Supplementary Table S3). The sarcocystid alignment was 706 bp long and contained 95 sequences, four representative of the nine from this study (Supplementary Table S4). Lastly, the eimeriid alignment was 619 bp long and included 91 sequences, two representative of the three from this study (Supplementary Table S5). The software jModelTest 2 was used to determine the best substitution model (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012): HKY + I + G for haemogregarines and sarcocystids, and GTR + I + G for eimeriids. Both Maximum Likelihood (ML) and Bayesian Inference (BI) phylogenetic analyses were conducted. ML analysis with random sequence addition (100 replicate heuristic searches) was used to assess evolutionary relationships in the software PhyML v3.0 (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010). Support for nodes was estimated using the bootstrap technique (Felsenstein, Reference Felsenstein1985) with 1000 replicates. Bayesian analysis was implemented using Mr. Bayes v3.2.6 (nbisweden.github.io/MrBayes/) with parameters estimated as part of the analysis. The analysis was run for 107 generations, saving one tree each 1000 generations. The log-likelihood values of the sample points were plotted against the generation time and all the trees prior to reaching stationarity were discarded, ensuring that burn-in samples were not retained. Remaining trees were combined in a 50% majority consensus tree (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001). For the haemogregarine alignment, Dactylosoma ranarum, Haemogregarina stepanowi and Haemogregarina balli were used as outgroups (based on Barta et al., Reference Barta, Ogedengbe, Martin and Smith2012). Eimeria maxima and Goussia janae were the outgroups for the sarcocystids (Wasserman et al., Reference Wassermann, Raisch, Lyons, Natusch, Richter, Wirth, Preeprem, Khoprasert, Ginting, Mackenstedt and Jäkel2017), while Eimeria steinhausi, Glossia noelleri and Gloussia neglecta were used for the eimeriid alignment (Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016).
Evolutionary divergences were estimated as the proportion of base substitutions per site between sequences (P-distances). These were calculated in MEGA 6 (www.megasoftware.net/), using the Maximum Composite Likelihood model (Tamura et al., Reference Tamura, Nei and Kumar2004). For the 18S rRNA gene, the sizes of the alignments used were 592 bp for haemogregarines, 543 bp for sarcocystids and 587 bp for eimeriids. Regarding sarcocystids, the genetic distances were also calculated for the 28S rRNA (608 bp alignment) and COI genes (981 bp).
Prevalence and host-specificity analyses
Differences in prevalence between types of parasites were calculated using the Fisher's exact test in R (fisher.test function, www.r-project.org). For haemogregarines only, further analyses were performed on prevalence differences across hosts and on host-specificity (sarcocystids and eimeriids were not considered given their low occurrence). To compare the levels of host-specificity between haemogregarine haplotypes, the STD and VarSTD indexes were calculated using the program TaxoBioDiv2 (Poulin and Mouillot, Reference Poulin and Mouillot2005). The STD considers the phylogenetic distance between host species to infer the specificity of parasites, and the lower its value the higher the specificity (equal to zero when the parasite occurs in a single host species). For this, the distance in a Linnaean taxonomic tree path length linking two host species (from Class, Order, Suborder, Family, Genus to Species) was used. The VarSTD measures the asymmetries in the distances between host species, for which the higher the value the higher the asymmetry.
Results
Parasite detection and prevalence differences between hosts
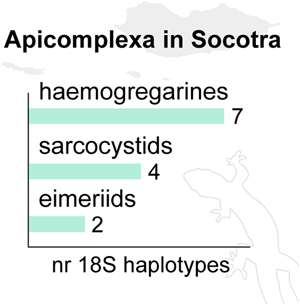
We detected three distinct apicomplexan parasites: haemogregarines, sarcocystids and eimeriids (Table 1), which significantly varied in their prevalence (P<0.001). From the 4718S rRNA sequences obtained, 35 were haemogregarines (75%), nine sarcocystids (19%) and three eimeriids (6%). Haemogregarines were thus the most common and widespread apicomplexan parasites (prevalence = 7.6%), infecting all host families analysed, except Colubridae and Leptotyphlopidae snakes (Table 1). Sarcocystids infections were detected in only three host families, namely Gekkonidae and Phyllodactylidae geckos, and the Scincidae skinks, with an overall prevalence of 2.0% (Table 1). Eimeriid infections were found in just two families (Phyllodactylidae and Lacertidae), corresponding to only 0.7% prevalence (Table 1). For haemogregarines, we detected differences in prevalence between the main host groups (snakes, geckos, chamaeleonids, lacertids and skinks) (P<0.001). Skinks were the most infected (36.6%), followed by chamaeleonids (20.0%), lacertids (8.5%), snakes (4.3%) and lastly geckos (3.7%) (Table 1). Within geckos, the prevalence was similar among the three families analysed (P = 0.262).
Genetic diversity and phylogenetic relationships
Among the 35 haemogregarine infections, we identified seven haplotypes based on the 18S rRNA sequences. Six of the seven haemogregarines (haplotypes 1, 2, 3, 4, 6 and 7) were closely related (P-distances ranging from 0.2 to 1.2%, Table 2), and clustered in a lineage containing species infecting rodents and reptiles from various geographical locations around the world, as well as several tick hosts (Fig. 2). This lineage also included haemogregarines from reptiles of Oman and nearby geographical areas. The seventh haemogregarine haplotype (haplotype 5 from the gecko Hemidactylus pumilio) diverged by more than 2% from the other six haplotypes (Table 2), and clustered with haemogregarines infecting lacertid lizards and snakes from Portugal and Morocco (Fig. 2).

Fig. 2. Tree derived from a Bayesian Inference analysis of haemogregarine 18S rRNA gene sequences. Bayesian Posterior probabilities are given above relevant nodes and bootstrap values for ML below them (only values over 70% are shown). New sequences from this study are identified in bold, larger font size and by different colours, the same as in Fig. 1. The full list of the sequences used can be found in Supplementary Table S3. Identification of the lineages marked with * was based on Karadjian et al. (Reference Karadjian, Chavatte and Landau2015), but see Maia et al. (Reference Maia, Harris, Carranza and Goméz-Díaz2016).
Table 2. Details on the seven haemogregarine haplotypes (h1–h7) detected in this study

The infected host species (and percentage of prevalence in each host in parenthesis), overall prevalence (P, in number of infected host individuals and %, percentage), host-specificity indexes (VarSTD and STD) and estimates of evolutionary divergence between the 18S rRNA sequences (P-distances) are given. Host species are coded by four letters, the first two capitals indicating the genus and the latter two letters the specific epithet (e.g. HAri = Haemodracon riebeckii).
Regarding the sarcocystids, four 18S rRNA haplotypes were identified, with P-distances varying between 0.1 and 2% (Table 3). Four 28S rRNA haplotypes were also found, while for the COI gene, only two haplotypes were retrieved. However, it should be noted that amplification of these two genes was not successful for all samples (see Material and methods section and Supplementary Table S2 for further details). The P-distance values for the 28S rRNA ranged from 0.2 to 17.1%, and was 6.5% between the two COI haplotypes (Table 3). According to the phylogeny based on the 18S rRNA gene, all four sarcocystids clustered with species of the genus Sarcocystis, but in different locations in the tree (Fig. 3). Haplotypes 1 and 4, infecting the skinks Hakaria simonyi and Trachylepis socotrana, were the most similar. Their closest relatives were sarcocystids found infecting lizards from Madagascar and snakes from Australia and Algeria (although in a poorly supported clade). In turn, haplotypes 2 and 3 (from the geckos Hemidactylus oxyrhinus and Haemodracon riebeckii, respectively) clustered in a well-supported clade, also containing species infecting reptiles, notably the lizard-infecting Sarcocystis lacertae (AY015113) and Sarcocystis gallotiae (AY015112). Other close relatives included sarcocystids infecting the snake Malpolon monspessulanus from Tunisia (KC696570), the lacertid Podarcis lilfordi from Spain (JQ762307) and the gecko Pristurus rupestris from Oman (KX453662, identical to haplotype 2).

Fig. 3. Tree derived from a Maximum Likelihood analysis of sarcocystid 18S rRNA gene sequences. Bayesian Posterior probabilities are given above relevant nodes and bootstrap values for ML below them (only values over 70% are shown). New sequences from this study are identified in bold, larger font size and by different colours, the same as in Fig. 1. The full list of the sequences used can be found in Supplementary Table S4.
Table 3. Estimates of evolutionary divergence between sarcocystid haplotypes (h) for three genes

The proportions of base substitutions per site are shown (P-distances).
For the eimeriids, only two 18S rRNA haplotypes were detected (6.3% divergence between the two sequences) (Fig. 4). Haplotype 1 from the gecko Haemodracon trachyrhinus grouped with sequences from Isospora isolated from skinks, geckos, agamids and lacertids (KU180238–KU180241 and KU180243–KU180245). Haplotype 2 from two Mesalina balfouri lacertids clustered with the representatives of the genus Lankesterella. Our sequence was identical to a parasite infecting the lacertid Acanthodactylus erythrurus from Spain (KJ131417). Other close relatives include lankesterellids of lizards from all over the world, including geckos from Oman (KX453651, KX453652, KX453655, KX453658 and KX453660, Fig. 4).

Fig. 4. Tree derived from a Bayesian Inference analysis of eimeriid 18S rRNA gene sequences. Bayesian Posterior probabilities are given above relevant nodes and bootstrap values for ML below them (only values over 70% are shown). New sequences from this study are identified in bold, larger font size and by different colours, the same as in Fig. 1. The full list of the sequences used can be found in Supplementary Table S5.
Prevalence, host-specificity and geographical distribution
Haemogregarine haplotypes differed in prevalence (P<0.001), with haplotypes 1 and 2 being more common (prevalence = 3.6 and 2.0%, respectively) than haplotypes 3–7 (prevalence between 0.2 and 0.4%; see Table 2). We detected three individuals with mixed infections between haplotype 1 and haplotypes 2, 3 and 7 (the only instance haplotype 7 was detected, Table 1). The seven haemogregarines also differed in their host-specificity (Table 2). Haplotypes 4, 5, 6 and 7 only occurred in a single host species, while infections by the other three haplotypes were detected in multiple reptile groups. On the other hand, haplotypes 1 and 2 infected eight species from different lizard groups, while haplotype 3 infected only two host species, each from a distinct group (see Table 2 for further details). Regarding geographical distribution, haplotypes 1 and 2 were found in multiple islands, in contrast with the other five haplotypes, all of which only occurred in one island (Fig. 1). The island of Socotra harboured six of the seven haemogregarine haplotypes detected, being three of them exclusive to this island (haplotypes 4, 5 and 6). We identified four haemogregarine haplotypes on the island of Adb al Kuri, two of them exclusive (haplotypes 3 and 7). Lastly, reptiles from the Samha and Darsa islands were only infected by haplotype 1.
As for sarcocystids and eimeriids, most haplotypes were also found on the island of Socotra (Fig. 1, Supplementary Table S1). The exceptions were the sarcocystid haplotype 2 and the eimeriid haplotype 2, which were found in Abd al Kuri and Darsa, respectively. Host ranges for both sarcocystids and eimeriids were also restricted to a single host species except the sarcocystid haplotype 1, which was found in the skinks H. simonyi and T. socotrana (Table 1). Overall, the reptile group hosting most parasite diversity were geckos, being infected by seven parasite haplotypes (four haemogregarines, two sarcocystids and one eimeriid), followed by skinks (four haemogregarines and two sarcocystids), lacertids (three haemogregarines and one eimeriid), and finally, chamaeleonids and snakes, each infected by a single haemogregarine haplotype (Table 1).
Discussion
Based on the results from the 18S rRNA, most of the haplotypes retrieved are exclusive to the archipelago. As so, it is highly probable these parasites are endemic, adding to the unique biodiversity of the Socotra Archipelago, just like their hosts. Moreover, the high genetic distance between them suggests that several parasite species might occur in Socotra (at least two haemogregarines, two Sarcocystis spp., one Isospora sp. and one Lankesterella sp.). There were, however, two haplotypes (sarcocystid haplotype 2 from H. oxyrhinus geckos and the eimeriid haplotype 2 from M. balfouri lacertids) that were identical to 18S rRNA sequences of parasites found infecting mainland reptiles, the P. rupestris geckos from Oman and A. erythrurus lacertids from Spain. Note that this latter genus is distributed from the Iberia Peninsula, across North Africa and Arabia to the Indian subcontinent (Tamar et al., Reference Tamar, Carranza, Sindaco, Moravec, Trape and Meiri2016). Interestingly, both hosts recently colonized the archipelago (5.7–3.0 and 7 Mya, respectively; Gómez-Díaz et al., Reference Gómez-Díaz, Sindaco, Pupin, Fasola and Carranza2012; Simó-Riudalbas et al., Reference Simó-Riudalbas, Tamar, Šmíd, Mitsi, Sindaco, Chirio and Carranza2019), which might explain this pattern. Close relatives of the remaining Socotran parasites were isolated from other reptile hosts, some also from Oman. Many Socotran reptiles are closely related to those of Arabia, which by vicariance or later long-dispersal colonization represent the major source of the biodiversity in the archipelago (e.g. Tamar et al., Reference Tamar, Simó-Riudalbas, Garcia-Porta, Santos, Llorente, Vasconcelos and Carranza2019). In the case of H. oxyrhinus, our findings support the ancestral biogeographic link between Omani and Abd al Kuri Hemidactylus geckos, which experienced intra-island speciation after the separation of the Socotra Archipelago from mainland Arabia (Gómez-Díaz et al., Reference Gómez-Díaz, Sindaco, Pupin, Fasola and Carranza2012).
Recently, Maia et al. (Reference Maia, Harris, Carranza and Goméz-Díaz2016) screened geckos and snakes from Oman, detecting 11 haemogregarine, four eimeriid and two sarcocystid haplotypes. We found no evidence of a significant reduction in parasite diversity from the mainland to the archipelago with the current data, considering that host species richness in Oman is higher than in Socotra. While the ancient separation of Socotra could have provided a relatively long time for new parasite diversity to accumulate (Cornuault et al., Reference Cornuault, Bataillard, Warren, Lootvoet, Mirleau, Duval, Mila, Thebaud and Heeb2012), vicariance might also have had a milder effect on parasite diversity than that predicted by the ‘parasite island syndrome’ (Nieberding et al., Reference Nieberding, Morand, Libois and Michaux2006). This syndrome was mostly based on volcanic islands, where species establish by oceanic dispersal, and thus being more susceptible to the founder effect and drift. In our case, although all haemogregarine haplotypes were unique to the archipelago, further sampling of mainland hosts and faster-evolving genetic markers will be needed to ascertain their phylogeography and endemicity.
Infections were found in all the four islands, but parasite diversity was not uniformly distributed, neither geographically nor among hosts. There were also differences in prevalence, with haemogregarines being the most frequent, as in previous works (e.g. Tomé et al., Reference Tomé, Maia and Harris2013; Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016). Prevalence values were based on the percentage of samples of each host species or host group for which we obtained a positive amplification identified as a parasite infection. Prevalence estimates could vary if other primer pairs would have been used (e.g. for haemogregarine detection; Perkins and Keller, Reference Perkins and Keller2001; Netherlands et al., Reference Netherlands, Cook, Du Preez, Vanhove, Brendonck and Smit2017). Based on our experience, the HEPF300 and HepR900 primer pair amplified a greater percentage of infected samples of reptile hosts and from a broader taxonomic range of parasites. Phylogenetic relationships and taxonomy within haemogregarines are still not clear (Karadjian et al., Reference Karadjian, Chavatte and Landau2015; Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016). However, current information shows different host-specificity and vectors according to the lineage (Karadjian et al., Reference Karadjian, Chavatte and Landau2015; Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016). Socotran haemogregarines fall in the Bartazoon lineage proposed by Karadijan et al. (Reference Karadjian, Chavatte and Landau2015), still under taxonomical debate. Within haemogregarines, some haplotypes were more common and widely distributed than others, according to their host-specificity and the biogeography of their hosts. Haplotypes 1 and 2 had the broader host-specificity and also the wider distribution in the archipelago. Haplotype 3 infected two distant species exclusive to Abd al Kuri Island. In contrast, haplotypes 4–7, which infected a single species, occurred in just one or two localities. This was supported by the host-specificity indexes (Table 2). However, it should be noted that these indexes take into account prevalence and, as sample size differed greatly across host species, these estimates might be biased.
Regarding the sarcocystid and eimeriid haplotypes, they were quite host-specific and geographically restricted. This pattern suggests that a lower host-specificity conferred some advantage to parasite geographical distribution, at least in this archipelago. The generally high host-specificity and the lower prevalence of haemogregarines on Socotra compared to continental Oman contradict the expectations of the island syndrome, but are in line with apicomplexan studies from other island systems (McAllister et al., Reference McAllister, Duszynski, Austin and Fisher2017; Tomé et al., Reference Tomé, Pereira, Jorge, Carretero, Harris and Perera2018). The drastic contrast in host-specificity between Socotran haemogregarines and between parasite groups could be due to many factors, including differences in life-cycle strategies and/or transmission pathways (more so than in Oman; Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016). However, further data are necessary to investigate this.
Across the four islands of the archipelago, Socotra harboured by far the highest parasite diversity. This is likely due to its much larger size (Jean et al., Reference Jean, Burnside, Carlson, Smith and Guégan2016) and its higher host species richness, important factors determining pathogen diversity. Additionally, the islands of Samha and Darsa were connected to Socotra as recently as 20 000 years ago (Van Damme, Reference Van Damme, Gillespie and Clague2009), which would have allowed gene flow. Our results support this, as the two small islands harboured parasite haplotypes also present on Socotra Island and the mainland. Interestingly, Abd al Kuri has never been connected to the other islands since they separated from the mainland, as it is isolated from them by deep seas (200–1000 m deep) that exceed the Pleistocene sea-level changes (Van Damme, Reference Van Damme, Gillespie and Clague2009). Accordingly, there we found two exclusive haemogregarine haplotypes, present in endemic lizard species of the island. Additionally, factors related to the parasite life-cycle should also be considered to explain these differences among islands, most notably the establishment success and abundance of non-reptile hosts, such as invertebrate vectors for haemogregarines (Poulin, Reference Poulin2011).
Regarding prevalence across reptile groups, skinks were the most infected hosts for the three apicomplexans (overall prevalence = 53.7%), what strongly contrasts with other studies, in which skinks were among the least infected reptiles for haemogregarines (e.g. Harris et al., Reference Harris, Maia and Perera2011; Maia et al., Reference Maia, Harris and Perera2011). It is particularly interesting to compare these results with the Canary Islands, where only 1.9% of the analysed skinks were infected (Tomé et al., Reference Tomé, Pereira, Jorge, Carretero, Harris and Perera2018). These sharp differences between the two archipelagos might be justified by differences in host diversity and ecology. Whereas Canary Island skinks are ground-dwelling (Carranza et al., Reference Carranza, Arnold, Geniez, Roca and Mateo2008), Socotran's are more generalist in diets and habitat use (Razzetti et al., Reference Razzetti, Sindaco, Grieco, Pella, Ziliani, Pupin, Riservato, Pellitteri-Rosa, Butikofer and Suleiman2011). As such, Socotran skinks have more chances to cross paths with other hosts and their parasites, and to be more exposed to vectors, such as ticks, mites or mosquitoes, increasing parasite encounter probability (Combes, Reference Combes2001).
Despite being the least infected reptile group, geckos harboured the highest haemogregarine diversity, as they are the most diverse reptile group in the archipelago, with over 16 described species. These include species that originated in situ from ancestors present at the islands when they separated from the mainland, as well as species that arrived as new colonizers (e.g. Gómez-Díaz et al., Reference Gómez-Díaz, Sindaco, Pupin, Fasola and Carranza2012; Martín et al., Reference Martín, Martínez, Pujol-Buxó, Vinolas, Llorente, Sanpera, Vasconcelos, Carranza and Santos2017; Tamar et al., Reference Tamar, Simó-Riudalbas, Garcia-Porta, Santos, Llorente, Vasconcelos and Carranza2019). Additionally, Socotran geckos have a wide range of activity pattern and habitat use, which further exposes them to a wider variety of parasites. For example, Hemidactylus and Haemodracon geckos are nocturnal, but are found under rocks or ground-dwelling, sharing habitat with lacertids and skinks (Razzetti et al., Reference Razzetti, Sindaco, Grieco, Pella, Ziliani, Pupin, Riservato, Pellitteri-Rosa, Butikofer and Suleiman2011; Tamar et al., Reference Tamar, Simó-Riudalbas, Garcia-Porta, Santos, Llorente, Vasconcelos and Carranza2019). On the other hand, Pristurus geckos are diurnal and, in many aspects, behave more like desert agamids than geckos (Badiane et al., Reference Badiane, Garcia-Porta, Červenka, Kratochvíl, Sindaco, Robinson, Morales, Mazuch, Price and Amat2014). Nonetheless, the absence of the gecko-exclusive lineage of haemogregarines is notorious, which was quite common in geckos from Oman (Maia et al., Reference Maia, Harris, Carranza and Goméz-Díaz2016). This suggests that this haemogregarine lineage might have ‘missed the boat’, arrived in Arabia posterior to the separation of Socotra, or remain undetected due to low prevalences and intensities of this particular lineage.
Similarly, snakes were also much more parasitized in Oman than in Socotra, where only one infected snake was detected with a host-specific haplotype. The diet of the Socotra snakes Hemerophis socotrae and Ditypophis vivax includes lizards (O'Shea, Reference O'Shea2018) and one of the transmission pathways of haemogregarines to snakes is by ingestion of infected prey (Smith, Reference Smith1996). Hence, a higher prevalence would be expected in these species than in the burrowing snakes Myriopholis spp. and Xerotyphlops socotranus. A reason for this might simply be the limited sampling size for some of the host species. Lastly, we also screened a few samples from the recently introduced Hemidactylus species, but no infections were found.
The data collected in this study show that, in parallel with their hosts, Socotra is also a centre of endemism for apicomplexan parasites. Genetically, most of the detected 18S rRNA haplotypes were closely related to samples from Arabia, indicating that this is likely their origin, as happens for most of their hosts. There were also some interesting and unexpected results regarding the contrasting levels of prevalence between hosts and of host-specificity between parasite haplotypes. These may be related to ecological factors, such as the behaviour and habitat use of the hosts and the life-cycle strategies of the parasites. As a whole, our results emphasize the importance of screening parasites in wild hosts from remote regions, in particular islands, and the need to consider host ecology and biogeography to better understand the dynamics of disease transmission across different taxa.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182020002000.
Acknowledgements
We are grateful to Ahmed Saeed, Salem Hamdiah, Yaya Saleh, Abubakar Salim, Xavier Santos, Mauro Fasola and Edoardo Razzetti for providing sampling materials. We thank the Environment Protection Authority (EPA) for the permits and logistic aid.
Financial support
Some authors were supported by ‘Fundação para a Ciência e Tecnologia’ (FCT) two Ph.D. and one postdoc grants (B.T., PD/BD/52601/2014; J.P.M., SFRH/BD/74305/2010; and R.V., SFRH/BPD/79913/2012, respectively), supported under the Programa Operacional Potencial Humano – Quadro de Referência Estratégico Nacional funds (POPH-QREN) from the European Social Fund (ESF) and Portuguese ‘Ministério da Educação e Ciência’. This study was part of B.T.'s thesis dissertation. Some authors were supported by FCT contracts (A.P., IF/01257/2012, and R.V., contract from FCT national funds from ‘Norma transitória’ DL57/2016/CP1440/CT0002); and by a grant from the Ministerio de Ciencia Innovación y Universidades co-funded by FEDER (S.C., PGC2018-098290-B-I00). This work was funded by projects from the Ministerio de Economía y Competitividad, Spain, co-funded by FEDER (CGL2012-36970, led by S.C.) with the support of Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement de la Generalitat de Catalunya (2017-SGR-991, led by S.C.); the Mohamed bin Zayed Species Conservation Fund (project 13055714, led by R.V.), and FCT (IF01257/2012/CP0159/CT0005 IF project, led by A.P.).
Conflict of interest
None.
Ethical standards
The Environmental Protection Authority (EPA) approved this study as it was in the scope of the agreement signed by an EPA representative and S.C. on 22 March 2010.