Introduction
The recent phylogenetic reconstruction of caryophyllidean tapeworms (Cestoda) (Scholz et al., Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021) has fundamentally changed the classification of species belonging to each of the four extant families of the order Caryophyllidea (i.e. Balanotaeniidae, Capingentidae, Caryophyllaeidae and Lytocestidae). This large-scale study again illustrated the importance of combining different methods and criteria in modern systematic research, but did not address karyology at all. However, a comparison of the cytogenetic characteristics of different species can also shed light on the evolution of species and significantly complement morphological and molecular phylogenetic analyses.
To date, the first cytogenetic review of the Cestoda, including the monozoic order Caryophyllidea (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011), has been supplemented by several later studies (Orosová and Oros, Reference Orosová and Oros2012; Bombarová and Špakulová, Reference Bombarová and Špakulová2015; Orosová et al., Reference Orosová, Provazníková, Xi and Oros2019, Reference Orosová, Marková, Provazníková, Oros, Radačovská, Čadková and Marec2021; Špakulová et al., Reference Špakulová, Bombarová, Miklisová, Nechybová and Langrová2019). However, the karyotypes of taxonomically related species studied within each family showed very inconsistent characteristics. This is due to the fact that the tapeworms were assigned to families according to the traditional – and now invalid – system. In addition, it is important to emphasize that some caryophyllidean genera that have been studied karyologically have been redefined using morphological and molecular evidence (Scholz et al., Reference Scholz, Oros, Bazsalovicsová, Brabec, Waeschenbach, Xi, Aydoğdu, Besprozvannykh, Shimazu, Králová-Hromadová and Littlewood2014, Reference Scholz, Oros, Choudhury, Brabec and Waeschenbach2015; Oros et al., Reference Oros, Uhrovič and Scholz2018). In total, reports on the chromosome number of caryophyllidean tapeworms have been published for 24 species from 15 genera (~18 and ~34% of known species and genera, respectively) belonging to all four families (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011). Therefore, it is worthwhile to verify the concordance of the position of individual species in the revised systematic structure of the order Caryophyllidea (Scholz et al., Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021) using karyological data.
In this study, we examined two caryophyllidean tapeworms common in Palearctic region, namely Caryophyllaeus laticeps from a chemically polluted locality and from an unpolluted locality and the cytogenetically unexplored species Paracaryophyllaeus gotoi, both belonging to the family Caryophyllaeidae. The tapeworm C. laticeps is the intestinal parasite of a wide range of freshwater cyprinid fishes. Recent comprehensive morphological (Hanzelová et al., Reference Hanzelová, Oros, Barčák, Miklisová, Kirin and Scholz2015; Barčák et al., Reference Barčák, Oros, Hanzelová and Scholz2017) and molecular phylogenetic (Bazsalovicsová et al., Reference Bazsalovicsová, Kráľová-Hromadová, Brabec, Hanzelová, Oros and Scholz2014; Xi et al., Reference Xi, Barčák, Oros, Chen and Xie2016) studies of C. laticeps specimens from different cyprinid hosts and different geographical locations have revealed the existence of several distinct morphotypes. All different forms represent five closely related genetic lineages and are mostly specific to different cyprinid hosts. In contrast, P. gotoi is strictly linked to loaches (Cobitidae) (Scholz et al., Reference Scholz, Oros, Bazsalovicsová, Brabec, Waeschenbach, Xi, Aydoğdu, Besprozvannykh, Shimazu, Králová-Hromadová and Littlewood2014) and no information on its chromosomes is yet available.
The main objectives of this study were to extend the basic chromosomal data and, at the same time, to investigate whether karyological data could also be useful for phylogenetic inferences about the Caryophyllidea at different taxonomic levels. This goal is especially achievable when suitable cytogenetic markers are available. Here, classical features as well as the distribution of GC/AT-rich heterochromatin were determined by chromomycin A3 (CMA3) and diamidino-2-phenylindole (DAPI) staining. The location of the nucleolar organizer region (NOR) was determined by silver and CMA3 staining, and the major rDNA loci were mapped by fluorescence in situ hybridization (FISH) in both tapeworm species. In addition, the karyotype of C. laticeps was of particular interest because the parasite originated from the extremely polluted water reservoir Zemplínska Šírava (eastern Slovakia), which is one of the most contaminated sites with polychlorinated biphenyls (PCBs) in Europe and worldwide (Šalgovičová and Zmetáková, Reference Šalgovičová and Zmetáková2006). Therefore, we compared this population with samples from unpolluted water and focused on possible chromosomal damage in C. laticeps due to the presence of environmental mutagens. In particular, the occurrence of structural chromosomal abnormalities and the possible presence of supernumerary chromosomes, i.e. B chromosomes, were investigated.
Materials and methods
Specimens of tapeworms
Cytogenetic analyses were performed on 15 specimens of C. laticeps isolated from the intestines of 13 out of 38 examined common breams (Abramis brama), collected from the polluted Zemplínska Šírava reservoir during the spring and autumn seasons in 2019 and 2020. This material was processed using complex cytogenetic methods. A small pond near Pozdišovce (eastern Slovakia) was defined as a reference site, an unaffected area for the investigation of chromosomal aberrations (CAs) in C. laticeps. This pond is fed by many small forest streams and rivulets and has no chemical plant nearby. It harmonizes perfectly with the natural environment and is considered unpolluted water. Three adult tapeworms of C. laticeps were dissected from the intestines of three out of 10 common breams sampled and all were examined cytogenetically. Sampling at both sites was carried out in accordance with Slovak legislation (No. 62/2019 and 34/2020, issued by the Ministry of Environment of the Slovak Republic). Individual parasites from both sites were rinsed several times in saline solution immediately after dissection from freshly killed fish hosts. The morphology of tapeworms was examined microscopically to determine their morphotype, as described previously (Hanzelová et al., Reference Hanzelová, Oros, Barčák, Miklisová, Kirin and Scholz2015). All specimens were identified as morphotype I. A body part of some individuals was fixed in 96% ethanol for DNA extraction.
Ten live specimens of P. gotoi were obtained from the intestine of 10 pond loaches (Misgurnus anguillicaudatus) out of 22 examined from the Yangtze river in Wuhan, Hubei province and Taihu lake in Wuxi, Jiangsu province, China, in September 2019. The specimens were processed similar to those of C. laticeps.
Chromosome preparation and cytogenetic techniques
Mitotic and meiotic chromosomes were obtained from testes, ovary and vitelline follicles using the ‘hot plate’ technique, as described previously (Orosová and Špakulová, Reference Orosová and Špakulová2018). For standard cytogenetic examination, chromosome preparations were stained with 5% Giemsa solution (Merck, New Jersey, USA) in phosphate buffer (pH 6.8) for 25 min and rinsed under running distilled water. The morphology of the chromosomes was determined using the centromeric indices according to the standard nomenclature method (Levan et al., Reference Levan, Fredga and Sandberg1964). The absolute length [AL, length of the long arm q (L) + length of the short arm p(S) in μm] of the chromosomes, the relative length (i.e. AL × 100%/half length of all chromosomes in metaphase) and the centromeric index [i.e. S/(L + S)] were measured and calculated in 10 best mitotic spreads with clearly distinguishable chromosomes. The mean and standard deviation were calculated using Microsoft Excel. Staining with silver nitrate (AgNO3) was performed according to the standard protocol commonly used for Cestoda (Orosová et al., Reference Orosová, Marková, Provazníková, Oros, Radačovská, Čadková and Marec2021). Fluorescent YOYO-1 staining to visualize the nucleoli was performed according to the previously described procedure (Orosová et al., Reference Orosová, Marec, Oros, Xi and Scholz2010b).
FISH experiments were performed with 18S rDNA probes obtained by polymerase chain reaction amplification from genomic DNA of C. laticeps, using two primers, 18S-WormA forward and 18S-WormB reverse, according to amplification conditions previously described by Littlewood and Olson (Reference Littlewood, Olson, Littlewood and Bray2001). Probes were labelled with biotin-16-dUTP (Roche Diagnostics, Mannheim, Germany) according to the improved nick translation procedure (Kato et al., Reference Kato, Albert, Vega and Birchler2006) with some modifications described in a previous study (Zrzavá et al., Reference Zrzavá, Hladová, Dalíková, Šíchová, Ounap, Kubíčková and Marec2018). The reaction time for labelling was 55 min at 15°C. Hybridization followed the standard protocol used for insects (Sahara et al., Reference Sahara, Marec and Traut1999) with slight modifications for Cestoda (Orosová and Špakulová, Reference Orosová and Špakulová2018). The preparations were treated with proteinase K (20 mg mL−1) in 1× phosphate-buffered saline (PBS) for 5 min at 37°C and incubated with 100 μg mL−1 RNase A in 2× SSC in a humid chamber for 1 h at 37°C. Chromosomes were denatured in 70% formamide in 2× SSC for 3 min and 30 s at 68°C. A probe mixture (10 μL; 50% deionized formamide, 10% dextran sulphate, 2× SSC, 25 μg of sonicated salmon sperm DNA) containing ~50 ng of biotinylated 18S rDNA probe was added to each slide and the preparation was left overnight (~20 h) at 37°C in a humid chamber. Hybridization signals were amplified and visualized by three-step detection with Cy3-conjugated streptavidin (Jackson ImmunoRes. Labs Inc., West Grove, PA, USA), biotinylated anti-streptavidin (Vector Labs Inc., Burlingame, CA, USA) and Cy3-conjugated streptavidin. The slides were then counterstained with 0.5 μg mL−1 DAPI in a ProLong Antifade (Invitrogen, Carlsbad, CA, USA) and sealed with nail polish. The stained slides were stored in the dark at 4°C for 2 days prior to examination.
Two-colour staining with CMA3 (GC-specific) and DAPI (AT-specific) was performed sequentially or in separate experiments (Rábová et al., Reference Rábová, Volker, Pelikánová, Ráb, Ozouf-Costaz, Pisano, Foresti and Toledo2015), and this approach was used for the first time in the class Cestoda. A stock solution of CMA3 (1 mg ml−1) was prepared by dissolving in MilliQ water for 5 days at 4°C in the dark. The working solution was prepared by dissolving in McIlvaine's buffer containing MgCl2 (1:1). Slides were either treated with CMA3/Methyl Green for 20 min in a wet chamber or by sequential banding with CMA3 for 15 min followed by DAPI staining (0.5 μg mL−1 in PBS containing 1% Triton X-100) for 15 min. Stained slides were observed with a light microscope (Giemsa staining) or a Leica DM 4000 B fluorescence microscope equipped with a DFC 450 C digital camera and photographed separately for each fluorescent dye. Black-and-white fluorescence images were pseudocoloured (light blue for DAPI, red for Cy3, green for YOYO-1 and CMA3) and merged using Adobe Photoshop, version 7.0.
Results
Karyotype constitution and meiosis in C. laticeps
Chromosome data were obtained from 207 dividing cells of 15 adult individuals of C. laticeps. The cells had diploid chromosome numbers ranging from 2n = 14 to 2n = 20. The vast majority of mitotic cells (81.2%) had 2n = 20 chromosomes, and the remaining cells lacked two, four or six chromosomes (Supplementary Table S1). These cells were found only sporadically on the preparations, so that the numerical discrepancies can be attributed exclusively to the artefacts of the chromosome preparation technique.
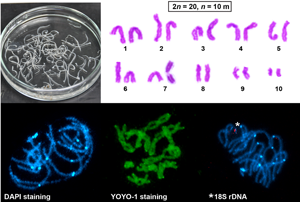
The basic karyotype of C. laticeps was described from 20 mitotic metaphases of testes, ovary and vitelline follicles and consisted of 10 chromosome pairs (2n = 20, FN = 40), with the karyotype formula interpreted as 2n = 20, n = 10m (Fig. 1). The chromosomes were large and the centromere positions were generally easy to identify after classical Giemsa staining. No secondary constrictions were observed on chromosomes in dividing cells. Chromosome lengths gradually decreased from the longest with about 13 μm to the shortest with about 4 μm (see Table 1 for complete measurements). The karyotype was symmetrical; all chromosomes were bi-armed, classified as metacentric. The total length of the haploid genome reached 91.86 μm. In addition, a clear size heteromorphism was observed in chromosome pair no. 10 in two of the 15 tapeworms examined (Fig. 1B and C); however, the chromosomes did not differ in their morphological shape, i.e. both were metacentric.

Fig. 1. Giemsa-stained chromosomes of Caryophyllaeus laticeps. (A–C) Mitotic chromosomes and karyotypes derived from mitotic metaphases, 2n = 20, n = 10 m. Arrows indicate NOR. Scale bar = 10 μm.
Table 1. Measurement (mean ± s.d.) and classification of chromosomes of Caryophyllaeus laticeps

a Mean ALs were calculated from 10 mitotic metaphases.
b m, metacentric chromosome.
Meiotic cells were analysed using different chromosome staining methods, namely conventional Giemsa staining (Supplementary Fig. S1), silver staining (Fig. 2) and fluorescent dye DAPI and YOYO-1 (Fig. 3), to determine the successive division stages. Meiotic spermatogenesis studied in the testes showed the standard steps of cell division characteristic of eukaryotic organisms. At the onset of meiosis, chromatin was arranged in long, thin strands (Supplementary Fig. S1C–E; Fig. 2A). Clumps of 10 bivalents per nucleus were regularly observed in the pachytene, diplotene and diakinesis (Supplementary Fig. S1F–I; Fig. 2B and C). The frequency of chiasmata of each chromosome pair was analysed in the well-spread diplotene bivalents (Supplementary Fig. S1G–I; Fig. 2C). The smallest chromosome pair always had one chiasma, while in longer pairs the chiasma frequency increased linearly with chromosome size. The largest chromosome pair (no. 1) had four to six chiasmata (most frequently six). The homeotypic division (meiosis II) proceeded as usual in the formation of normal sperm. In meiotic metaphase II (Supplementary Fig. S1J and K; Fig. 2D), each secondary spermatocyte consisted of a haploid set of 10 chromosomes. After separation of each chromosome in its centromere, haploid spermatids were formed (Supplementary Fig. S1L; Fig. 2E). The subsequent spermiogenesis (Supplementary Fig. S1M–O; Fig. 2F) ended with the production of normal mature sperm (Supplementary Fig. S1P).

Fig. 2. Silver nitrate-stained chromosomes of C. laticeps showing nucleoli and NOR location (arrows). (A) Interphase nucleus with one large nucleolus. (B) Pachytene nucleus with nucleolus residue on one chromosome bivalent. (C) Diplotene stage showing a single NOR site on the bivalent no. 7. (D) Metaphase II. (E) Anaphase II, two cells with a single NOR signal on four chromatids. (F) Haploid spermatid. Scale bar = 10 μm.

Fig. 3. FISH with the 18S rDNA probe (red) on C. laticeps chromosomes. (A) Two mitotic metaphases; note a pair of chromosomes with clusters of 18S rDNA signals on each chromatid near the centromere (arrowhead). (B) Interphase nucleus. (C) Two leptotene nuclei. (D) Zygotene. (E) Pachytene bivalents showing a cluster of interstitial 18S rDNA signals associated with heterochromatic DAPI block. (F) Pachytene complement showing YOYO-1-stained nucleolar residues that co-localize with cluster of hybridization signals of the 18S rDNA probe. N, nucleolus. (G) Pachytene (stained only with DAPI) with distinct interstitial and terminal DAPI bands on the smallest bivalent (arrow). (H) Pachytene. (I) Diplotene, three cells. Asterisks indicate the seventh bivalent with rDNA cluster. (J) Eight metaphase II nuclei, each showing one chromosome with a cluster of interstitial 18S rDNA signals. (K) Graphical interpretation of metaphase II. (L) Early anaphase II with the smallest and biggest bivalents already separated (arrows). The chromosomes were counterstained with DAPI except for (F) (counterstained with YOYO-1). Scale bar = 10 μm.
A single large nucleolus was observed after YOYO-1 and Ag-staining in the early stages of spermatocyte division (Figs 2A, B and 3F), which disappeared before entering the diplotene stage (Fig. 2C). At the pachytene stage, an obvious nucleolus site was located interstitially in the middle of one bivalent in all spermatocyte cells examined (Fig. 2B). A single active nucleolus was also found in all interphase nuclei after Ag-staining (Fig. 2A).
Localization of NORs and heterochromatin
The AT-specific DAPI staining showed bright centromeric bands on all chromosomes (Fig. 3A, E–J, L). In addition, distinct interstitial and terminal DAPI bands were observed on the bivalent of the smallest chromosome pair at the pachytene stage in some cells examined (Fig. 3G). However, these bands were never identified on the two smallest chromosomes in mitotic metaphase spreads. FISH with the 18S rDNA probe showed that the rRNA genes (NORs) are located interstitially, postcentromerically on the short arms of the small metacentric chromosome pair no. 7 (7p cen) (Fig. 3A). The location of the rDNA cluster near the centromere was confirmed by its association with DAPI-highlighted centromeric heterochromatin. Hybridization signals of the probe were always associated with one bivalent during meiotic prophase I (Fig. 3C–I). Also in metaphase II (Fig. 3J and K) and anaphase II (Fig. 3L), chromosome no. 7 showed a clear rDNA signal. Sequential YOYO-1 staining and FISH with the 18S rDNA probe showed that the major rDNA is usually co-localized with the YOYO-1-stained nucleolar residues associated with the NOR-bivalent in pachytene spermatocytes (Fig. 3F).
Silver impregnation confirmed the presence of the interstitial NOR on the short arms of metacentric chromosome no. 7 (Fig. 2C–E). CMA3 staining, indicating the presence of GC-rich heterochromatic blocks, showed bright fluorescence at the site previously identified by silver impregnation and FISH. This implies that the major rDNA is associated with GC-rich bands. Comparison of the same chromosomes after CMA3/DAPI staining revealed CMA3+/DAPI− bands in the pericentromeric regions of the short arms of pair no. 7 (Fig. 4A–F). In addition, weak CMA3+ bands were observed in the telomeric regions of all chromosomes during meiotic prophase I (Fig. 4A, C) and in anaphase II (Fig. 4I). These bands were not observed in either meiotic metaphase II or in mitotic metaphase (Fig. 4D and H).

Fig. 4. Chromosomes of C. laticeps sequentially stained with CMA3/DAPI (A–F) and only with CMA3 (G–I). (A) Diplotene and (D) metaphase II after CMA3. (B) The same diplotene and (E) metaphase II after DAPI. (C, F) Merged images demonstrates that CMA3 positive region (red dot) is DAPI negative. In the small insets, the NOR-bearing bivalents showing CMA3+/DAPI− bands in the pericentromeric region. (G) Pachytene nucleus with CMA3+ band (yellow). (H) Metaphase I nuclei showing chromosomes with GC-rich heterochromatic blocks. (I) Anaphase II nucleus. Arrows indicate the location of NOR (red dots). Scale bar = 10 μm.
Chromosomal aberrations
Figure 5 shows different types of CAs found in mitotic metaphases of C. laticeps from the polluted water reservoir Zemplínska Šírava. Four types of CAs were identified in the CA assay with conventional Giemsa staining: SCG – single chromatid gap (appears as achromatic region; Fig. 5A), CF – centric fragmentation (Fig. 5B), ISCG – isochromatid gap (Fig. 5C) and SCB – single chromatid break (appears as true break; Fig. 5D). The total number of CAs and the number of cells with CA were 10 and 9, respectively, from the polluted water reservoir (Table 2). The most common types of CAs were ISCGs (Fig. 5C). No B chromosomes were observed. Only one ISCG was detected in C. laticeps samples from the unpolluted pond near Pozdišovce (Table 2). However, statistical analysis by Fisher's exact test showed that the difference in the number of CAs in the samples of C. laticeps from the polluted Zemplínska Šírava compared to the samples from the unpolluted pond is not statistically significant (P = 0.0677).

Fig. 5. Four types of CAs in mitotic metaphase cells of C. laticeps. (A) SCG; the inset shows enlarged SCG from a different metaphase. (B) CF; the inset shows enlarged CF from the same metaphase. (C) ISCG; the inset shows enlarged ISCG from a different metaphase. (D) SCB; the inset shows enlarged SCB from the same metaphase. Scale bar = 10 μm.
Table 2. Number of CAs in mitotic metaphase nuclei of C. laticeps from Zemplínska Šírava and unpolluted pond

CA, chromosomal aberrations; ISCG, isochromatid gap; SCG, single chromatid gap; SCB, single chromatid break; CF, centric fragment.
a The difference in the number of CAs between localities is not statistically significant (P = 0.0677) – compared by one-tailed Fisher's exact test using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA).
Meiosis in P. gotoi and localization of NORs
Chromosome data were obtained from 31 dividing meiotic cells of 10 adult individuals of P. gotoi, and mitotically dividing cells were found only exceptionally (2n = 20, Fig. 6E). Clumps of 10 bivalents (n = 10) were regularly observed in pachytene, diplotene/diakinesis and meiotic metaphase II nuclei (Fig. 6A–C and I). Based on the number and metacentric morphology of chromosomes in the haploid sets, which were clearly identifiable at different stages of meiotic division, the karyotype formula of P. gotoi was determined to be 2n = 20, n = 10 m. No differences in the number or type of chromosomes were detected. Measurements of chromosome length could not be made as not enough well-spread mitotically dividing cells were found. This phenomenon is probably related to the autumn period in which the fish hosts were collected; the seasonality of mitotic cell division is relatively well known in most Cestoda (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011). FISH with the 18S rDNA probe identified only one bivalent with an interstitial cluster of rRNA genes (Fig. 6D–I). The interstitial location near the centromere was also confirmed in mitotic metaphases (Fig. 6E) and metaphase II nuclei (Fig. 6I). A large compact nucleolus was usually observed after YOYO-1 staining (Fig. 6H).

Fig. 6. Chromosomes of Paracaryophyllaeus gotoi stained with AgNO3 (A–C) and FISH with the 18S rDNA probe (red) (D–I). (A, B) Pachytene cells with a large nucleolus. (C) Diakinesis. (D) Interphase nucleus. (E) Mitotic metaphase. (F, G) Pachytene bivalents showing a cluster of interstitial rDNA signals associated with a heterochromatic DAPI block of one bivalent. (H) Bouquet stage (zygotene) counterstained with YOYO-1; N, remnants of nucleolus. (I) Metaphase II. The chromosomes in (D–G) and (I) were counterstained with DAPI. Scale bar = 10 μm.
Discussion
The karyology of parasitic flatworms still receives little attention, although there is an obvious need to further diversify the criteria for parasite systematics. Traditional morphological methods in combination with molecular approaches (sequence data) are commonly used to solve taxonomic and evolutionary problems in Cestoda. However, there are several reports indicating a conflict between morphological and genetic data (Doanh et al., Reference Doanh, Shinohara, Horii, Yahiro, Habe, Vannavong, Strobel, Nakamura and Nawa2009; Scholz et al., Reference Scholz, Brabec, Kráľová-Hromadová, Oros, Bazsalovicsová, Ermolenko and Hanzelová2011, Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021). Traits obtained by cytogenetic techniques, as well as ultrastructural traits, are an important part of systematic studies in many groups of organisms, as they are among the basic characteristics of the species (Dobigny et al., Reference Dobigny, Ozouf-Costaz, Bonillo and Volobuev2002; Petkevičiūtė et al., Reference Petkevičiūtė, Stunžėnas and Stanevičiūtė2018). The question often arises as to what extent karyotype characteristics are useful for phylogenetic inference. In this study, we plotted all available karyotype traits for the order Caryophyllidea over a phylogenetic tree reconstructed on the basis of molecular data (Supplementary Table S2; Fig. 7) and attempted to answer whether these data support the newly formulated phylogenetic relationships.

Fig. 7. Overview of available data on chromosome number 2n/3n, chromosome morphology (a, acrocentric; m, metacentric; sm, submetacentric; t, telocentric; min, minute elements) and chromosomal pattern of rDNA in haploid genome (p, short chromosomal arm, q, long chromosomal arm) of caryophyllidean tapeworms along with their latest phylogenetic relationships based on Oros et al. (Reference Oros, Uhrovič and Scholz2018) and Scholz et al. (Reference Scholz, Brabec, Kráľová-Hromadová, Oros, Bazsalovicsová, Ermolenko and Hanzelová2011, Reference Scholz, Oros, Choudhury, Brabec and Waeschenbach2015, Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021). *Data obtained in this study.
Karyotype characteristics in the family Caryophyllaeidae
According to the new family classification, members of the genera Caryophyllaeides and Khawia, which previously belonged to the Lytocestidae, were moved to the family Caryophyllaeidae (Scholz et al., Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021). The chromosome set is now known for nine species from five genera of this family (Supplementary Table S2). The karyotypes are characterized by two diploid numbers, 2n = 20 (Caryophyllaeides fennica, P. gotoi and C. laticeps, which all have metacentric chromosomes only) and 2n = 16 (five Khawia spp., which all have metacentric and acrocentric chromosomes). A special triploid species Atractolytocestus huronensis escapes this rule (3n = 24, n = 4m + 3a + 1 min). All members of the family Caryophyllaeidae have similarly high values of absolute chromosome lengths, but these are generally considered to be relatively large in all caryophyllidean species and do not represent a good distinguishing feature (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011).
Three species, C. fennica, P. gotoi and C. laticeps, show a very close karyological relationship with a constant diploid number of 2n = 20 (except for the rare occurrence of triploid individuals in C. laticeps; Fig. 7) and a stable karyotype morphology with long, gradually decreasing and exclusively bi-armed metacentric chromosomes. The high similarity of their karyotypes was discussed a decade earlier (Orosová et al., Reference Orosová, Kráľová-Hromadová, Bazsalovicsová and Špakulová2010a).
The karyotypes of five studied species of the genus Khawia are also very similar. All have 2n = 16, similar absolute and relative lengths, two types of chromosomes (metacentric and acrocentric) in their complements and the presence of the typical large metacentric chromosome pair no. 1. Nevertheless, a comparison of their karyotypes revealed distinct interspecific differences in the chromosomal morphology of the corresponding pairs, making the differentiation of these species relatively straightforward (discussed in detail in Orosová et al., Reference Orosová, Provazníková, Xi and Oros2019). Taken together, all these traits show a strong evolutionary relationship between the Khawia species and it is certain that these chromosome sets arose from a common karyotype. Compared to C. fennica, P. gotoi and C. laticeps, all studied species of the genus Khawia have less symmetrical karyotypes with the presence of acrocentric chromosomes in their complements.
By mapping the basic chromosomal features of caryophyllidean species to the most recent phylogenetic tree (Scholz et al., Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021), four subgroups can be identified (1–4, Fig. 7). The first two consist of species with the diploid number 2n = 20 and only bi-armed chromosomes. The third subgroup includes species of the genera Atractolytocestus and Breviscolex, the latter formerly placed in the family Capingentidae but now belonging to the Caryophyllaeidae. The only karyologically studied species within this third clade is the well-known triploid A. huronensis. However, nothing is known about its congener Atractolytocestus sagittatus or about the genus Breviscolex. The fourth subgroup consists of all Khawia species. It is obvious that subgroups 1 plus 2 and subgroup 4 are characterized by karyotypic constancy between species, in particular by the absence of interspecific variability in chromosome number. On the other hand, the striking differences in chromosome number and morphology between Khawia spp. and all other cytogenetically known members of the Caryophyllaeidae (C. fennica, P. gotoi and C. laticeps) indicate a progressive reduction in chromosome number and severe chromosome rearrangements within the family. Symmetrical karyotypes, similar chromosome morphology (all metacentric) and occurrence in both main sister clades (in the clade C. fennica + P. gotoi and the clade comprising three other groups) suggest that n = 10 may be the ancestral haploid chromosome number of this family. The decrease in chromosome number in Khawia spp. to n = 8 could hypothetically be due to extensive chromosome rearrangements, multiple fissions/fusions, in the early stages of speciation of these species. Consequently, intrachromosomal changes such as pericentromeric inversions could be involved in the divergence of chromosomal morphology within the Khawia clade. The markedly divergent karyotype of the only species studied from the third subclade, A. huronensis, is apparently a consequence of polyploidy and not the result of chromosome fusions and fissions.
Karyotype characteristics in the family Capingentidae
The phylogenetic tree mentioned above (Scholz et al., Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021) showed that the family Capingentidae is a sister group of the family Caryophyllaeidae. The Capingentidae are the most species-rich and karyologically best-studied family among the Caryophyllidea. Chromosome numbers are now known for 12 representatives of seven genera, and chromosome structure data are available for seven species (Supplementary Table S2). All these taxa were previously placed in the family Caryophyllaeidae.
Literature data suggest that greater variation in diploid number, from 2n = 14 to 2n = 20, and more asymmetrical karyotypes with shorter mono-armed chromosomes are characteristic of the Capingentidae. However, the close relationship of species belonging to the same genus and sharing the same diploid number is obvious even in this family (Fig. 7). Thus, cytogenetics has clearly confirmed the separation of a recently established genus Pseudoglaridacris from the original genus Glaridacris (Oros et al., Reference Oros, Uhrovič and Scholz2018). Indeed, the karyotypes of the two original species Glaridacris catostomi and Glaridacris vogei have 2n = 20 chromosomes, while the two members of the new genus Pseudoglaridacris laruei and Pseudoglaridacris confusa have karyotypes with 2n = 16 (Supplementary Table S2). In total, the cytogenetically studied capingentid species belong to five intra-family subgroups (Motomura, Reference Motomura1929; Grey and Mackiewicz, Reference Grey and Mackiewicz1974, Reference Grey and Mackiewicz1980; Grey, Reference Grey1979; Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011; Scholz et al., Reference Scholz, Waeschenbach, Oros, Brabec and Littlewood2021). The diploid numbers within these clades are always the same, but they usually differ from each other. Furthermore, triploid individuals or populations have occurred within two species (G. catostomi and Isoglaridacris bulbocirrus), and this trend is characteristic of the entire order Caryophyllidea (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011).
The strong conservatism in diploid number at the genus level and the correlation between a cladistic event and a change in chromosome number is thus also evident in the family Capingentidae. In all species, the difference in centromere position between the corresponding chromosomes within genera is probably due to intrachromosomal changes such as pericentric inversions. This pattern of chromosomal rearrangements is common in all major animal groups, including some tapeworm genera (Petkevičiūtė, Reference Petkevičiūtė1996, Reference Petkevičiūtė2003; Petkevičiūtė and Bondarenko, Reference Petkevičiūtė and Bondarenko2001; Naseeb et al., Reference Naseeb, Carter, Minnis, Donaldson, Zeef and Delneri2016; Hooper and Price, Reference Hooper and Price2017; Wellenreuther and Bernatchez, Reference Wellenreuther and Bernatchez2018). Similar to the family Caryophyllaeidae, the haploid chromosome number of n = 10 occurs in both main sister clades of the family Capingentidae (in the clade Glaridacris + Promonobothrium and the clade comprising the other groups). This suggests that also in this family the ancestral karyotype consisted of n = 10 chromosomes. The high number of species with a lower haploid number (n = 7–9) thus indicates an evolutionary trend towards a reduction in chromosome number.
Karyotype characteristics in the families Balanotaeniidae and Lytocestidae
Due to the sparse information on the structure of karyotypes within these two caryophyllidean families, it is not possible to assess the above-mentioned trend towards conservatism of diploid numbers at the genus level. Basic cytogenetic information is known for three species of the family Balanotaeniidae, and two of them (Caryoaustralus sprenti and Notolytocestus minor) were previously placed in the Lytocestidae. All three species have different chromosome numbers (Supplementary Table S2) (Grey, Reference Grey1979). Caryoaustralus sprenti has the lowest chromosome number (2n = 6) of the Caryophyllidea studied to date and all chromosomes are metacentric. In contrast, the karyotype of N. minor (2n = 12) consists entirely of acrocentric chromosomes, and the morphology of the chromosomes of Balanotaenia bancrofti (2n = 12) is unknown. These differences in chromosome number and morphology suggest that karyotype evolution in these species, all native to Australia, may have occurred through a series of centric fusions or fissions. Only one species from the family Lytocestidae, Lytocestus indicus, has been studied cytologically. Its karyotype consists of eight pairs of chromosomes (2n = 16), but nothing is known about their size and morphology (Vijayaraghavan and Subramanyam, Reference Vijayaraghavan and Subramanyam1977).
Arrangement of rDNA in chromosomes of caryophyllidean tapeworms
The collected data on tapeworms indicated the prevalence of chromosome complements with only one locus of rDNA (NOR), which was always interstitially located on a small metacentric chromosome and associated with C-DAPI+ heterochromatin (Orosová et al., Reference Orosová, Marec, Oros, Xi and Scholz2010b, Reference Orosová, Provazníková, Xi and Oros2019, Reference Orosová, Marková, Provazníková, Oros, Radačovská, Čadková and Marec2021; Orosová and Oros, Reference Orosová and Oros2012). However, in C. fennica, two clusters of different sizes were observed on two chromosome pairs, and in triploid A. huronensis, two clusters were found on one trivalent (Kráľová-Hromadová et al., Reference Kráľová-Hromadová, Štefka, Špakulová, Orosová, Bombarová, Hanzelová, Bazsalovicsová and Scholz2010; Orosová et al., Reference Orosová, Kráľová-Hromadová, Bazsalovicsová and Špakulová2010a; Špakulová et al., Reference Špakulová, Bombarová, Miklisová, Nechybová and Langrová2019). It has been hypothesized that one rDNA locus represents ancestral status, while karyotypes with multiple clusters represent derived status (Orosová et al., Reference Orosová, Provazníková, Xi and Oros2019). This hypothesis is also supported by our finding of a single interstitial rDNA locus in the karyotypes of C. laticeps and P. gotoi (this study), located in the pericentromeric region of metacentric chromosomes (Fig. 7). However, we have no information on the number and position of NORs from other families of Caryophyllidea. The available data thus support the hypothesis that the presence of a single interstitial NOR is the ancestral feature only for the family Caryophyllaeidae.
Sequential CMA3+/DAPI/Ag-NOR staining in the same metaphases revealed that NOR is DAPI− and CMA3+, suggesting that the major rDNA in C. laticeps is associated with GC-rich DNA. CMA3-positive sites were also identified in the telomeric regions of all chromosomes, suggesting that not all GC-rich heterochromatin matches NOR. AT-rich heterochromatin bands were found in the centromeric regions of all chromosomes (C-DAPI+) in C. laticeps. In contrast, C. fennica, which is karyotypically very similar, had two additional interstitially located positive bands on the short and long arms of chromosome pair no. 7 (Orosová et al., Reference Orosová, Kráľová-Hromadová, Bazsalovicsová and Špakulová2010a), which serve as a very good marker to distinguish C. laticeps from C. fennica.
Intraspecific variability of C. laticeps
The karyotype of C. laticeps, a relatively common Palaearctic tapeworm, has been studied thrice so far. In addition to the present work, two geographically distant populations of C. laticeps were studied. The first C. laticeps sample was from the Rybinsk reservoir on the Volga river in Russia (Petkevičiūtė and Kuperman, Reference Petkevičiūtė and Kuperman1992) and the second from the Tisa river (south-eastern Slovakia) (Bombarová and Špakulová, Reference Bombarová and Špakulová2015). All three studies were based on parasites of the same fish host, the common bream (A. brama), which is associated with the same morphological type of the tapeworm C. laticeps, morphotype I. Most cytogenetic characteristics of all geographically distant parasite populations were also consistent. The diploid chromosome number was always 2n = 20, all karyotypes consisted exclusively of bi-armed chromosomes, and the values of relative and AL of the corresponding chromosomes were almost identical. Moreover, the chiasma frequency on the bivalents was in the same range. Nevertheless, there were certain differences characterizing the local populations. First, the studied sample of Russian tapeworms included 25 diploid and one triploid specimen, and a rare trisomy of the smallest chromosome no. 10 was detected in the diploid tapeworms. Such a phenomenon was not observed in any of the Slovak populations. There were also slight differences in the mean total length of haploid complements (TLCs), which were, however, close to each other (the Russian, Tisa river and Zemplínska Šírava populations: 87.8, 80.6 and 91.8 μm, respectively). These TLCs are the highest documented so far in Cestoda. Some variability also applies to the chromosomes of the last pair (no. 10), which are clearly different from the other elements of the genome in all three populations. As mentioned above, the smallest chromosomes varied in number in the Russian population, while they varied in length in our analysis. Another apparent variation in chromosome morphology was seen in the centromere position in chromosome pair no. 3, which was classified as submetacentric in the Slovak population from the Tisa river (I c = 36.50), but metacentric in the other two populations (I c = 44.12 from Rybinsk reservoir, Russia; I c = 46.81, this study). The reason for this could be either true geographical variability or methodological differences. The presence of such a small difference in the morphology of the corresponding chromosomes can be explained by a small chromosome deletion or a nonreciprocal translocation, but most likely the differences in the centromeric index are due to a difference in chromosome contraction. In other words, chromosome spiralization is a dynamic process (Bajer, Reference Bajer1959) and not all evaluated chromosomes in a diploid cell contract at the same rate during mitosis (Proffitt and Jones, Reference Proffitt and Jones1969). It is also worth noting that such small differences between populations in the length of the corresponding chromosomes could be attributed to the high degree of subjectivity in the metric evaluation of chromosomes.
Effects of PCB contamination on chromosomes of C. laticeps: a pilot study
The Zemplínska Šírava reservoir is known for high levels of PCBs, which are among the most dangerous environmental pollutants due to their extremely high environmental persistence (Ogura, Reference Ogura2004). Fish parasites, including tapeworms, are known to be able to tolerate highly toxic pollutants in the aquatic environment (Sures et al., Reference Sures, Nachev, Selbach and Marcogliese2017). Recently, the cestode C. laticeps has been used as a model to study the bioaccumulation of PCBs and has been found to have an exceptional ability to accumulate PCBs (200 times higher compared to host fish tissues) (Brázová et al., Reference Brázová, Miklisová, Barčák, Uhrovič, Šalamún, Orosová and Oros2021). Chromosomal damage is among the first to occur. We used the intestinal parasite as a model for the first time to investigate the possible occurrence of chromosomal abnormalities in mitotic cells. However, the results of our study are only partially consistent with what is known about the effects of PCBs on the induction of CAs (Sandal et al., Reference Sandal, Yilmaz and Carpenter2008). The cytogenetic study conducted revealed an increased level of CAs in C. laticeps originating from this unusually contaminated site compared to the uncontaminated site area, but without statistical significance, probably due to insufficient sample size. B chromosomes, which are often associated with environmental pollution, were not detected at all. As we mentioned above, Petkevičiūtė and Kuperman (Reference Petkevičiūtė and Kuperman1992) recorded the occurrence of aneuploid and polyploid cells and the existence of triploid forms, as well as the presence of small additional chromosomes (three homologues of the 10th pair) in the Russian population of C. laticeps. The authors did not link these observations to pollution at the time, but the Rybinsk reservoir on the Volga river was also one of the chemically polluted sites with elevated PCB levels (Kozlovskaya and German, Reference Kozlovskaya and German1997; Chuiko et al., Reference Chuiko, Tillitt, Zajicek, Flerov, Stepanova, Zhelnin and Podgornaya2007). It is clear that fish tapeworms are sensitive to water pollution and could serve as indicators, but karyological analyses are relatively challenging.
Conclusions
It is evident that the available karyological data are consistent with the delineation of five genera (Caryophyllaeus, Khawia, Glaridacris, Isoglaridacris and Pseudoglaridacris) and are consistent with the new phylogenetic reconstruction of the order Caryophyllidea. New original data on the karyotypes of two caryophyllidean species, C. laticeps and P. gotoi, have further supported this reconstruction. Thus, the inclusion of classical and molecular cytogenetic characters in phylogenetic analyses will obviously help to improve the accuracy and reliability of such analyses. Mapping the available cytogenetic data to the most recent phylogenetic tree then allowed us to formulate a hypothesis about the ancestral haploid number of n = 10 chromosomes for the family Caryophyllaeidae, which could also be the ancestral number for its sister family Capingentidae. Furthermore, a comparison of two populations of C. laticeps from waters with and without PCB contamination showed a slightly increased rate of chromosomal abnormalities in the contaminated site.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022000622.
Author contributions
M. O. and F. M. conceived the study; M. O., Mi. O., D. B. and T. B. performed the field-collection and examined the fishes. M. O. and A. M. performed experiments and data analysis. M. O. created the figures. M. O. and F. M. prepared a draft of the manuscript. All authors discussed the results and commented on the manuscript. All authors approved the manuscript.
Financial support
This work was supported by the Slovak Research and Development Agency (No. APVV 18-0467), Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and the Slovak Academy of Sciences (VEGA, No. 2/0126/20) and Bilateral Mobility Plus Project (No. SAV-AV ČR-21-03).
Conflict of interest
The authors declare there are no conflicts of interest.