INTRODUCTION
Cyathostomins are a complex group of parasitic nematodes that infect equids. The group consists of 50 species, although 10 common species usually predominate in horses (Ogbourne, Reference Ogbourne1976; Bucknell et al. Reference Bucknell, Gasser and Beveridge1995; Lyons et al. Reference Lyons, Tolliver, Drudge, Stamper, Swerczek and Granstrom1996, Reference Lyons, Tolliver and Drudge1999; Lichtenfels et al. Reference Lichtenfels, Kharchenko, Krecek and Gibbons1998, Reference Lichtenfels, Kharchenko and Dvojnos2008; Chapman et al. Reference Chapman, French and Klei2002). Due to a high prevalence and a propensity to develop anthelmintic resistance, cyathostomins have been considered the principle parasitic pathogens of horses for a number of decades (Love et al. Reference Love, Murphy and Mellor1999). Several clinical syndromes have been reported, including larval cyathostominosis; a severe colitis presumed to result from the synchronous emergence of large numbers of developing larvae from the large intestinal mucosa and submucosa (Giles et al. Reference Giles, Urquhart and Longstaffe1985; van Loon et al. Reference van Loon, Deprez, Muylle and Sustronck1995; Love et al. Reference Love, Murphy and Mellor1999). Many horses are susceptible to cyathostomin infection all of their lives and, as a result, animals can develop clinical disease at any age (Chapman et al. Reference Chapman, French and Klei2002; Kornaś et al. Reference Kornaś, Cabaret, Skalska and Nowosad2010); however most cases of severe clinical disease occur in horses of less than 5 years (Reid et al. Reference Reid, Mair, Hillyer and Love1995).
For cyathostomin species, morphological identification is restricted to the anatomical characteristics of the head and tail of adult worms (Lichtenfels et al. Reference Lichtenfels, Kharchenko, Krecek and Gibbons1998). The accessible pre-parasitic stages, such as eggs and infective third-stage larvae (L3), cannot be identified to species based on morphological parameters. As a result, several molecular tools have been developed to identify cyathostomin species at all stages of the life cycle (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998; Hodgkinson et al. Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001, Reference Hodgkinson, Lichtenfels, Mair, Cripps, Freeman, Ramsey, Love and Matthews2003; Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007). All of these have focussed on ribosomal (r) DNA sequences, specifically the internal transcribed spacer (ITS) and the intergenic spacer (IGS) sequences, which are present in the genome in multiple copies (Elder and Turner, Reference Elder and Turner1995; Gasser and Newton, Reference Gasser and Newton2000). Amplification and subsequent analysis of the IGS DNA region from 16 cyathostomin species (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998) led to the development of a PCR-ELISA method that was used to identify the following common species: Cyathostomum catinatum, Cylicocyclus ashworthi, Cylicocyclus insigne, Cylicocyclus nassatus, Cylicostephanus goldi, Cylicostephanus longibursatus (Hodgkinson et al. Reference Hodgkinson, Lichtenfels, Mair, Cripps, Freeman, Ramsey, Love and Matthews2003), Cyathostomum tetracanthum and Cylicocyclus leptostomum (Hodgkinson et al. unpublished data). This PCR-ELISA technique was used to identify morphologically indistinct pre-parasitic stages in 2 studies that addressed important issues in cyathostomin biology; these being, the identification of fourth-stage larvae recovered from the feces of cases of larval cyathostominosis (Hodgkinson et al. Reference Hodgkinson, Lichtenfels, Mair, Cripps, Freeman, Ramsey, Love and Matthews2003) and the identification of eggs present in feces after anthelmintic treatment (Hodgkinson et al. Reference Hodgkinson, Freeman, Lichtenfels, Palfreman, Love and Matthews2005). The reverse line blot hybridization method was also developed to exploit the variation observed within IGS DNA sequences (Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007). Until now, identification of 13 species has been described using this method: 7 of the species named above (excluding Cya. tetracanthum), as well as Coronocyclus coronatus, Coronocyclus labiatus, Coronocyclus labratus, Cyathostomum pateratum, Cylicostephanus calicatus and Cylicostephanus minutus. The advantage of this method is that it enables several DNA probes to be used simultaneously for a number of species and has been applied to explore the biology of anthelmintic resistance in cyathostomins (Cernanská et al. Reference Cernanská, Paoletti, Kralova-Hromadova, Iorio, Cudekova, Milillo and Traversa2009; Traversa et al. Reference Traversa, Iorio, Otranto, Giangaspero, Milillo and Klei2009, Reference Traversa, Milillo, Barnes, Von Samson-Himmelstjerna, Schurmann, Demeler, Otranto, Lia, Perrucci, Frangipane Di Regalbono, Beraldo, Amodie, Rohn, Cobb and Boeckh2010; Ionita et al. Reference Ionita, Howe, Lyons, Tolliver, Kaplan, Mitrea and Yeargan2010; van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010).
Critical to the success of each of these methodologies is a sound knowledge of the level of intra- and inter-specific sequence variation within the IGS DNA region. Previous studies have demonstrated levels of intra- and inter-specific polymorphism for a number of cyathostomin species; however, often sequence is only generated from small numbers of individuals and for a limited number of species (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998; Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007). The utility of these DNA probes requires careful and rigorous evaluation with new subsets of worms from the same species (referred to herein as ‘homologous’ species) and from different species (referred to herein as ‘heterologous’ species) for robust methods of molecular species identification to be maintained. Here, we report sequences of the IGS DNA region for 21 different cyathostomin species to reveal further insights into the utility and accuracy of the previously reported DNA probes (Hodgkinson et al. Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001, Reference Hodgkinson, Lichtenfels, Mair, Cripps, Freeman, Ramsey, Love and Matthews2003; Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007; van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010). We also explore the implication that these new data have for the validation of new and existing identification tools for these important equine parasites.
MATERIALS AND METHODS
Collection of parasites and isolation of genomic DNA
Individual adult worms were collected from the dorsal colon, ventral colon and caecum of horses at a local abattoir, washed in phosphate-buffered saline (pH 7·4), the parasite heads removed for morphological identification and the remainder stored in liquid nitrogen as described previously (Hodgkinson et al. Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001). Identification to genus and species was according to Lichtenfels et al. (Reference Lichtenfels, Kharchenko, Krecek and Gibbons1998). Genomic DNA was extracted from individual adult cyathostomins following an overnight incubation at 56°C, using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's instructions, with a final elution volume of 30 μl of distilled water. L3 were lysed in preparation for the PCR step as described previously (van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010).
Polymerase chain reaction amplification
PCR reactions were carried out in 50 μl reaction volumes containing 50 mM KCl, 10 mM Tris–HCl (pH 8·3), 1·5 mM MgCl2, 200 μM of each dNTP, 1·25 U AmpliTaq Gold (Life Technologies). The amount of template varied according to the source of genomic DNA; 1 μl of L3 lysate or 2 μl of adult DNA. The IGS DNA region was amplified using a combination of the conserved primers CY1 and CY26 in the forward orientation, with either CY4 or CY18 in reverse (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998). The cycling conditions were an initial 10 min at 94°C, followed by 40 cycles at 94°C for 1 min, 60°C for 1 min and 72°C for 2 min, with a final extension at 72°C for 10 min. Negative and positive control samples were included where appropriate: negative samples did not include DNA, whilst positive control samples contained genomic DNA from an adult cyathostomin of known species. PCR products were analysed by agarose gel electrophoresis using SYBR® Safe DNA stain (Life Technologies).
Cloning and sequence analysis of the IGS DNA region
The IGS DNA region was amplified from cyathostomin species using the PCR conditions above in a direct or semi-nested approach (see Tables 1 and 2). Once amplified, IGS PCR products were cloned into the pGEM T-Easy vector (Promega), according to manufacturer's instructions. Derived clones of the expected size (as well as ‘variant’ clones of unexpected size) from an individual parasite or species were sent for sequencing to a commercial service (GATC Biotech). The sequences were analysed using ClustalW2 and BLAST (http://www.ebi.ac.uk/Tools/msa/clustalw2/ and http://blast.ncbi.nlm.nih.gov/Blast.cgi). Consensus sequences were compiled using ClustalW2 and this programme was used to generate all alignments and subsequent identity scores. The latter were used to compile sequence identity matrices for all cyathostomin IGS DNA sequence data generated (Table 3). IGS DNA sequence for 10 species is reported for the first time here (defined as ‘novel’ IGS DNA sequence), along with sequence from 8 species for which the IGS DNA region has been reported previously (defined as ‘additional’ IGS DNA sequence). All additional sequences were compared to the original sequence submissions of Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998) to produce more accurate consensus sequences.
Table 1. Novel sequence of the intergenic spacer region for 10 cyathostomin species

| Species | Primers used | Size of IGS bp | No. of individuals sequenced | Intra-specific variation |
| Cor. coronatus * | ||||
| Variant A | CY1/CY18† | 1834 | 3‡(4) | 10% |
| Variant B | CY1/CY18† | 1782 | 4‡(4) | 23% |
| Cor. labratus | CY1/CY18† | 1312 | 2 | 12% |
| Cra. acuticaudatum | CY1/CY18 | 946 | 1 | – |
| Cya. tetracanthum | CY1/CY18 | 969 | 7 | 5% |
| Cyc. elongatus | CY1/CY18 | 921 | 1 | – |
| Cyd. bicoronatus | CY1/CY18 | 803 | 3 | 7% |
| Cys. calicatus * | CY1/CY4 | 422 | 3 | 5% |
| Cys. minutus * | ||||
| Variant A | CY1/CY18† | 1688 | 3(4) | 3% |
| Variant B | CY1/CY18† | 1911 | 3(4) | 21% |
| Para. mettami | CY1/CY18 | 622 | 3 | 1% |
| Pot. imparidentatum | CY1/CY18 | 796 | 2 | 1% |
* Corrected IGS sequence.
† Semi-nested PCR using CY26 as forward primer.
‡ Two of the individuals were used for both variants.
(), no. of clones sequenced.
Table 2. Additional sequencing of the intergenic spacer region for 8 cyathostomin species

| Species | Primers used | Size of IGS (bp) | No. of individuals sequenced | Intra-specific variation |
| Cya. catinatum | CY1/CY4 | 619 | 3+5 | 3% |
| Cya. pateratum | CY1/CY4 | 693 | 3+3 | 1% |
| Cyc. ashworthi | CY1/CY4 | 603 | 3+6 | 6% |
| Cyc. insigne | CY1/CY4(CY18*) | 605 (927) | 3+3* | 2% |
| Cyc. leptostomum | CY1/CY4(CY18*) | 616 (936) | 1+3 (3*) | 10% |
| Cyc. nassatus | CY1/CY4 | 632 | 3+3 | 3% |
| Cyc. radiatus | CY1/CY4(CY18*) | 599 (921) | 1+1* | 2% |
| Variant A | CY1/CY18 | 931 | 0+3 | 5% |
| Cor. labiatus | CY1/CY18 | 1708 | 1+2 | 1% |
* Subset of individuals amplified with CY1 and CY18 primers; no. of individuals sequenced by Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998) + no. of individuals sequenced by present study.
Table 3. IGS sequence identity matrix for 23 cyathostomin species, showing the levels of inter-specific variation amongst species

Design and validation of species-specific probes
To design new species-specific probes, all available IGS DNA sequence data, including new consensus sequences, were compared by ClustalW2 alignments. New and previously published DNA probes were also analysed using ClustalW2, BLAST and MyBLAST (http://mybioweb.nhri.org.tw), a web-base tool to BLAST against sequences in a personal data base. Specificities of the newly designed probes were confirmed by hybridization with IGS PCR products derived from morphologically identified adult worms of homologous and heterologous cyathostomin species (Lichtenfels et al. Reference Lichtenfels, Kharchenko, Krecek and Gibbons1998, Reference Lichtenfels, Kharchenko and Dvojnos2008).
Reverse line blot (RLB) hybridization
Conditions and primers for amplification of the IGS DNA fragment were as described above, except that the reverse primers were 5′ biotinylated as described (Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007; van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010). The RLB for species-specific hybridization of IGS amplicons from cyathostomins is a modification of the method of Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007). Specific DNA probes with 5′C6-aminolinker were coupled to a Biodyne C membrane, with each probe being coupled at 2 different concentrations, 800 and 200 pmol per lane. Ten μl of the IGS amplicon per lane were used for hybridization. Hybridized products were detected with streptavidin-POD conjugate (Roche), followed by incubation in ECL detection reagent and exposure to Amersham Hyperfilm ECL (both GE Healthcare).
RESULTS
Analysis of IGS DNA sequences
The novel IGS DNA sequences generated for the 10 cyathostomin species are shown in Table 1 and the sequences for those species previously investigated are shown in Table 2. Comparison of these sequences indicated that some of the original sequences reported were inaccurate; these being sequence submissions of Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998), for 3 species, Cys. calicatus (AJ223338), Cor. coronatus (AJ223340) and Cys. minutus (AJ223347). The conclusion that these sequences are inaccurate is based on several observations made by the authors: hybridization experiments using probes designed from these species did not bind to homologous IGS sequence (data not shown; Hodgkinson et al. Reference Hodgkinson, Lichtenfels, Mair, Cripps, Freeman, Ramsey, Love and Matthews2003; van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010). On BLAST analysis, the sequences showed higher levels of sequence identity to heterologous species in the Genbank™ database and sequencing of a second cohort of morphologically identified parasites of Cys. calicatus (3 individuals), Cor. coronatus (7 individuals) and Cys. minutus (6 individuals) provided new consensus sequences that did not show significant identity to the IGS sequences generated by Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998). Finally, as reported in this present paper, not all cyathostomin species have the CY4 primer site sequence, including Cor. coronatus and Cys. minutus. Although Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998) did amplify a product from these 2 species with the CY1 and CY4 primers, initially the PCR was carried out with the CY26 and CY18 primer combination; where a larger product was observed. At the time this discrepancy in size was suggested to occur between the CY4 and CY18 primers; however, the study reported here supports that the CY1 and CY4 PCR amplification from Cor. coronatus and Cys. minutus was due to contamination with DNA from other species. The accurate IGS DNA sequences for these species have now been submitted to GenBank™ under the following Accession numbers: Cys. calicatus HM142928, Cor. coronatus variant A HM142938, variant B HM142939 and Cys. minutus variant A HM142940, variant B HM142941.
To develop robust species-specific probes it was necessary to obtain IGS DNA sequence from as many individual adult worms as possible. For this reason, the IGS DNA region was amplified from at least 3 individuals of each species to examine levels of intra-specific variation (Tables 1 and 2). Previously, for some species, such as Cylicocyclus radiatus, IGS DNA sequence was reported for only 1 individual (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998). The more detailed sequencing reported here revealed further insights into the complexity of the IGS DNA region not previously observed. For example, of the IGS DNA sequences amplified from 4 morphologically identified Cyc. radiatus individuals, only 1 showed high levels of sequence identity to the sequence previously published for this species (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998, 98%); whilst the remaining 3 sequences displayed lower levels of identity (ranging between 70 and 71%), suggesting that there are ‘sequence variants’ for some species. This was also the case for Cys. minutus and Cor. coronatus, for which a highly variable middle section of the IGS DNA region was flanked by conserved 5′ and 3′ ends (Fig. 1). For all 3 species, the degree of variation was too great to allow a consensus sequence to be generated.

Fig. 1. Schematic representation of Coronocyclus coronatus and Cylicostephanus minutus rDNA variants (A and B) showing location of conserved regions, variable region and repeat regions. Black boxes represent 99% conserved IGS region, unfilled boxes represent variable regions, 48% and 55% for Cor. coronatus and Cys. minutus respectively. The grey box represents a 240 bp IGS repeat region found in Cor. coronatus individuals. Dotted arrows indicate position of probe sequences and the bold arrow indicates a site for an insertion of sequence in Cys. minutus variant B when compared with variant A.
For all 21 species, intra-specific sequence variation was shown to range from 1 to 23%, excluding the degree of variation within species for which the variants were reported above. Four species showed only 1% variation (Tables 1 and 2) and, as expected, the greatest intra-specific variation was observed for those species where variants have been described, specifically Cor. coronatus variant B and Cys. minutus variant B (23% and 21%, respectively, Table 1). The species that had the third highest level of intra-specific variation was Cor. labratus; this appeared to be due to the presence of repeat regions within the IGS DNA region in this species. Sequence identities amongst different species ranged from 38 to 97% (Table 3) with Cyc. insigne:Cyc. radiatus and Cya. pateratum:Cya. catinatum showing the lowest levels of inter-specific variation, at only 3% and 5%, respectively. However, for each species, the level of intra-specific variation was always lower than that observed for inter-specific variation for those individuals examined, illustrating that, despite its relative complexity, this region supports molecular species identification providing that well-validated tools are employed.
An important finding from our comparative analysis of the IGS DNA sequence data showed that the sequence of the 23-mer, CY4 primer (Kaye et al. Reference Kaye, Love, Lichtenfels and McKeand1998) was not conserved in all species examined. For example, this sequence was not present in the following species: Craterostomum acuticaudatum, Cor. coronatus, Cyc. minutus, Parapoteriostomum mettami and Poteriostomum imparidentatum and showed a degree of nucleotide variation for 4 species: Cor. labiatus (2 bp differences), Cor. labratus (1 bp difference), Cylicocyclus elongatus (1 bp difference) and Cylicodontophorus bicoronatus (4 bp differences). It was possible to amplify a PCR product from Cor. labiatus, Cor. labratus and Cyc. elongatus, but on several occasions PCR amplification was unsuccessful or was considered suboptimal.
RLB validation and application
Here, we report on 14 newly designed and validated RLB probes and the re-validation of 9 previously described RLB probes (Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007; van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010) against a greater number of cyathostomin species (Table 4). To validate the RLB probes, the IGS DNA region was amplified from at least 3, morphologically identified adult worms and hybridized to each DNA probe sequence as represented in Fig. 2. Three probes designed to hybridize to DNA derived from Cylicocyclus auriculatus, Cylicostephanus bidentatus and Tridentoinfundibulum gobi were shown not to cross-hybridize to DNA from 20 heterologous species but, because DNA derived from these species was not available at the time of the RLB study, these probes require further validation with DNA derived from the homologous species before they can be applied as diagnostic tools.
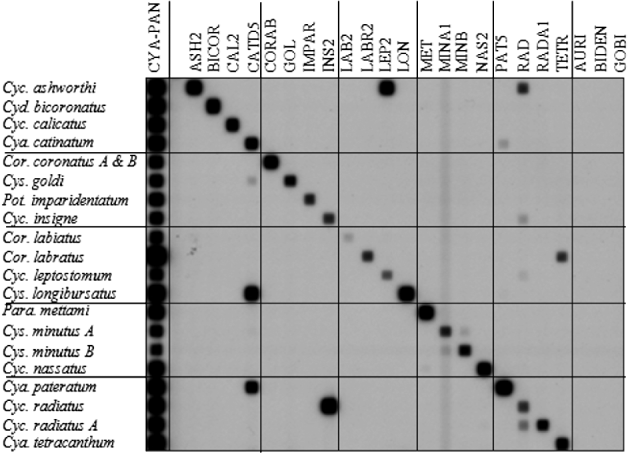
Fig. 2. Figure representing the validation of IGS RLB probes (800 pmol/lane) against 18 species of cyathostomin. Each species is represented by IGS PCR products amplified from 3 individuals. See Table 4 for probe descriptions.
Table 4. Probe sequences validated in RLB technique

| Species | Probe sequence 5′−3′ | Reference |
| Cor. Coronatus variants A & B | CORAB - TTCTCAAAAGCAAGGGGGACTTC | Present paper |
| Cor. labiatus | LAB2 - GTTCTATTAGGTTGTCTAAGAA | Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007) |
| Cor. labratus | LABR2 - GCTGAAATGCCGTGTTAGT | Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007) |
| Cya. catinatum | CATD5 - CGACTAGGCGTACATCATA | van Doorn et al. (Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010) |
| Cya. pateratum | PAT5 - CATACAGTTGTAACATTCTCG | van Doorn et al. (Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010) |
| Cya. tetracanthum | TETR - TGGCATCCTTCAAGGTTTCAA | Present paper |
| Cyc. ashworthi | ASH2 - GTTCTACTTTATGCAGTGTAAA | Present paper |
| Cyc. auriculatus ** | AURI - CATAATGGCTGTGAATTATGGG | Present paper |
| Cyc. bidentatus ** | BIDEN - CATTGCAGCGATTTACAAATAG | Present paper |
| Cyc. insigne | INS2 - GTATGTATATGTATCAATGTCTTAA | Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007) |
| Cyc. leptostomum | LEP2 - ATGTATGCCATTCTTTTATATGTA | Present paper |
| Cyc. nassatus | NAS2 - GCAAGAACTTCGCTGAAATG | Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007) |
| Cyc. radiatus | RAD - AGACAGCACTTGCTGTGCCAAT | Present paper |
| Cyc. radiatus variant A | RADA1 - GGGAGGGGTTTCAAATAATCGAC | Present paper |
| Cyd. bicoronatus | BICOR - GCTTTCTGATGCGATAAATGACAT | Present paper |
| Cys. calicatus | CAL2 - ACATGCAACACCCTGTTCAA | van Doorn et al. (Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010) |
| Cys. goldi * | GOL - TCTTAGCATCAGGAGAAAT | Hodgkinson et al. (Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001) |
| Cys. longibursatus * | LON - GGAGAAATTGGTGGCGACT | Hodgkinson et al. (Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001) |
| Cys. minutus variant A | MINA1 - GGTCACGCTCGATTAACATGCC | Present paper |
| Cys. minutus variant B | MINB - GATTTCGCAATTCAACATACG | Present paper |
| Para. mettami | MET - GTCTTCTACTCGAGAGGGGG | Present paper |
| Pot. imparidentatum | IMPAR- GGCTTGATTCACGCGCTAGCTAAA | Present paper |
| Tri. gobi ** | GOBI - CCACATTTATGTACGAAACATC | Present paper |
| Cyathostomin PAN | Cya-PAN - GAGACTATCCTATGATCGGGTG | Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007) |
* Probe sequence first reported by Hodgkinson et al. (Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001), with subsequent validation for RLB by Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007).
** Require validating with homologous species.
To demonstrate the application of the new probes developed herein, IGS DNA sequence was PCR amplified from a population of L3 cultured from the feces of a group of donkeys kept at the Donkey Sanctuary (Devon, UK) and subsequently identified to species by RLB (Fig. 3). Seven different cyathostomin species were identified, representing 94% of the 50 L3 analysed. The remaining 3 L3 that could not be identified to species hybridized with the cyathostomin PAN probe, indicating that the larvae were cyathostomins but were not species detected by the RLB. Of the 7 species identified, Cya. tetracanthum and Cya. pateratum were the most frequently detected in the population of 50 parasites, at 42% and 22%, respectively.

Fig. 3. Species identification of 50 L3 using the modified RLB method. * L3 that fall into this category react with the cyathostomin PAN probe but not with one of the 23 species-specific probes.
DISCUSSION
The work presented here illustrates the complexity of sequences within the IGS DNA region of cyathostomins and highlights the potential of this region for distinguishing between species, providing that the sequences used are validated properly for intra-specific and inter-specific variation.
Here, we report a greater degree of intra-specific variation than reported previously for some species; for example, Cyc. leptostomum and Cyc. nassatus, 10% and 3%, respectively, for which Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998) reported a range between 0 and 2·2%. Furthermore, Traversa et al. (Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007) sequenced the IGS DNA region from 13 cyathostomin species, as well as 3 Strongylus (large strongyle) species, and did not report any intra-specific variation for all species examined. However, in the latter study, the exact number of individuals examined for each species was not reported and the associated sequences were not submitted to GenBank™. The current study highlights the importance of using as many individuals as possible to generate consensus IGS DNA sequences for a given species. This will ensure that any downstream molecular species identification studies will be as accurate as possible. That said, it should be noted that, for the 5 species discussed here (Cra. acuticaudatum (1 individual), Cyc. elongatus (1 individual), Cor. labratus (2 individuals), Pot. imparidentatum (2 individuals) and Cyc. radiatus (2 individuals), the derived IGS DNA sequences represent material obtained from fewer than 3 individual worms. This is because these species are relatively rare and our findings will require confirmation in future studies once we obtain further members of these species for our archive.
For many species, the IGS DNA region contains several areas of sequence repeats, which are thought to be involved in the control of ribosomal transcription and/or processing (Baker and Platt, Reference Baker and Platt1986; Dewinter and Moss, Reference De Winter and Moss1986; Labhart and Reeder, Reference Labhart and Reeder1986; Harrington and Chikaraishi, Reference Harrington and Chikaraishi1987). Variation in the number of repeating sequences results in variability in the overall length of the IGS DNA region between individuals and between species (Moss and Stefanovsky, Reference Moss and Stefanovsky1995). Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998) reported a lack of large repeating sequences within the IGS DNA region. Subsequent studies have shown that Cya. catinatum (Hodgkinson et al. Reference Hodgkinson, Love, Lichtenfels, Palfreman, Ramsey and Matthews2001) and Cya. pateratum (Hodgkinson et al. unpublished data) individuals have a variable number of repeats just upstream of the CY4 primer site. Analysis of the novel sequence data generated here suggests that additional cyathostomin species also possess sequence repeat regions, though these differ in pattern from those observed previously in Cya. catinatum and Cya. pateratum. For example, Cor. coronatus variant A, has a CATA repeat that varies in length up to ∼ 200 bp, as well as another ∼100 bp fragment that is repeated towards the 3′ end of the IGS DNA region and Cor. labratus has a repeated ∼100 bp sequence towards the 5′ end of the IGS DNA region. The repeats from Cor. coronatus and Cor. labratus do not bear any resemblance to each other. Interestingly, repeats in other organisms, primarily plant species, are found to be between 100 and 200 bp, consistent with our observations in cyathostomins and are often species specific (Appels and Dvorak, Reference Appels and Dvorak1982; Flavell et al. Reference Flavell, O'Dell, Thompson, Vincentz, Sardana and Barker1986). The function of these repeat sequences requires further investigation.
For several cyathostomin species, we have reported ‘variant’ IGS DNA sequences, which were amplified more than once from several individuals of a given species. Similar variation in ITS rDNA sequences has been described for Cys. minutus, resulting in this being referred to as a ‘cryptic’ species (Hung et al. Reference Hung, Chilton, Beveridge, Zhu, Lichtenfels and Gasser1999a). From our analysis, it appears that several other species, including Cyc. ashworthi and, in particular, Cyc. radiatus that shows 24% intra-specific variation when comparing the Cyc. radiatus IGS to that of Cyc. radiatus variant A, may follow a similar trend of cryptic complexes within the IGS DNA region. It is interesting that 2 of these species, Cys. minutus and Cor. coronatus, have similar-sized ITS-2 rDNA sequences, that are 100 bp shorter than other cyathostomin ITS-2 rDNA sequences (Hung et al. Reference Hung, Gasser, Beveridge and Chilton1999b). Observations from Cor. coronatus are interesting as both variant IGS DNA sequences were amplified from the same individual using the same primer set. As IGS DNA sequences are represented in the genome in multiple copies (Elder and Turner, Reference Elder and Turner1995; Gasser and Newton, Reference Gasser and Newton2000), our findings may represent amplification of different copies or paralogues. Alternatively, as the sex of the individual adult worms that were sequenced here was not recorded, it is possible that the DNA sequences were generated from female worms that contained eggs. The sex of the worm should be recorded in future studies and, where possible, only male worms used.
Despite a greater number of species being analysed here, the levels of inter-specific variation observed (38–97% sequence identity) is similar to the range of 40–97% reported previously by Kaye et al. (Reference Kaye, Love, Lichtenfels and McKeand1998). The lack of sequence conservation for the CY4 primer sequence in some species is an important observation given the use of this primer sequence in the RLB technique used previously to differentiate 13 cyathostomin species (Traversa et al. Reference Traversa, Iorio, Klei, Kharchenko, Gawor, Otranto and Sparagano2007). The fact that some species either lack this sequence or have a variable sequence at this site indicates that those species would be missed in analysis, and so we recommend that the more conserved CY18 primer sequence be used in subsequent identification studies to capture as many cyathostomin species as possible. This is supported by the work of Ionita et al. (Reference Ionita, Howe, Lyons, Tolliver, Kaplan, Mitrea and Yeargan2010) who reported the use of a CY18-biotin primer in the RLB technique.
The RLB is a useful method for species identification of cyathostomins that is less time-consuming and costly than other methods. Inaccuracies in some IGS DNA sequences published previously, along with the observed increased levels of intra-specific variation for several species as well as the novel IGS DNA sequences reported here, necessitated re-validation of the RLB for use in identifying cyathostomins. This newly validated method can identify 18 species, including all the major species previously observed in equines (Ogbourne, Reference Ogbourne1976; Bucknell et al. Reference Bucknell, Gasser and Beveridge1995; Lyons et al. Reference Lyons, Tolliver, Drudge, Stamper, Swerczek and Granstrom1996, Reference Lyons, Tolliver and Drudge1999; Lichtenfels et al. Reference Lichtenfels, Kharchenko, Krecek and Gibbons1998, Reference Lichtenfels, Kharchenko and Dvojnos2008; Chapman et al. Reference Chapman, French and Klei2002). Due to an inaccuracy in the original sequence submission for Cys. calicatus IGS DNA, van Doorn and colleagues (2010) re-designed molecular RLB probes for Cya. catinatum, Cya. pateratum and Cys. calicatus. They described cross-hybridization between Cys. longibursatus individuals and the Cya. catinatum RLB probe. This cross-hybridization was also observed in the current experiments but, as reported by van Doorn et al. (Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010), the lack of reciprocal hybridization ensures that the probe sequences are specific when identifying individual parasite DNA. Similarly, DNA from Cor. labratus individuals cross-hybridizes with the Cya. tetracanthum probe, where the sequence differs by only 2 bp, but the Cor. labratus probe does not bind to DNA derived from Cya. tetracanthum worms, allowing for differentiation between these species. Some species show low levels of inter-specific variation; 2 such examples are Cyc. radiatus and Cyc. insigne and Cya. catinatum and Cya. pateratum, showing 97% and 95% sequence identity, respectively. In the case of Cyc. radiatus, it was not possible to design a probe exclusive to this species, as DNA from a subset of Cyc. insigne individuals hybridized to this probe and so, in some cases, these two species cannot be distinguished. A similar situation was also observed for a small number of Cyc. ashworthi and Cyc. leptostomum individuals. Low levels of cross hybridization were also observed for the Cys. minutus variants, with the Min B probe cross-hybridizing with some Cys. minutus variant A individuals. It should also be noted that, due to high levels of intra-specific variation for Cys. minutus variant B, the Min B probe may not always hybridize to DNA derived from homologous variants. As additional Cys. minutus individuals are sourced and the IGS DNA sequenced in the future, this probe will be further optimized. The observation that cross-hybridization occurs between some species suggests that this method is only really viable for identification of individual L3 rather than pools of larvae.
This study has highlighted that several species may comprise cryptic complexes and as intra-specific variation may vary according to geographical location, combined with the fact that cross-hybridization occurs for some species, the authors recommend continuous refinement and routine testing of this method with material from different locations and always to include positive controls for all molecular probes. The subfamily Cyathostominae consists of 50 species and the RLB as presented here is able to identify 18 species. Some species are particularly rare and are not often observed and hence are not often available for molecular study. As and when any remaining species are obtained, the IGS sequence should be amplified and new probes developed and tested to increase the potential of this method for molecular identification of cyathostomins.
The RLB has been applied to identify cyathostomin eggs and larvae and the findings here support the use of robustly designed probes to identify pre-parasitic developmental stages to species level. The majority of molecular studies investigating the species composition of cyathostomin infections have used parasites derived from horses or ponies (Cernanská et al. Reference Cernanská, Paoletti, Kralova-Hromadova, Iorio, Cudekova, Milillo and Traversa2009; Traversa et al. Reference Traversa, Iorio, Otranto, Giangaspero, Milillo and Klei2009, Reference Traversa, Milillo, Barnes, Von Samson-Himmelstjerna, Schurmann, Demeler, Otranto, Lia, Perrucci, Frangipane Di Regalbono, Beraldo, Amodie, Rohn, Cobb and Boeckh2010; Ionita et al. Reference Ionita, Howe, Lyons, Tolliver, Kaplan, Mitrea and Yeargan2010; van Doorn et al. Reference van Doorn, Kooyman, Eysker, Hodgkinson, Wagenaar and Ploeger2010). So far, all studies on cyathostomin species prevalence in donkeys have relied on morphological identification (Eysker and Pandey, Reference Eysker and Pandey1989; Matthee et al. Reference Matthee, Krecek and Milne2000, Reference Matthee, Krecek and Gibbons2002, Reference Matthee, Krecek and McGeoch2004; Kharchenko et al. Reference Kharchenko, Kuzmina, Trawford, Getachew and Feseha2009). The data here describe for the first time that DNA probes have been used to identify cyathostomin L3 derived from donkey populations. Two species of cyathostomin, Cya. tetracanthum and Cya. pateratum, were found to be the most abundant in the group of L3 analysed. Cya. tetracanthum has previously been reported to be prevalent in cyathostomin populations of donkeys, particularly those in Africa (Eysker and Pandey, Reference Eysker and Pandey1989; Matthee et al. Reference Matthee, Krecek and Milne2000, Reference Matthee, Krecek and McGeoch2004). The ability to determine the species of L3 in donkeys and parasites of horses at the molecular level facilitates comparative species analysis in live equid hosts in the future. Importantly, macrocyclic lactone (ML) resistance has been reported for cyathostomins in donkeys (Trawford et al. unpublished data) and the results reported here will allow investigations into species associations with ML resistance in donkeys in the future. Indeed, we are currently analysing the species composition of donkey-derived cyathostomin L3 populations that display differing sensitivities to ivermectin in vivo and in vitro.
ACKNOWLEDGEMENTS
We would like to thank The Donkey Sanctuary (Devon, UK) for providing fecal material. We would like to acknowledge Dr J.R. Lichtenfels, Patricia A. Pilitt (Parasite Biology, Epidemiology and Systematics Laboratory, Agricultural Research Service, US Department of Agriculture, Beltsville, MD 20705, USA), Sharon Tolliver (Department of Veterinary Science, Gluck Equine Research Center, University of Kentucky, USA) and Vitaly A. Kharchenko (Department of Parasitology, Schmalhausen Institute of Zoology NAS of Ukraine, Kyiv, Ukraine) for expert identification of nematodes. In the design of the RLB probes we would like to acknowledge the large strongyle IGS DNA sequences generated by Dr Elli Wright.
FINANCIAL SUPPORT
The work described here was generously supported by The Horse Trust (K.C., J.B.M., J.E.H., Project Reference Number: G708) and The Horserace Betting Levy Board, UK (J.B.M and J.E.H: Veterinary Project 671).









