Published online by Cambridge University Press: 07 August 2001
A new method has been established to define the limits on a spontaneous mutation rate for a gene in Plasmodium falciparum. The method combines mathematical modelling and large-scale in vitro culturing and calculates the difference in mutant frequencies at 2 separate time-points. We measured the mutation rate at 2 positions in the dihydrofolate reductase (DHFR) gene of 3D7, a pyrimethamine-sensitive line of P. falciparum. This line was re-cloned and an effectively large population was treated with a selective pyrimethamine concentration of 40 nM. We detected point mutations at codon-46 (TTA to TCA) and codon-108 (AGC to AAC), resulting in serine replacing leucine and asparagine replacing serine respectively in the corresponding gene product. The substitutions caused a decrease in pyrimethamine sensitivity. By mathematical modelling we determined that the mutation rate at a given position in DHFR was low and occurred at less than 2.5×10−9 mutations/DHFR gene/replication. This result has important implications for Plasmodium genetic diversity and anti-malarial drug therapy by demonstrating that even with low mutation rates anti-malarial resistance will inevitably arise when mutant alleles are selected under drug pressure.
During the asexual Plasmodium life-cycle, nucleotide substitutions, deletions, sequence insertions, slippage and gene amplification arise both in vivo and in vitro under selective pressure (reviewed by Kemp, Cowman & Walliker, 1990; Shirley et al.1990). These mutational events constitute the first step in the generation of genetic diversity in Plasmodium species.
The mutation rate is determined by the difference in mutant frequencies at a minimum of 2 time-points. To date, numerous studies have reported the selection of Plasmodium mutants under anti-malarial pressure (reviewed by Peters, 1998); only a few have reported estimated mutation frequencies. Metachloridine-resistant P. gallinaceum parasites were selected at a frequency of 1 mutant per 109 parasites (Bishop, 1958). A 10-fold lower frequency was estimated for the selection of pyrimethamine-resistant P. chabaudi and P. berghei (Walliker, Carter & Morgan, 1973; Walliker, Carter & Sanderson, 1975). Multidrug-resistant P. falciparum parasites resistant to 5-fluoroorotate were present at a frequency of 1 mutant per 106 parasites (Gassis & Rathod, 1996; Rathod, McErlean & Lee, 1997). These are estimated frequencies at a single time-point. Thus an actual mutation rate at the nuclear DNA level in Plasmodium has not been determined. Such a rate would provide a fundamental basis to our growing knowledge of genetic diversity in natural and in vitro propagating malaria populations.
The purpose of this work was to design and implement a method to define the upper and lower limits of a spontaneous mutation rate for a nuclear gene in P. falciparum. We studied the gene dihydrofolate reductase (DHFR). DHFR mutants are, to a degree, resistant to the anti-folates, pyrimethamine and cycloguanil (Peterson, Walliker & Wellems, 1988; Peterson, Milhous & Wellems, 1990). The substitution from serine to asparagine at codon-108 (S108N) in the DHFR gene is described as the key mutation conferring pyrimethamine resistance (Peterson et al.1990). We designed a mathematical model to first assess the feasibility of estimating a mutation rate in vitro; secondly, to determine the power (number of replicates required) for given mutation rates; and thirdly, to specify the culturing parameters required to detect a mutant phenotype.
We designed the experiment to observe DHFR mutant frequencies from a Plasmodium culture, when the complete history of the culture since cloning was known. A mutation rate was calculated from the difference in mutant frequencies at 2 time-points. Upper and lower limits on the estimated mutation rate were determined by stochastic modelling. We describe the use of this method to calculate the limits on the mutation rate at 2 positions in the DHFR gene of a cloned P. falciparum line: leucine to serine at codon-46 (a novel mutation) and serine to asparagine at codon-108.
To measure mutant frequencies at 2 separate time-points an effectively large Plasmodium population size and a significant number of micro-culture well replicates were required. A selective pyrimethamine concentration greater than the wild-type minimum inhibitory concentration (MIC), was added to ‘control’ wells on Day 0. This determined the frequency of wells with pre-existing pyrimethamine-resistant mutants. After n parasite cycles the same pyrimethamine concentration was added to ‘test’ wells. This was to select for DHFR pyrimethamine-resistant mutants that had arisen spontaneously during the experimental period between Day 0 and Day N. To validate our experimental design and ensure that the detection of a mutant was feasible in vitro, we used mathematical modelling and spiked a population of wild-type pyrimethamine-sensitive parasites with a single pyrimethamine-resistant DHFR mutant.
We designed a virtual culture to simulate the probability of detecting a mutant phenotype by modelling theoretical mutation rates, the parasite growth rate in vitro and the effect of subculturing. The model predicted the number of replicates (power) required to detect a mutant phenotype. By simulating the culturing conditions used in the experiments, the model was then used to predict the limits on the mutation rate for the mutant frequencies.
The mathematical model is implemented by a computer program written in Pascal (version 7.0). A stochastic series of events occur. These events determine the number of mutants arising from the wild-type population, the probability of a mutant invading an erythrocyte for the start of the next cycle and the probability of the mutant progeny being retained at each subculture. The expected number of parasites after each event is predicted from either binomial or Poisson distributions, which are determined by binomial or Poisson random number generators respectively. In this program a parasite generation over approximately 48 h for P. falciparum is defined as the parasite cycle. Within each cycle, the program models 4 rounds of DNA replication and assumes that a single trophozoite divides asexually into 16 merozoites during schizogony (Coatney et al.1971).
A detailed version of the mathematical model is available on the QIMR web-site at: http://www.qimr.edu.au / research / labs / allans / mutation_model.pdf. Briefly, the program is initiated with a pre-defined number of wild-type and mutant parasites (zero mutants). The expected number of mutants is assigned after each round of DNA replication by a Poisson distribution from the theoretical probability of a mutation occurring and the total number of wild-type parasites. This allows a range of probabilities to be examined. A mutation can occur at any of the 4 rounds of DNA replication within a single parasite cycle. The probability of a mutation occurring independently at different replication rounds within the same cycle is the product of the mutation rates, which is very low. After completion of the first cycle the numbers of mutants and wild-type parasites are tracked separately.
The expected numbers of mutants and wild-types which successfully invade red blood cells (RBCs) for the next cycle is calculated using a binomial random number generator from the number of existing mutants or wild-types and the proportions of each subpopulation that invade. This models the multiplication factor for each cycle. The addition of fresh RBCs and medium to the culture is modelled after a defined number of cycles or when the total parasite population reaches a pre-defined threshold. This is modelled in 3 different ways. First, the addition of RBCs results in a 100% probability that all mutants will remain in culture; secondly, the removal of a proportion of the culture (split) results in a probability, equal to the proportion discarded, that any particular mutant will be lost; and thirdly, an even split of the culture into a new vessel (subculturing) results in an equal probability of a particular mutant in either culture vessel. The expected number of wild-type and mutant parasites remaining in the culture is determined by a binomial random number generator.
For modelling a mutation rate and predicting the expected number of replicates which contain at least 1 mutant parasite, the model is run for a specific number of cycles. Thus the time modelled depends upon the length of the parasite cycle (48 h for P. falciparum, 24 h for P. berghei and other rodent malarias).
Pyrimethamine-sensitive 3D7, a cloned line of NF54 (Walliker et al.1987) was spiked with pyrimethamine-resistant HB3 parasites (DHFR S108N mutants) (Walliker et al.1987). The HB3 parasites were serially diluted into micro-culture wells to give, on average, 3.3, 0.7 or 0.3 parasites per well. Each well also contained 1.2×106 3D7 parasites and 48.8 nM pyrimethamine (>MIC for 3D7) in 100 μl of medium at 3% haematocrit (HCT). The 50% inhibitory concentration (IC50) of HB3 is between 1000 and 1600 nM (Cowman et al.1988; Peterson et al.1990). The cultures were maintained under standard conditions in complete RPMI-1640 medium (see below) and assayed on Day 22 by the parasite-specific lactate dehydrogenase (pLDH) assay (Makler et al.1993; Goodyer & Taraschi, 1997). This was to validate the experimental design by determining whether a single DHFR mutant could propagate to a detectable level in vitro under drug pressure with non-viable pyrimethamine-sensitive parasites.
The 3D7 line was cultured in vitro at 37 °C by standard methods (Trager & Jensen, 1976) in tissue culture flasks gassed with 5% CO2/5% O2/90% N2. Fresh O+ RBCs and standard RPMI-1640 medium were added approximately every 2 days. The line was re-cloned by limiting dilution (Rosario, 1981) to an average of 0.3 parasites per well in 96-well plates. Viable parasites were detected after 24 days by the pLDH assay and 3D7 clone-6 was isolated. The clone propagated in vitro for a further 27 days to a maximum of 6×107 parasites of which approximately 2.4×107 ring forms were frozen in glycerolyte (Baxter, USA) and stored in liquid N2. Comparison of the packed cell volume (PCV) before and after thawing showed that about 50% (1.2×107) viable parasites were recovered after thawing. This population expanded to 3.6×108 rings after 14 days with subculture every 2 or 3 days. Three aliquots, each with 1.2×108 ring parasites, were frozen in glycerolyte and labelled the #2/2 stabilate. One of these aliquots was used for Experiment 1. Another aliquot was cultured for 17 more days (8.5 cycles), grown in 3×75 cm2 flasks and used for Experiment 2. A MIC of 39 nM pyrimethamine in standard RPMI-1640 medium was measured for 3D7 clone-6 (data not shown).
In preparation for the mutation experiments, parasite cultures were synchronized to ring forms by isolating the mature schizont stages on a 63% Percoll gradient in phosphate-buffered saline, pH 7.4 (v/v) (Saul et al.1982; Dluzewski et al.1984). The schizonts were returned to culture and ring-stage parasites were isolated 12–15 h later after suspension of culture to 10% HCT in a solution of 5% (w/v) sorbitol (Lambros & Vanderberg, 1979; Myler et al.1982).
The experiments, which were set up in micro-culture 96-well plates, are summarized in Fig 1. Experiment 1, using 1020 wells, aimed to detect a DHFR mutant genotype at a mutation rate of 1×10−9 events/DHFR gene/replication (derived from the power calculations). Briefly, on Day 0 each of the ‘control’ wells contained at least 6×104 3D7 clone-6 ring-stage parasites and 40 nM pyrimethamine in 100 μl at 3% HCT. Each of the 900 ‘test’ wells contained 150 3D7 clone-6 ring-stage parasites. On Day 20 after 10 parasite cycles, 6×105 parasites were estimated in each ‘test’ well. These parasites were then treated with 40 nM pyrimethamine. Throughout the culturing period, medium with or without pyrimethamine replaced the old medium every 2 days. On Days 6 and 34, fresh RBCs were added to each well. On Day 46, 26 days after pyrimethamine was added to the ‘test’ wells, the cultures were resuspended and 100 μl were incubated with 0.5 μCi [3H]hypoxanthine for 48 h. Viable parasites were assayed as described (Chulay, Haynes & Diggs, 1983). The cultures were harvested onto glass fibre filter mats, air dried, coated in lipophilic scintillation fluid and counted in the BetaplateTM automated liquid scintillation counter (Wallac Oy, Finland). Results were expressed as counts per minute (cpm) and compared against a standard curve of parasitaemia versus cpm.

Fig. 1. Flow chart for the selection protocol of DHFR mutants and determination of the mutant frequency at 2 separate time-points.
Experiment 2 aimed to detect a 10-fold lower mutation rate of 1×10−10 events/DHFR gene/replication using 8280 wells (derived from the power calculations). This design incorporated a higher number of 3D7 clone-6 parasites per ‘test’ well on Day 0 and required fewer parasite cycles before the addition of pyrimethamine. On Day 0, each well contained approximately 37500 3D7 clone-6 ring-stage parasites in 100 μl at 3% HCT. The 4140 ‘control’ wells also had 40 nM pyrimethamine. On Day 4 100 μl of fresh medium at 3% HCT were added to all wells. On Day 8 (after 4 parasite cycles) the ‘test’ wells, which contained between 1×106 and 1.5×106 parasites, were treated with 40 nM pyrimethamine. Fresh medium with or without pyrimethamine replaced the old medium every 48 h. On Day 14, the 200 μl of culture in each well were resuspended, 100 μl were removed and 100 μl of fresh medium at 3% HCT with pyrimethamine were added to each well. The removed 100 μl fraction from each well in 7 plates was pooled into a single 75 cm2 flask. This gave 10×75 cm2 flasks for each of the ‘control’ and ‘test’ groups. The flasks were checked routinely for parasites by Giemsa-stained blood films. On Day 35, 100 μl from each well were incubated for 48 h with 0.5 μCi [3H]hypoxanthine to test for viable parasites as described.
In parallel, 3D7 clone-6 was maintained as stock culture and re-cloned on Day 8 to an average of 0.3 parasites per well. This was cultured under the same conditions, but without pyrimethamine, to determine the required number of days before parasites were detectable.
Sample DNA from all flasks was isolated from 100–300 μl of packed RBCs as described (Lahiri & Nurnberger, 1991; Cheng et al.1997). Fifty reactions containing 1.25 U proof reading Taq-Polymerase (Pfu), 75 ng each of the forward (5′-ATG ATG GAA CAA GTC TGC A) and reverse (5′-CTT TGT CAT CAT TCT TTA AAG GCA) primer, 0.2 mM dNTPs (Promega, USA), 3.5 mM MgCl2 solution (Perkin–Elmer) and 1×PCR reagent buffer (Perkin–Elmer, USA) were heated in a DNA engine under the cycling conditions 93 °C for 40s, 55 °C for 50s, 70 °C for 60s over 30 cycles. PCR products were loaded onto a 2% agarose gel, electrophoresed and stained with ethidium bromide. The DHFR PCR product was gel purified using the QIAGEN Gel extraction kit and sequenced with ABI automated sequencing following the manufacturer's instructions.
The 50% inhibitory growth concentrations (IC50) of pyrimethamine, cycloguanil and methotrexate were tested on the wild-type and mutant 3D7 parasites. Stock solutions of 10 mM pyrimethamine (Sigma, USA) prepared in 0.5% lactic acid, 10 mM of cycloguanil hydrochloride (Imperial Chemical Industries plc, UK) in 50% methanol and 1 mM methotrexate hydrate (Sigma) in 1.12% dimethyl sulphoxide (Sigma) in double distilled H2O were all 0.22 μm filter sterilized. Further dilutions of each of the drugs were made in culture medium. Sensitivity assays were set up in triplicate with 2-fold serial dilutions from 100000 nM to 0.75 nM (Desjardins et al.1979). After a 96 h incubation, wells positive for viable parasites were detected by the pLDH assay. The IC50 value was calculated using a four parameter non-linear logistic regression model.
Estimates on the mutation rate limits were modelled mathematically from the difference in the number of mutant-positive replicates occurring at the end of the pre-experimental and the experimental periods. The pre-experimental period for Exps 1 and 2 modelled the culturing history of 3D7 clone-6 which ended on Day 0 before pyrimethamine was added (see Fig. 1). It was modelled according to the subculturing parameters, the number of cycles, the loss of parasites through freezing and thawing, the number of parasites estimated to be in the stock culture on Day 0 and the proportion of stock culture used in each experiment. The growth rates of the wild-types and mutants were assumed to be equal and were calculated from the number of wild-types and parasite cycles during this period. To determine the frequency of mutant-positive wells, the number of pre-existing mutants was distributed binomially amongst the ‘test’ and ‘control’ wells (1020 wells for Exp. 1 and 8280 wells for Exp. 2). This was determined for defined theoretical mutation rates and each simulation was run for 1000 iterations.
The experimental period was defined as the time between Day 0 and Day N. Day N was the day when pyrimethamine was added to the ‘test’ wells. Both wild-type and mutant growth rates were assumed to be 2.3 per cycle. To predict the number of culture replicates carrying mutants, theoretical mutation rates were modelled for 10 cycles for Exp. 1 and 4 cycles for Exp. 2. Each simulation at a defined mutation rate was run for 1000 iterations. For each iteration within the simulation set, the mutant frequency difference was calculated from the number of mutant-positive replicates on Day N minus the number of mutant-positive replicates on Day 0. From the 1000 iterations the mean and the 95% confidence limits were then calculated.
The power calculations showed that a mutation rate of 1×10−9 events/DHFR gene/replication could be measured with 900 replicates. A 10-fold lower rate could be measured using 4140 replicates. The experimental design was then validated by spiking 3D7 cultures with HB3 parasites grown in 48.8 nM pyrimethamine. Viable HB3 parasites were detected after 22 days of culture. The number of positive wells correlated with the expected values of a Poisson distribution. This demonstrated that, on average, either 3.3, 0.7 or 0.3 HB3 parasites had been spiked per well and could grow in vitro with dying 3D7 parasites and the associated products of cell death. Both these results confirmed that the experimental technique would work and measuring a mutation rate in vitro was feasible.
In Exp. 1, no ‘control’ wells were positive and 1/900 ‘test’ wells were positive for viable mutant parasites. Sequencing of the DHFR gene fragment showed a nucleotide mutation had occurred at codon-108, where AGC was substituted for AAC, resulting in a change from the wild-type serine to an asparagine in the gene product. This was the expected S108N mutant.
In Exp. 2, 0/8280 wells (‘control’+‘test’) were positive for viable pyrimethamine-resistant parasites tested by [3H]hypoxanthine incorporation. However, 2 of the ‘control’ flasks (labelled A and I), for detecting pre-existing mutants, and 2 of the ‘test’ flasks (labelled K and L) for detecting spontaneous mutants, yielding 4/20 flasks, were positive for viable parasites resistant to at least 40 nM pyrimethamine after 46 days (23 cycles) of continual culturing. Sequencing of the DHFR gene fragment from these parasite populations from each flask showed the same transitional mutation had occurred at codon-46 where wild-type TTA was replaced by TCA. This resulted in a change from wild-type leucine to serine in the corresponding protein. No other mutations, including S108N, were detected in the sequenced DHFR product, which covered the known mutations from codon-16 to codon-164 within the DHFR active site from codon-11 to -200 (Bzik et al.1987). DNA extracted from a third ‘control’ flask revealed a mixed peak of T and C bases at codon-46, but no viable parasites were isolated. Amplification and size comparison of the standard genotypic markers merozoite surface protein-1 (MSP-1), MSP-2 and glutamate rich protein (GLURP) run on a 2% agarose gel confirmed that the both the S108N and L46S mutants were 3D7-type parasites at these 3 loci (data not shown).
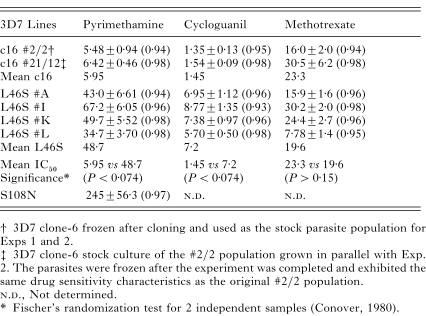
Both single-point mutations at codon-46 and codon-108 caused a marked decrease in sensitivity to pyrimethamine and cycloguanil, but not to methotrexate (Table 1). The pyrimethamine IC50 for the 3D7 S108N mutant was at least 4-fold less than the reported IC50 for HB3 (1000–1600 nM). The IC50 data showed intra-experimental variation between the wild-type and L46S mutants for the 3 anti-folates, reflecting systemic variation in the pLDH assay. There was a significant difference between the mean IC50 values of 3D7 clone-6 and 3D7 L46S for pyrimethamine (5.95 vs 48.7, P<0.074 and cycloguanil (1.45 vs 7.2, P<0.074) (Table 1) using Fischer's randomization test for 2 independent samples (Conover, 1980). Due to the small control sample size this was the lowest significance level applicable by the test (Conover, 1980). Thus the P-value could be lower. No significant difference was calculated for methotrexate (23.3 vs 19.6, P>0.15) (Table 1).
Table 1. The pyrimethamine, cycloguanil and methotrexate IC50 values for wild-type 3D7 and the L46S mutants and the pyrimethamine IC50 for the 3D7 S108N mutant. (Viable L46S mutants were detected in flasks labelled A, I, K and L. The IC50 of each anti-malarial drug and parasite line was calculated by 4 parameter non-linear logistic regression analysis. The standard error (S.E.) for the IC50 value and the R-value for the regression analysis are also shown.)
The upper and lower limits on the mutation rate for detecting at least 1 mutant-positive culture from 900 replicates and the upper limit on the mutation rate for detecting no mutants in 4140 wells was estimated from the 95% confidence limits on the mean difference of the mutant frequencies.
In Exp. 1, we observed that 1/900 wells contained S108N mutant parasites. No other wells were positive for DHFR mutants. We estimated that the S108N mutation rate occurred between the limits of 3×10−11 and 3×10−9 mutations/DHFR gene/replication (Fig. 2). In Exp. 2, 0/4140 ‘test’ cultures contained detectable S108N mutant parasites. Thus an upper limit on the S108N mutation rate could be estimated at 2.5×10−9 mutations/DHFR gene/replication (Fig. 3). The real mutation rate was likely to lie between 3×10−11 and 2.5×10−9 muations/DHFR gene/replication. The most probable rate was 7×10−10 mutations/DHFR gene/replication, which correlated with 1/900 mutant-positive wells (Fig. 2).

Fig. 2. Mathematical simulation of the mean difference between ‘test’ and ‘control’ mutant-positive wells for Exp. 1. In total 900 wells, each with 150 wild-type parasites, were modelled for 10 cycles. Wild-type and mutant growth rates were assumed to be 2.3 per cycle. The upper and lower 95% confidence limits (_._._) on the mean ([squf ]) were predicted from 1000 iterations. The mutation rate range for a 1/900 mutant positive wells (....) was determined.

Fig. 3. Mathematical simulation of the mean difference between ‘test’ and ‘control’ mutant-positive wells for Exp. 2. In total 4140 replicates, each with 37500 wild-type parasites, were modelled for 4 cycles. Wild-type and mutant growth rates were assumed to be 2.3 per cycle. One thousand iterations were run. From the lower 95% confidence limit (_._._) on the mean ([squf ]), the upper mutation rate limit for 0/4140 mutant positive wells (....) was determined.
The L46S DHFR mutants were growing in a selective pyrimethamine concentration close to their IC50 values (Table 1). The frequency of wells carrying the L46S mutants was unknown. Since 5 culture flasks contained L46S mutants and each of these cultures had been pooled from the wells of 7 plates, there must have been at least 2.5 mutant-positive wells from 4140 ‘control’ wells on Day 0 before pyrimethamine was added. Thus the lower limit on the mutation rate which allowed at least 2.5 wells to be carrying pre-existing mutants was 7×10−12 mutations/DHFR gene/replication (Fig. 4). The real mutation rate could be higher since the survival probabilities of the L46S mutants in 40 nM pyrimethamine was unknown.

Fig. 4. Mathematical simulation of the mean number of positive wells carrying pre-existing 3D7 L46S mutants. This modelled the culture history of 3D7 clone-6 before Exp. 2 was set up. In total 1000 iterations were run. From the upper 95% confidence limit (_._._) and the mean ([squf ]), the mutation rate limits for 2.5/4140 mutant-positive replicates were determined (....).
To confirm these low mutation rates we mathematically simulated the published observation of pyrimethamine-resistant P. berghei arising in 1/50 mice and 1/20 hamsters (Diggens, Gutteridge & Trigg, 1970). We predicted that the mutation causing pyrimethamine resistance most likely occurred between the limits of 1×10−13 and 2×10−11 mutations/DHFR gene/replication (data not shown). However, this resistance may have represented a gross chromosomal mutation rather than a single-point mutation, since the authors indicated that the resistance was associated with an increase in the level of DHFR (Diggens et al.1970).
The chance of observing a mutant phenotype, is proportional to the size of the parasite population under selective pressure. Although Plasmodium mutants have been selected against a wide range of anti-malarial drugs that target both nuclear and mitochondrial protein encoding genes, only a few of these studies reported estimated mutation frequencies. Using a new approach that combined both mathematical modelling and large-scale in vitro culturing, we measured the limits on mutation rates in the P. falciparum DHFR gene from the difference in mutant frequencies at 2 separate time-points after validating our experimental design.
The novel L46S mutation in DHFR has never been described in any other human (Eldin de Pécoulas et al.1998) or rodent (Cheng & Saul, 1994) malaria species or in other eukaryotic or prokaryotic DHFR (Bzik et al.1987; Lemcke, Christiansen & Jørgensen, 1999). Remarkably we detected this mutant from Exp. 2 after it had grown at its IC50 value in 40 nM pyrimethamine. This implied that only half the mutant population could have survived after each parasite cycle, resulting in a less than optimal reproduction ratio. Thus the detection of the L46S mutant demonstrated that a S108N mutant would have been detected if this mutation had occurred in the parasites distributed amongst the 4140 ‘test’ wells.
The frequency of wells carrying L46S mutants could not be determined. It is unknown whether any of the ‘test’ wells actually contained L46S mutants that had arisen spontaneously before pyrimethamine treatment. We speculate that stochastic variation on the Day 14 subculture and different environmental conditions between the micro-culture wells and flasks resulted in the L46S mutants surviving to detection only in the flasks. Since viable L46S mutants were detected in both ‘control’ and ‘test’ flasks, the mutants must have been present in the wells prior to the addition of pyrimethamine on Day 0. Thus the simplest explanation is that the L46S mutant was pre-existing in the 3D7 cloned wild-type population before Exp. 2 was set up. This conclusion validated our theoretical modelling approach for measuring the difference in mutant frequencies and demonstrated that the risk of pre-existing mutants is real. If experiments are not carefully designed to counter the effect of pre-existing mutants, an over-estimation of the mutation rate of the gene under study could result.
Since the frequency of mutants was low (1/900, 0/4140 wells and 5/20 flasks), mathematical modelling was essential to estimate the limits on the mutation rate. We concluded that the mutation rates of the L46S and S108N substitutions were <2.5×10−9 mutations/DHFR gene/replication. This rate is consistent with frequencies of pyrimethamine resistance estimated in animal models (Walliker et al.1973, 1975). Both the S108N and L46S DHFR mutations arose spontaneously in culture. It is probable that the L46S mutation occurred in the last few days before synchronization of the parasite stages, since this mutation was not selected from the #2/2 stabilate which gave rise to the S108N mutant nor was it selected from the stock #2/2 3D7 culture flask grown in parallel with Exp. 2. If it had arisen earlier in the pre-experimental culturing period we would have expected to see more flasks carrying L46S mutant parasites.
The point mutations at codon-46 and codon-108 represent transitional pyrimidine (thymine to cytosine) and purine (guanine to adenine) substitutions respectively. In P. falciparum the incidence of transitional and transversional nucleotide substitutions can be variable (Escalante, Lal & Ayala, 1998) and rates of base pair substitutions can vary by several orders of magnitude even within the same gene (Kunz, Ramachandran & Vonarx, 1998; Drake, 1999). Thus point mutation rates in other genes could be higher or lower than what we have estimated in DHFR.
Evidence for low mutation rates in P. falciparum 3D7 and other lines naturally resistant to only 1 anti-malarial has been shown indirectly from the study by Rathod and colleagues who were unable to detect drug-resistant mutants in parasite populations[les ]108 parasites (Rathod et al.1997). Mutations in the cytochrome b gene of P. falciparum mitochondrial DNA, which is expected to undergo mutations at a higher rate than nuclear genes (reviewed by Avise, 1991) were also estimated to occur at a low rate of 2×10−9 mutations/parasite/cycle (Korsinczky et al.2000). From field isolates, the incidence of nucleotide substitutions in the apical membrane antigen-1 and MSP-1 genes of P. vivax was consistent with low mutation rates (Figtree et al.2000).
This study has important consequences for the development and testing of any new anti-malarial drugs. It describes methodology in measuring mutation rates in a nuclear gene which is targeted by a commonly used anti-malarial and can be applied to study mutation rates in vivo. We have confirmed that mutation rates in Plasmodium genes appear to be low, occur at random and can alter the drug sensitivity of the parasite. The mutated alleles then become selected under drug pressure. Since malaria patients may typically have 1010–1012 parasites circulating at the time of treatment, even at a low mutation rate it would be expected that drug-resistant mutants will arise during a single infection. However, there must be significant selection pressure against the mutants by the wild-type parasites. This has been recently demonstrated in vivo, where the frequency of DHFR mutations were low before prophylaxis and then increased rapidly under sustained drug pressure (Doumbo et al.2000).
Drug resistance will inevitably arise, due to the probability of a spontaneous mutation occurring in the gene under selective pressure, the subsequent probability of the mutant becoming fixed in the within-host parasite population and then later fixed in the global parasite population within an endemic region. Combination drug therapy for malaria is envisioned to be the most efficient method of reducing the frequency of mutant drug-resistant parasites since the probability of a parasite surviving is the product of the specific gene mutation rates for each drug.
This work was funded with grants from the Australian National Health and Medical Research Council and the Queensland State Government.

Fig. 1. Flow chart for the selection protocol of DHFR mutants and determination of the mutant frequency at 2 separate time-points.

Table 1. The pyrimethamine, cycloguanil and methotrexate IC50 values for wild-type 3D7 and the L46S mutants and the pyrimethamine IC50 for the 3D7 S108N mutant.

Fig. 2. Mathematical simulation of the mean difference between ‘test’ and ‘control’ mutant-positive wells for Exp. 1. In total 900 wells, each with 150 wild-type parasites, were modelled for 10 cycles. Wild-type and mutant growth rates were assumed to be 2.3 per cycle. The upper and lower 95% confidence limits (_._._) on the mean ([squf ]) were predicted from 1000 iterations. The mutation rate range for a 1/900 mutant positive wells (....) was determined.

Fig. 3. Mathematical simulation of the mean difference between ‘test’ and ‘control’ mutant-positive wells for Exp. 2. In total 4140 replicates, each with 37500 wild-type parasites, were modelled for 4 cycles. Wild-type and mutant growth rates were assumed to be 2.3 per cycle. One thousand iterations were run. From the lower 95% confidence limit (_._._) on the mean ([squf ]), the upper mutation rate limit for 0/4140 mutant positive wells (....) was determined.

Fig. 4. Mathematical simulation of the mean number of positive wells carrying pre-existing 3D7 L46S mutants. This modelled the culture history of 3D7 clone-6 before Exp. 2 was set up. In total 1000 iterations were run. From the upper 95% confidence limit (_._._) and the mean ([squf ]), the mutation rate limits for 2.5/4140 mutant-positive replicates were determined (....).